Abstract
p300/CBP-associated factor (PCAF) is a coactivator of the tumor suppressor, p53. PCAF participates in p53's transactivation of target genes through acetylation of both bound p53 and histones within p53 target promoters. Using microarrays, we discovered that PCAF itself is induced by p53 in a panel of breast tumor cell lines. Two p53 mutant breast tumor cell lines, BT-549 and UACC-1179, were chosen for further study of PCAF induction by wild-type p53. PCAF induction following adenoviral transduction of p53 expression was confirmed with real-time polymerase chain reaction in a time course experiment. Chromatin immunoprecipitation experiments then showed that PCAF induction was associated with increased p53 binding to the PCAF promoter, which contains p53 consensus-binding sites. PCAF induction by p53 activity was further demonstrated in wild-type p53 MCF10A cells when PCAF expression was induced following activation of endogenous wild-type p53 with doxorubicin in a dose- and time-dependent manner. Furthermore, the doxorubicin-induced increase in PCAF expression was blocked by pretreatment of the MCF10A cells with siRNA (small interfering RNA) targeted against p53 mRNA. Taken together, the results show that PCAF expression can be induced by wild-type p53.
Keywords: Microarray, p53, PCAF, p300/CBP-associated factor, acetyltransferase
Abbreviations: ChIP, chromatin immunoprecipitation; PCAF, p300/CBP-associated factor; RT-PCR, reverse transcription-polymerase chain reaction; siRNA, small interfering RNA; GFP, green fluorescent protein
Introduction
The tumor suppressor, p53, functions primarily through transcriptional transactivation of its target genes to block proliferation of damaged cells [1,2]. p53 transactivation activity is stimulated by a variety of cellular insults, including infection, DNA damage, and oxidative damage. Studies have linked p53's transactivation activity to acetylation of both p53 itself [3–5] as well as histones, including those within p53 target promoters [6–8]. Acetylation of both p53 and histones appears to occur through recruitment of histone histone acetyltransferases (HATs), which include p300, CREB-binding protein (CBP), and p300/CBP-associated factor (PCAF) [4,9,10]. In addition, all three coactivators acetylate p53 in response to DNA damage, thus increasing p53's sequence-specific DNA-binding ability at target promoters [4,5].
The significance of PCAF in p53's control of cell growth is also suggested by studies investigating the early region 1 (E1) of the adenovirus genome. The adenovirus genome produces E1A RNA, which is spliced into 12S and 13S transcripts to produce proteins of 243 and 289 amino acids, respectively. The E1B region produces two transcripts with overlapping reading frames: one for a 19-kDa protein that inhibits apoptosis and the other for a larger 55-kDa protein. Several studies have reported that the larger of the E1B proteins functions to inhibit p53 acetylation by PCAF, whereas the E1A protein inhibits nucleosomal acetylation by PCAF [6,10,11]. The fact that the adenoviral E1 proteins specifically target PCAF's interactions with p53 and its coactivators to transform cells indicates the importance of PCAF in p53's function.
In a prior study, we used cDNA microarrays to investigate the response of breast epithelial cell lines to adenovirally transduced wild-type p53 [8]. Eleven breast epithelial cell lines, nine p53 mutants and two wild type for p53, were compared to untreated MCF10A cells following 24 hours of either adenoviral infection with empty vector or infection with adenoviral vector expressing wild-type p53. Multidimensional scaling identified a cluster of eight cell lines that were p53-responsive. The responsive cell lines were then used to identify genes induced by adenoviral transduction of wild-type p53, but not adenoviral infection with empty vector or no treatment. Selection of responsive genes was by similarity (Pearson's correlation coefficient of 0.99) of their expression pattern to that of a previously identified p53-inducible gene quinone oxidoreductase (PIG3) [12]. The responsive genes selected in this manner included PCAF, and led to the hypothesis that PCAF was a p53 target gene.
Two cells lines, UACC-1179 and BT-549, which had induced PCAF following adenoviral transduction of wild-type p53 as measured by microarray, were selected for further study. To test PCAF induction further in UACC-1179 and BT-549, the time course of both PCAF and p53 induction following adenoviral transduction of wild-type p53 was determined using real-time reverse transcription-polymerase chain reaction (RT-PCR). Direct activation of PCAF expression by p53 was investigated by measuring p53 binding to the PCAF promoter in the area of a p53 consensus-binding site using chromatin immunoprecipitation (ChIP). Further experiments using MCF10A cells, which are wild type for p53, measured the dose and time dependence of PCAF induction following activation of endogenous p53 activity by doxorubicin. To further confirm the role of p53 in PCAF induction, siRNA directed against p53 mRNA was used to reduce activated p53 levels in MCF10A cells prior to doxorubicin treatment. A subsequent decrease in PCAF induction as a result of pretreatment with siRNA against p53 mRNA was demonstrated. Taken together, the results support PCAF as a p53 target gene.
