Abstract
Recognition of poly(A) sites in yeast pre-mRNAs is poorly understood. Employing an in vitro cleavage system with cleavage and polyadenylation factor (CPF) and cleavage factor IA we show that the efficiency and positioning elements are dispensable for poly(A)-site recognition within a short CYC1 substrate in vitro. Instead, U-rich elements immediately upstream and downstream of the poly(A) site mediate cleavage-site recognition within CYC1 and ADH1 pre-mRNAs. These elements act in concert with the poly(A) site to produce multiple recognition sites for the processing machinery, since combinations of mutations within these elements were most effective in cleavage inhibition. Intriguingly, introduction of a U-rich element downstream of the GAL7 poly(A) site strongly enhanced cleavage, underscoring the importance of downstream sequences in general. RNA- binding analyses demonstrate that cleavage depends on the recognition of the poly(A)-site region by CPF. Consistent with in vitro results, mutation of sequences upstream and downstream of the poly(A) site affected 3′-end formation in vivo. A model for yeast pre-mRNA cleavage-site recognition outlines an unanticipated high conservation of yeast and mammalian 3′-end processing mechanisms.
Keywords: mRNA/polyadenylation signals/pre-mRNA 3′-end processing/RNA–protein interactions
Introduction
Eukaryotic messenger RNA precursors (pre-mRNAs) must be modified at the 3′-end to serve as efficient templates for protein synthesis. The 3′-end processing reaction involves endonucleolytic cleavage followed by polyadenylation of the upstream cleavage product (for recent review see Zhao et al., 1999). Sequence elements in the 3′ untranslated regions (3′-UTRs) of pre-mRNAs specify the site at which adenosines are added [poly(A) site]. The mammalian 3′-end formation complex selects the poly(A) site (usually the dinucleotide CA) through recognition of the conserved upstream AAUAAA sequence and a degenerate downstream element (reviewed in Zhao et al., 1999). These interactions are mediated by CPSF-160 (Murthy and Manley, 1995) and CstF-64 proteins (MacDonald et al., 1994), respectively. Bridging of CPSF and CstF factors across the poly(A) site to form a stable ternary complex on the pre-mRNA is achieved through interactions of CstF-77 with CPSF-160 (Murthy and Manley, 1995) and other CstF subunits (Takagaki and Manley, 1994).
In contrast to higher eukaryotes, Saccharomyces cerevisiae uses degenerate and complex signals to direct the reaction. The definition of yeast polyadenylation signals started with the analysis of a mutant CYC1 allele that has a severe 3′-end formation defect in vivo (cyc1-512; Zaret and Sherman, 1982). The 3′-UTR region of this allele carries a deletion of elements that were also required for processing of synthetic RNA in extracts (Butler and Platt, 1988). Subsequently, these elements were defined as efficiency element (EE) and positioning element (PE), and equivalent elements were analysed in many yeast pre-mRNAs (for reviews see Guo and Sherman, 1996a; Zhao et al., 1999). The poly(A) site itself, which is in most cases Py(A)n, also acts as a polyadenylation signal (Heidmann et al., 1992). Since sequences downstream of the poly(A) site are generally not thought to be significant for 3′-end formation (reviewed in Zhao et al., 1999), complex assembly and cleavage site selection in yeast are believed to differ significantly from mammals.
Protein factors can be separated biochemically that are sufficient to reconstitute cleavage and polyadenylation in vitro (Chen and Moore, 1992). Site-specific cleavage requires cleavage and polyadenylation factor IA (CF IA), cleavage and polyadenylation factor IB (CF IB) and cleavage factor II (CF II). Specific polyadenylation occurs when CF IA, CF IB, poly(A)-binding protein (Pab1p) and polyadenylation factor I (PF I) are present. A factor harbouring PF I and CF II activities (designated cleavage and polyadenylation factor, CPF) was isolated from yeast extracts by affinity purification (Ohnacker et al., 2000). Stable and stoichiometric association of CF II and PF I subunits in CPF suggest that this factor forms a functional unit in vivo. Analysis of yeast polyadenylation factors identified a complex subunit composition that includes at least 15 different polypeptides (reviewed in Zhao et al., 1999).
RNA-binding proteins involved in pre-mRNA cleavage are predicted to mediate poly(A)-site recognition. The CF IB protein Nab4p/Hrp1p contains RNP-type RNA recognition motifs (Kessler et al., 1997). RNA-binding and SELEX analyses suggest that this protein interacts with EE sequences (Chen and Hyman, 1998; Valentini et al., 1999). Interestingly, pre-mRNAs are cleaved at the poly(A) site and at additional positions in the absence of Nab4p/Hrp1p, and selection of the correct cleavage site depends on Nab4p/Hrp1p in a concentration-dependent manner (Minvielle-Sebastia et al., 1998). The CF IA subunit Rna15p contains an RNA recognition motif (Minvielle-Sebastia et al., 1991). No specific interactions between Rna15p and pre-mRNA substrates are reported (Minvielle-Sebastia et al., 1991), but SELEX experiments identified U-rich consensus sequences as high-affinity binding ligands (Takagaki and Manley, 1997). UV-crosslinking experiments with purified CF II implicated the Ydh1p/Cft2p subunit in RNA binding (Zhao et al., 1997). Furthermore, the Yth1p subunit of CPF preferentially binds RNA near the poly(A) site (Barabino et al., 2000).