Materials and Methods
Cell Culture
The MCF10A, MDA-MB-435, MDA-MB-231, MDA-MB-157, MDA-MB-468, BT-549, and Hs578T breast cancer cells were obtained from the American Type Culture Collection (Rockville, MD). The remaining cell lines were early passage sporadic breast cancer cell lines developed and maintained at the Arizona Cancer Center Cell Culture Shared Service (Tucson, AZ). UACC-1179 and UACC-2087 cell lines were derived from pleural effusions, whereas UACC-893 was derived from a primary tumor [13–15]. MDA-MB-435 and MDA-MB-231 were maintained in RPMI 1640 containing 5% fetal bovine serum supplemented with 50 µg/ml penicillin/streptomycin. MDA-MB-468, BT-549, and Hs578T were maintained in RPMI 1640 containing 10% fetal bovine serum supplemented with 50 µg/ml penicillin/streptomycin. MDAMB-453, MDA-MB-157, UACC-1179, and UACC-2087 were maintained in M15 containing 5% fetal bovine serum supplemented with 50 µg/ml penicillin/streptomycin. MCF10A was maintained in mammary epithelial growth media (Cell Applications, Inc., San Diego, CA).
Adenoviral Infection
Recombinant adenovirus serotype 5 containing wild-type p53 with a green fluorescent protein (GFP) detection marker (p53) or with GFP alone (empty vector) were a kind gift of Bert Vogelstein [16] and were propagated at the Gene Transfer Vector Core, University of Iowa (Iowa City, IA). Breast cancer cell lines were grown, counted, and then infected with 200 pfu/cell of either GFP adenovirus 5 or p53/GFP adenovirus 5. Infection efficiency was monitored by scoring cells for GFP production using flow cytometry and fluorescence microscopy; each virus resulted in >95% infection efficiency.
Microarray Analysis
The probe, target, and microarray fabrication were performed as previously described [17]. Briefly, 5184 sequence-validated IMAGE consortium bacterial clones were purchased from Research Genetics (Huntsville, AL; catalog no. gf200). Probes were produced by PCR amplification of the cDNA inserts directly from bacterial cultures. Primers and unincorporated nucleotides were removed using a 96-well PCR clean-up kit from Qiagen (Valencia, CA). After quantitation, the purified PCR products were dried and resuspended in 10 µl of 2x SSC and printed onto aminoalkylsilane-modified glass slides (catalog no. S4651; Sigma, St. Louis, MO) using four quill-type pins (catalog no. SMP4; Telechem International, San Jose, CA) mounted onto an OmniGrid robot (GeneMachines, San Carlos, CA). A list of the clones on the array is available upon request. After printing, the slides were fixed, washed, and stored in the dark at room temperature and <40% humidity until use.
Fluorescent cDNA was made from reverse transcription of 40 µg of total RNA in the presence of 50 µM Cy5-dCTP or Cy3-dCTP (Amersham Pharmacia Biotechnology, Piscataway, NJ) in a 25-µl volume containing the following: 500 ng of oligo dT [12–18]; 1x Superscript Buffer; 400 U of Superscript II; 3.3 U of RNAse inhibitor (all from Gibco BRL, Grand Island, NY); 400 µM each of dGTP, dATP, and dTTP; 100 µM dCTP; and 10 mM dithiothreitol. The cDNA target was purified, lyophilized to dryness, resuspended in 10 µl of hybridization buffer (2x SSC, 0.1% sodium dodecyl sulfate [SDS], 100 ng/µl CotI DNA, 100 ng/µl oligo dA), denatured by boiling for 2.5 minutes, and hybridized to a microarray overnight at 62°C for 18 hours. Following hybridization, slides were washed and scanned for Cy3 and Cy5 fluorescence using an Axon GenePix 4000 microarray reader (Axon Instruments, Foster City, CA) and quantitated using GenePix software. Results were loaded into GeneSpring (Silicon Genetics, Redwood City, CA) and normalized using Lowess intensity-dependent normalization, and PCAF expression ratios for all 11 cell lines were examined.
Real-Time RT-PCR Analysis
UACC-1179 and BT-549 cells were infected with recombinant adenovirus serotype 5 containing GFP detection marker only (empty vector) or wild-type p53 and GFP detection marker (p53). RNA was isolated at 0, 12, 18, 24, and 36 hours followed by real-time RT-PCR. Reverse transcription used TaqMan Reverse Transcription Reagents (Roche Molecular Systems, Branchburg, NJ) and 200 ng of total RNA in a 50-µl reaction. Reverse transcription was primed with random hexamers and incubated at 25°C for 10 minutes followed by 48°C for 30 minutes, 95°C for 5 minutes, and a chill at 4°C. Each PCR reaction consisted of 3.75 µl of cDNA added to 12.5 µl of TaqMan Universal PCR Master Mix (Roche Molecular Systems), 1.25 µl of gene-specific primer/probe mix (Assays-by-Design; Applied Biosystems, Foster City, CA) and 7.5 µl of PCR water. PCR conditions were: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, alternating with 60°C for 1 minute using an Applied Biosystems 7000 SDS and Applied Biosystems' Assays On Demand primers specific to PCAF (assay ID Hs00187332 m1) and GAPDH (assay ID Hs99999905 m1). PCAF-specific product was normalized to GAPDH and quantitated using the comparative (ΔΔCt) Ct method as described in the ABI Prism 7000 sequence detection system user guide. PCAF and p53 expression values were averaged across three independent experiments in each cell line and standard error was calculated for graphing.
ChIP/Real-Time PCR Analysis
The ChIP assay for p53 bound to target promoters was performed as described previously [8]. Briefly, intact living cells were treated with 1% formaldehyde to form DNA-protein cross-links. Cells were then collected by centrifugation and resuspended in an SDS lysis buffer. DNA-protein complexes were sonicated to approximately 700 bp as determined by gel electrophoresis. One tenth of the sample was set aside for input control and the remaining sample was precleared with A/G PLUS Agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Following preclearing, the sample was split in half and one portion was incubated with 30 µl of anti-p53 antibody conjugated to agarose beads (clone DO1; Oncogene Research Products, Boston, MA) or 30 µl of protein A/G PLUS agarose. The other half of the sample was mock-incubated by leaving out the anti-p53 antibody and provided a no-antibody control for nonspecific protein-DNA binding. After incubation, beads were washed and chromatin-antibody complexes were eluted. DNA-protein cross-links in all samples, including the input DNA control, were reversed and DNA-purified for amplification.
Real-time PCR was used to analyze chromatin-immunoprecipitated DNA and input control DNA, using an ABI Prism 7000 sequence detection system (Applied Biosystems) detector. Real-time PCR was carried out in triplicate on 5 ng of DNA at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute, and 72°C for 1 minute. PCAF promoter-specific primers (Assays by Design; Applied Biosystems) were used at a final concentration of 500 nM for both forward and reverse primers (forward primer: GCACAATTTTGAAAACTACGTAACG; reverse primer: ATGCAGTGGAAAAAGTTTGAATACAA; TaqMan 6-FAM dye-labeled probe: CCAGAAACGAATCCT). Primers were designed to probe the ∼500-bp 5′ region of the PCAF transcription start site; the amplicon was designed to overlap specifically with a region rich in p53 consensus-binding sites (5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′ separated by 0–13 bp [18]). The presence of a single PCR product of correct size was confirmed by agarose gel electrophoresis. Quantitation was determined by applying the comparative Ct method. Briefly, fold enrichment was calculated by subtracting the Ct value of the ChIP DNA from the Ct value of the input DNA fraction and by using this value as the power to which 2 is raised (i.e., 2Ct(Input)Ct(ChIP)).
Activation of p53 by Doxorubicin and Knock Down with siRNA
MCF10A cells were mock-transfected for 6 hours and treated with doxorubicin, or transfected with a pool of five siRNAs (SMARTPool siRNA; Dharmacon RNA Technologies, Lafayette, CO) directed against p53 RNA for 6 hours followed by treatment with doxorubicin (Sigma) in six-well plates. RNA was isolated using an RNeasy Kit (Qiagen) and analyzed for PCAF expression by reverse transcription coupled with real-time PCR on an Applied Biosystems 7000 sequence detection system using the primers and probe described above. For the doxorubicin, dose-response RNA was isolated 18 hours after 0, 0.2, 0.5, 0.75, and 1 µM doxorubicin treatment; for the time course, RNA was isolated at 0, 12, 18, 24, 36, and 48 hours after a doxorubicin dose of 0.5 µM. PCAF PCR product was normalized to GAPDH and quantitated using the comparative Ct method as described above. PCAF expression was expressed relative to control (0 µMdoxorubicin in the dose response and time 0 in the time course). The dose-response experiment was averaged across three independent experiments, and graphed along with the standard error. The time course experiment was run twice with similar results.