We show that in vitro cleavage of a short CYC1 substrate does not depend on EE and PE. Instead, mutation of U-rich elements upstream and downstream of the poly(A) site and mutation of the poly(A) site itself reduced correct cleavage. RNA-binding analyses with the CPF subunit Ydh1p/Cft2p and purified CPF demonstrate that interactions of this factor with the poly(A)-site region determine the cleavage site. These findings suggest that the topological arrangement of processing factors across the poly(A) site is conserved between yeast and mammals.
Results
Efficiency and positioning elements are dispensable for specific poly(A)-site recognition within a short CYC1 substrate in vitro
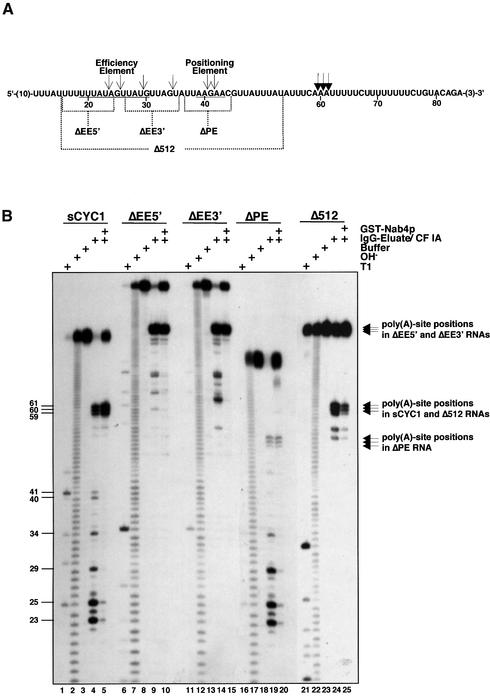
Previous work showed that endonucleolytic cleavage of mutant CYC1-512 RNA with CF IA and CF II factors in vitro can occur in the absence of CF IB (Minvielle-Sebastia et al., 1998). To corroborate these observations we designed a short 87 nucleotide (nt) CYC1 RNA substrate (sCYC1), which allowed analysis of cleavage products at nucleotide resolution (Figure 1A). Initially, we also produced mutant RNAs that contained deletions in 5′ and 3′ regions of the EE (ΔEE5′ and ΔEE3′), the PE (ΔPE) and both EE and PE sequences (Δ512; Figure 1A). To avoid length constraints in 5′ regions of ΔEE5′, ΔEE3′ and Δ512 RNAs, 38 nt of additional CYC1 upstream sequence were included in these substrates. RNAs were transcribed in vitro and 5′-end labelled. For in vitro cleavage we produced an IgG eluate derived from a ProtA–Pfs2p expressing strain (MO20; Ohnacker et al., 2000). Extract prepared from strain MO20 was incubated with IgG-agarose and the fusion protein bound to the agarose via the protein A tag. After extensive washes of the matrix, bound proteins were eluted by cleavage with TEV protease (Carrington and Dougherty, 1988). Silver-stain analysis of the IgG eluate after SDS–PAGE revealed a polypeptide band pattern typical for the CPF complex (not shown; Ohnacker et al., 2000). The IgG eluate was supplemented with purified CF IA and incubated with RNA under conditions that allowed cleavage only (see Materials and methods).
Fig. 1. Specific cleavage of sCYC1 by CPF and CF IA in vitro does not require efficiency and positioning elements. (A) Sequence of the sCYC1 transcript. Only CYC1-derived nucleotides are shown and positions relative to the transcription start site are indicated by numbers. Vector-derived nucleotides are indicated by numbers in brackets. EE and PE are underlined and indicated. Nucleotides deleted in ΔEE5′, ΔEE3′, ΔPE and Δ512 transcripts are indicated. ΔEE5′, ΔEE3′ and Δ512 RNAs contain 38 nt of additional 5′ CYC1 sequence. Filled arrowheads mark poly(A)-site cleavage positions and thin arrowheads mark cryptic cleavage sites. (B) In vitro cleavage assay with 1.0 µl of affinity-purified CPF and 0.4 µl of purified CF IA and indicated 5′-32P-labelled transcripts in the absence (lanes 4, 9, 14, 19 and 24) or presence of 100 ng of GST–Nab4p (lanes 5, 10, 15, 20 and 25). Also shown are T1 nuclease reactions (T1; lanes 1, 6, 11, 16 and 21), alkaline hydrolysis ladders (OH–; lanes 2, 7, 12, 17 and 22) and mock-treated samples (Buffer; lanes 3, 8, 13, 18 and 23). RNAs were resolved on 8.3 M urea/10% polyacrylamide sequencing gels. Arrows mark poly(A)-site cleavage positions within the indicated RNA substrates on the right of the panel. Length of cleavage products is indicated by numbers on the left of the panel.