Western Blot Analysis
Total cell lysate was separated on a 10% SDS-PAGE gel and transferred to PVDF membrane by standard methods. An anti-p53 mouse monoclonal antibody (clone DO1; Oncogene Research Products) diluted 1:1000 followed by a 1:10,000 dilution of goat anti-mouse HRP was used to detect p53. The β-actin used as a loading control was detected with a 1:2000 dilution of β-actin antibody (clone AC-40; Sigma) followed by a 1:10,000 dilution of goat anti-mouse horseradish peroxidase. The horseradish peroxidase -conjugated antibodies were visualized by exposure of the blot to enhanced chemiluminescence (ECL) staining (Amersham Pharmacia Biotechnology). The experiment was performed independently three times with similar results. Densitometry was used to quantitate β-actin and p53 bands. Each p53 band was normalized to the β-actin band in the same lane, and then normalized to the untreated control to calculate the fold induction shown.
Results
Table 1 shows the breast cell lines studied, PCAF induction following adenoviral transduction of empty vector or wild-type p53, p53 status/type of mutation, and p53 responsiveness. Each of the 11 cell lines was infected with an empty adenoviral vector or a vector expressing wild-type p53 and compared in a two-color hybridization to untreated MCF10A cells. The ratio of PCAF expression relative to MCF10A following the two treatments is shown in the second and third columns. To express PCAF induction within each cell line relative to treatment rather than MCF10A, the p53-infected ratio was divided by the empty vector ratio. The resulting PCAF induction ranged from 1.2 to 9.5 and is shown as (p53/empty vector). The average increase in PCAF expression in wild-type p53-infected cells over empty vector-infected cells was 3.5.
Table 1.
Breast Cell Lines Studied, PCAF Induction Following Adenoviral Transduction of Empty Vector or Wild-Type p53, p53 Status/Type of Mutation, and p53 Responsiveness.
| Cell Line | (Empty Vector/MCF10A) | (p53-Transduced/MCF10A) | (p53/Empty Vector) | p53 Mutation | p53 Responsive? |
| MCF10A | 0.6 | 1.5 | 2.7 | Wild type | Yes |
| UACC-1179 | 0.7 | 2.5 | 3.4 | R213X | Yes |
| UACC-2087 | 0.4 | 1.1 | 2.9 | V216M | No |
| UACC-893 | 0.4 | 1.2 | 2.9 | Wild type | Yes |
| BT-549 | 1.1 | 4.0 | 3.7 | R249S | Yes |
| HS-578T | 0.5 | 1.0 | 2.2 | V157T | No |
| MDA-MB-435 | 2.7 | 3.2 | 1.2 | G226E | No |
| MDA-MB-231 | 0.8 | 2.7 | 3.3 | R280K | Yes |
| MDA-MB-157 | 0.7 | 3.2 | 4.7 | Del codons 2–4 | Yes |
| MDA-MB-468 | 0.5 | 1.3 | 2.4 | R273H | Yes |
| MDA-MB-453 | 0.6 | 5.4 | 9.5 | Del codons 368 | Yes |
| Average | 0.8 | 2.5 | 3.5 |
PCAF expression was measured by cDNA microarray. Ratios in the first two columns show PCAF expression in the cell lines relative to untreated MCF10A cells following adenoviral transduction with empty vector (empty vector/MCF10A) or wild-type p53 (p53-transduced/MCF10A). To express PCAF induction within each cell line relative to treatment rather than MCF10A, the MCF10A reference was determined by dividing the (p53-transduced/MCF10A) ratio by the (empty vector/MCF10A) ratio. The resulting PCAF induction is shown as (p53/empty vector). The p53 status of each cell line was determined from the literature (http://p53.curie.fr) or as described previously [8]. Responsiveness to p53 was determined by multidimensional scaling as described previously [8].
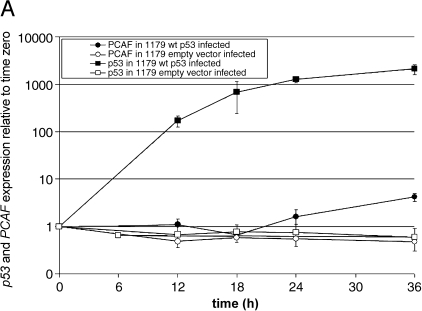
The UACC-1179 and BT-549 cell lines, which were both p53-mutant and responsive to transduced wild-type p53, were selected to further study PCAF induction by p53. First, the microarray results were expanded into a time course of p53 and PCAF induction following infection with wild-type p53 using real-time RT-PCR (Figure 1, A and B). The results confirmed PCAF induction following wild-type p53 infection and showed that induction of PCAF mRNA was time-dependent, whereas infection with empty adenoviral vector had no affect. In UACC-1179 cells at 36 hours, the average induction across three independent replicates was 4.2, whereas the BT-549 cells were more responsive to p53 infection with an average induction of 5.9 in three independent infections. At 24 hours, the time used in the microarray experiments, the measured induction in UACC-1179 cells was 1.6, compared to 3.4 when measured by microarray. In BT-549 cells, PCAF induction measured by real-time RT-PCR was 4.5, compared to 3.7-fold when measured by microarray.
Figure 1.
(A and B) p53 induces PCAF expression in p53 mutant breast cancer cells infected with wild-type p53. RNA from UACC-1179 and BT-549 cells was collected 6, 12, 18, 24, and 36 hours after infection with wild-type p53 and used to quantitate PCAF and p53 expression relative to time zero using real-time RT-PCR. Closed circles and squares represent p53 and PCAF expression in wild-type p53-infected UACC-1179 and BT-549 cells, whereas open circles and squares represent p53 and PCAF expression in empty vector-infected UACC-1179 and BT-549 cells. Each cell line was independently infected in triplicate; average expression is shown with one standard error (SE).
Because p53 activates transcription through binding to target promoters, the PCAF promoter was analyzed for tandem p53 consensus-binding sites. The PCAF promoter was found to contain multiple p53 consensus-binding sites; however, primers and probe were designed to query two regions at -400 and -580 bp from transcription start because they contained multiple overlapping potential p53-binding sites (Figure 2A). To test if p53 bound directly to the PCAF promoter, UACC-1179 and BT-549 cells infected with wild-type p53 were subjected to ChIP followed by real-time PCR amplification of the PCAF promoter. Intact wild-type p53-infected and control cells were treated with formaldehyde to cross-link genomic DNA to any bound proteins. The genomic DNA was isolated, sonicated into approximately 700-bp fragments, and a portion was immunoprecipitated with antibody to wild-type p53. Subsequent PCR amplification using primers specific to the PCAF promoter allowed the relative amount of p53-immunoprecipitated PCAF promoter in infected and control cells to be compared. The results in Figure 2, B and C show representative amplification plots from UACC-1179 and BT-549 cells. Comparison of the PCAF amplification curves from p53-infected and untreated control cells without immunoprecipitation revealed nearly identical amounts of PCAF promoter DNA in the samples (compare open squares to open circles). This result showed that equal amounts of input genomic DNA result in equal amounts of PCAF PCR product in the absence of enrichment using p53 antibody. In contrast, when the genomic DNA was immunoprecipitated with p53 antibody prior to amplification, the PCAF promoter amplified earlier from p53-infected cells rather than from untreated cells (compare closed squares to closed circles). Across three independent experiments, the average increase in p53 binding to the PCAF promoter in p53-infected cells was 8 ± 4 in UACC-1179 cells and 4 ± 2 in BT-549 cells compared to control. A negative control was done by performing the ChIP assay with a mock immunoprecipitation (no anti-p53 antibody); no significant product was amplified from these samples and therefore the amplification curves are not plotted in Figure 2, B and C. These results demonstrate that the wildtype p53 expressed from the adenoviral vector bound to the PCAF promoter in vivo, and support direct p53 transactivation of the PCAF promoter.
Figure 2.
(A) Diagram of the PCAF promoter region analyzed for p53 binding. The bent arrow represents the transcription start site, whereas the filled rectangle shows the first exon. The vertical arrows represent tandem p53 consensus-binding sites with greater than 80% match to the published consensus sequence. The open rectangles show the location of the primers used to amplify the PCAF promoter, and striped rectangle shows the position of the probe. The numbers indicate nucleotide position relative to transcription start. (B and C) p53 binds the PCAF promoter in vivo in wild-type p53-infected breast cancer cells. p53 binding to the PCAF promoter was measured in UACC-1179 and BT-549 after 24 hours of no treatment or after 24 hours of infection with wild-type p53 using ChIP coupled to real-time PCR analysis. Chromatin was immunoprecipitated using a p53 antibody (clone DO1) and PCAF promoter-specific real-time PCR was performed on input control DNA and DNA from the immunoprecipitated chromatin. The threshold bar for Ct determination was set within the linear range of PCR amplification and fold enrichment of p53 binding was determined by the comparative Ct method. The experiment was performed independently three times in each cell line with similar results; representative real-time PCR graphs are shown. Average fold enrichment of p53 binding was 8 ± 4 in UACC-1179 cells and 4 ± 2 in BT-549 cells compared to control.
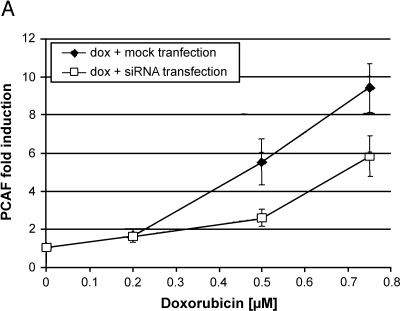
To verify the connection between wild-type p53 activity and PCAF expression, the endogenous wild-type p53 within MCF10A breast epithelial cells was activated and PCAF induction was measured using real-time PCR. Doxorubicin, a DNA-damaging agent, was used to stimulate p53 activity at concentrations of 0.2 to 0.75 µM. To show that PCAF induction following activation of p53 was, in fact, p53-dependent, some cells were pretreated with siRNA directed against p53 in order to block the accumulation of activated p53 following doxorubicin treatment. Figure 3A shows that after 18 hours after doxorubicin treatment, PCAF expression was induced in a dose-dependent manner, with the maximal induction at 0.75 µM (closed diamonds). To demonstrate that the induction of PCAF was p53-dependent, MCF10A cells were treated with a pool of five siRNAs targeted against p53 RNA for 6 hours prior to the 18 hours of doxorubicin treatment. Compared to the mock-transfected cells, cells transfected with siRNAs against p53 RNA induced less PCAF expression at similar doses of doxorubicin (Figure 3A, open squares).
Figure 3.
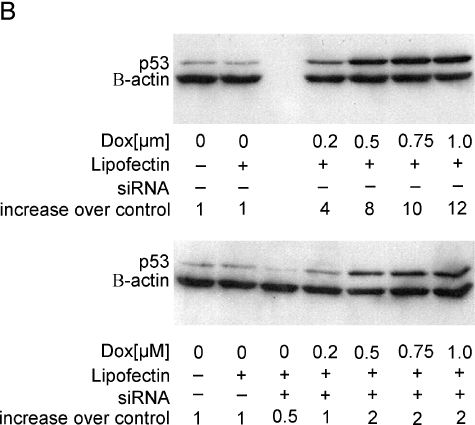
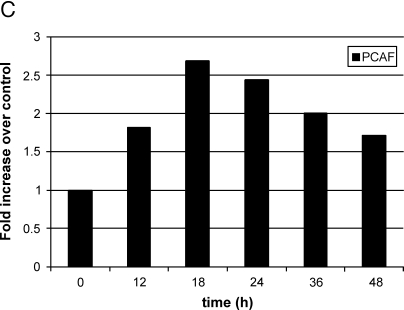
(A) Activation of endogenous wild-type p53 in MCF10A cells using doxorubicin induces PCAF expression, whereas loss of p53 prior to doxorubicin treatment reduces PCAF induction. MCF10A cells were mock-transfected for 6 hours and treated with doxorubicin or transfected with a pool of five siRNAs directed against p53 RNA for 6 hours followed by treatment with doxorubicin. At 24 hours, RNA was harvested and PCAF expression was quantitated using real-time RT-PCR. The average of three independent experiments is shown ±1 SE. (B) Western blot analysis showing accumulation of p53 protein following treatment with doxorubicin, or doxorubicin and siRNAs directed against p53. The top panel shows p53 accumulation following doxorubicin treatment; the bottom panel shows the same after pretreatment with siRNAs against p53. Bands were quantitated using densitometry and normalized first to the β-actin loading control and then to the untreated control (left lane of each panel) to calculate the increase over control shown below each western. (C) Representative time course of PCAF induction following doxorubicin treatment of MCF10A cells. Cells were treated with 0.5 µM doxorubicin and RNA were harvested at the times indicated. PCAF mRNA was quantitated using real-time RT-PCR, normalized to GAPDH, and expressed relative to time zero.
To confirm that the reduced PCAF induction in siRNA-treated cells was due to a loss of active p53, a Western blot analysis was performed to compare p53 protein levels following doxorubicin treatment or siRNA transfection and doxorubicin treatment. Figure 3B shows that the amount of p53 protein increased with doxorubicin dose (top panel), whereas pretreatment with siRNAs directed against p53 mRNA decreased p53 protein accumulation (bottom panel). For example, when normalized to the β-actin control, p53 protein at the 0.75 µM doxorubicin dose was 10-fold increased over control, but in siRNA-pretreated cells, p53 protein was induced only two-fold over control. The lower p53 protein accumulation in siRNA-pretreated cells after doxorubicin treatment correlates with the reduced PCAF induction observed in Figure 3A.
Lastly, in support of direct p53 activation of the PCAF promoter, a time course of PCAF induction was performed following treatment of MCF10A cells with 0.5 µM doxorubicin. This was the dose of doxorubicin shown to cause an eight-fold accumulation of p53 protein after 18 hours in Figure 3B; if PCAF was a direct target of p53 transactivation, then PCAF induction should occur soon after p53 accumulation. As seen in Figure 3C, PCAF mRNA was increased nearly two-fold at 12 hours following doxorubicin treatment and continued to rise up to 18 hours at which point PCAF mRNA declined slowly. This result confirms that PCAF mRNA increased rapidly following stimulation of p53 activity by doxorubicin and, along with the ChIP results, suggests that PCAF is a direct target of p53.
Discussion
The purpose of the studies presented here was to investigate the unexpected finding that PCAF was induced in breast cells transduced with wild-type p53. This observation was first made from the microarray data presented in Table 1. By calculating the PCAF induction relative to the empty vector control in Table 1, we insured that the calculated increase in PCAF expression was p53-specific, and not a result of the adenoviral infection itself. It is important to note that the p53 responsiveness listed in Table 1 was determined by multidimensional scaling based on the response of many genes, not PCAF alone. This distinction becomes important when considering that the three cell lines that were categorized as unresponsive to p53, of which two still induced PCAF. These three cell lines did not show a strong-enough response across the ∼5000 genes queried to be grouped with the cluster of responsive cell lines and thus are labeled unresponsive. Most importantly, this result should not be interpreted as indicating that PCAF was induced independently of p53 expression. Instead, PCAF was induced specifically by expression of wild-type p53 even when many other genes were not; this observation provides further evidence that PCAF was a direct target of p53. Interestingly, the lone cell line, MDA-MB-435, that did not induce PCAF may actually be a melanoma [19,20].
The induction of PCAF by p53 was confirmed and expanded by a time course study in which PCAF and p53 expression in UACC-1179 and BT-549 cells was shown to increase with time following infection with wild-type p53. Although both cell lines induced PCAF, BT-549 showed a more robust response than UACC-1179 despite having a smaller induction of p53 mRNA. These results likely reflect differences in the efficiency with which the adenoviral vector can express p53 mRNA in the two cell lines. Having confirmed the microarray results, ChIP using p53 antibody and real-time PCR amplification of the PCAF promoter showed that the induction of PCAF expression was associated with increased p53 binding to the PCAF promoter in vivo. The exact sequence to which wild-type p53 bound in the PCAF promoter was not identified by the ChIP assay; however, the primers and probe used to amplify the PCAF promoter likely recognize one of the p53-binding sites located in the 500-bp surrounding the primer probe upstream of the transcription start site. A search for tandem p53 consensus-binding sites in the 2-kb upstream of the PCAF transcription start site revealed two regions, at approximately -400 and -580, that contained multiple overlapping tandem consensus-binding sites for p53. The primers and probe used to amplify the PCAF promoter were designed to overlap the -580 region (Figure 2A), but would likely have detected p53 binding to the -400 region as well. Regardless of whether p53 was binding to one or both of these regions, the ChIP assay showed conclusively that in vivo p53 binding to the PCAF promoter increased when cells were infected with wild-type p53.
Recognizing that PCAF induction may have resulted from artificially high expression of wild-type p53 by the adenoviral vector, we switched to a more physiologically representative model for further studies. The model consisted of wild-type p53-expressing MCF10A cells in which p53 activity was induced by treating the cells with doxorubicin. PCAF induction by endogenous p53 in doxorubicin-treated MCF10A cells was measured using real-time RT-PCR. The results showed that PCAF induction increased with doxorubicin dose (Figure 3A), whereas a Western blot analysis confirmed concomitant accumulation of p53 protein (Figure 3B, top panel). Additionally, siRNA knockdown of p53 prior to doxorubicin treatment showed that reducing activated p53 protein (Figure 3B, lower panel) resulted in the reduction of PCAF expression (Figure 3A). In addition to demonstrating that PCAF induction was p53-dependent, these results showing induction of PCAF by endogenous p53 confirmed that the results seen with adenoviral transduction were not an artifact of overexpression of wild-type p53. Lastly, we performed a time course of PCAF induction in the MCF10A doxorubicin model, which showed that PCAF was induced as early as 12 hours following doxorubicin treatment and supported the results from the ChIP analysis suggesting that PCAF is a direct p53 target.
Stimulation of p53 by cellular insult, such as treatment with the DNA-damaging agent doxorubicin, leads to transactivation of gene expression by p53 and its coactivators such as PCAF. Because PCAF can acetylate p53 and histones, the results presented here suggest that a type of signal amplification may be possible. It is possible that the induced PCAF expression could lead to increased PCAF recruitment at p53-bound target promoters where PCAF could acetylate p53 and/or histones, thus increasing p53's transactivation activity. Such a series of events is suggested by recent studies in which p53 binding and acetylation appear to increase the recruitment of HATs to p53 target promoters with a subsequent increase in histone acetylation acetylation and transcription [8,21,22]. Additionally, PCAF has recently been shown to have a substrate preference for histone H3 N-terminal residues and histone H3 acetylation in two tumor-suppressor gene promoters increased following expression of p53 [7]. Finally, MDM2 was recently shown to inhibit PCAF acetylation of p53 [23]. Taken together with the results reported here, it appears possible that transactivation of p53 target genes may be controlled by a balance between the competing effects of PCAF and MDM2 in their interactions with p53 at the gene and protein levels.
Acknowledgements
We thank the Arizona Cancer Center Genomics Shared Service and Cell Culture Shared Service, and the Gene Transfer Vector Core, University of Iowa, for technical assistance.
Footnotes
This work was supported by grants CA65662 to B.W.F., NIH 73612 to F.E.D., 3P30 CA23074-19 to the Arizona Cancer Center, and ES06694 to the Southwest Environmental Health Sciences Center. M.O. was supported by a Cancer Biology Training grant from the National Institutes of Health. R.J.W. received support from a NIEHS Toxicology Training grant.
References
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 4.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 7.Trievel RC, Li FY, Marmorstein R. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal Biochem. 2000;287:319–328. doi: 10.1006/abio.2000.4855. [DOI] [PubMed] [Google Scholar]
- 8.Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez L, Domann J, Futscher FE. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene. 2003;22:3624–3634. doi: 10.1038/sj.onc.1206545. [DOI] [PubMed] [Google Scholar]
- 9.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Colosimo AL, Yang XJ, Liao D. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol Cell Biol. 2000;20:5540–5553. doi: 10.1128/mcb.20.15.5540-5553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 13.Meltzer P, Leibovitz A, Dalton W, Villar H, Kute T, Davis J, Nagle R, Trent J. Establishment of two new cell lines derived from human breast carcinomas with HER-2/neu amplification. Br J Cancer. 1991;63:727–735. doi: 10.1038/bjc.1991.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson F, Emerson J, Dalton W, Yang JM, McGee D, Villar H, Knox S, Massey K, Weinstein R, Bhattacharyya A. Clonal chromosome abnormalities in human breast carcinomas: I. Twenty-eight cases with primary disease. Genes Chromosomes Cancer. 1993;7:185–193. doi: 10.1002/gcc.2870070402. [DOI] [PubMed] [Google Scholar]
- 15.Trent J, Yang JM, Emerson J, Dalton W, McGee D, Massey K, Thompson F, Villar H. Clonal chromosome abnormalities in human breast carcinomas: II. Thirty-four cases with metastatic disease. Genes Chromosomes Cancer. 1993;7:194–203. doi: 10.1002/gcc.2870070403. [DOI] [PubMed] [Google Scholar]
- 16.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts GS, Futscher BW, Isett R, Gleason-Guzman M, Kunkel MW, Lu H. cDNA microarray analysis of multidrug resistance: doxorubicin selection produces multiple defects in apoptosis signaling pathways. Pharmacol Exp Ther. 2001;299:434–441. [PubMed] [Google Scholar]
- 18.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 19.Ellison G, Klinowska T, Westwood RFR, Docter E, French T, Fox JC. Further evidence to support the melanocytic origin of MDAMB-435. J Clin Pathol. 2002;55:294–299. doi: 10.1136/mp.55.5.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, Scudiero DA, Eisen MB, Sausville EA, Pommier Y, Botstein D, Brown PO, Weinstein JN. A Gene Expression database for the molecular pharmacology of cancer. Nature. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 21.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. cetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 22.Espinosa JM, Emerson BM. ranscriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 23.Jin Y, Zeng SX, Dai MS, Yang XJ, Lu H. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J Biol Chem. 2002;277:30838–30843. doi: 10.1074/jbc.M204078200. [DOI] [PubMed] [Google Scholar]