Figure 1B shows that sCYC1 is cleaved efficiently at the poly(A) site after each adenosine residue (positions 59, 60 and 61) and at positions 23 and 25 in the EE (lane 4). Minor cleavage sites were observed at positions 24, 26, 29, 34, 40 and 41. Multiple products did not arise through exonuclease trimming since the use of internally labelled substrates resulted in the accumulation of corresponding upstream and downstream fragments (results not shown). When recombinant glutathione S-transferase (GST)– Nab4p (CF IB) was included in the assay, cleavage at major upstream sites in the EE was strongly reduced and completely inhibited at minor sites, while a significant amount of substrate was not cleaved at all (lane 5). Thus, Nab4p/Hrp1p was not required for cleavage but acted to restrict cleavage to poly(A)-site positions consistent with previous results (Minvielle-Sebastia et al., 1998).
ΔEE5′ and ΔEE3′ RNAs were cleaved efficiently at the poly(A) site (Figure 1B, lanes 9, 10, 14 and 15). Deletion of EE sequences harbouring cleavage sites observed in sCYC1 disturbed cleavage at these positions. Deletion of the PE did not affect upstream sites; however, cleavage at the poly(A) site was strongly reduced (lanes 19 and 20). Concomitantly, increased cleavage at site 29 was observed in the absence of GST–Nab4p. Δ512 RNA, in contrast, was cleaved efficiently at the poly(A) site in the absence of GST–Nab4p and this cleavage was strongly reduced in the presence of GST–Nab4p (lanes 24 and 25). Therefore, EE sequences were dispensable for cleavage of sCYC1 at the poly(A) site. In ΔPE RNA, poly(A)-site cleavage was severely reduced. This is, however, likely to be due to preferred cleavage at cryptic sites (position 29) since Δ512 RNA, lacking both EE and PE elements, was processed efficiently. We conclude that site-specific cleavage of sCYC1 at the poly(A) site can occur in the absence of classical yeast polyadenylation signals (EE and PE).
U-rich elements upstream and downstream of CYC1 and ADH1 poly(A) sites contribute to cleavage-site recognition
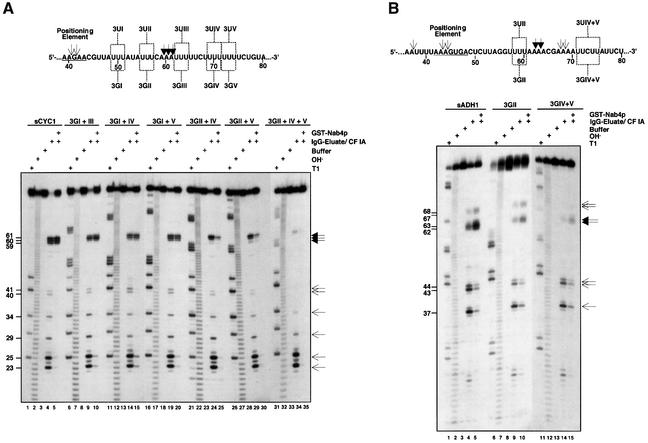
To identify RNA elements that specify cleavage at the sCYC1 poly(A) site we mutated sequence elements downstream of EE and PE sequences. A feature of these sequences is the predominance of uridines (Us). Initially, U-triplets (designated 3UI to 3UV; Figure 2A) both upstream and downstream of the poly(A) site were changed to guanosines (Gs; designated 3GI to 3GV) and the mutant RNAs were tested for in vitro cleavage. We found that these mutations only moderately influenced cleavage at poly(A)-site positions compared with sCYC1 (data not shown).
Fig. 2. Mutational analysis of U-rich elements upstream and downstream of sCYC1 and sADH1 poly(A) sites. Sequences of sCYC1 and sADH1 poly(A)-site regions are shown in (A) and (B), respectively. Only gene-derived nucleotides are shown and positions relative to the transcription start site are indicated by numbers. Filled arrowheads mark the major poly(A)-site cleavage positions and thin arrowheads mark cryptic cleavage sites. Also indicated are positions of 3UI to 3UV sequences as well as corresponding uridine to guanosine changes (3GI to 3GV). In vitro cleavage reactions in (A) and (B) were performed as described in the legend of Figure 1B with the 5′-32P-labelled transcripts indicated. Lane order of in vitro cleavage reactions in the absence or presence of GST–Nab4p, T1 digest, alkaline hydrolysis ladder and mock-treated reactions is as in Figure 1B. On the right of the panels filled arrowheads mark the major poly(A)-site cleavage positions and thin arrowheads mark minor cryptic cleavage sites. Length of cleavage products is indicated by numbers on the left of the panels.
We reasoned that combinations of 3G mutations upstream and downstream of the poly(A) site might enhance the phenotypes. Figure 2A shows processing of substrates that carry combinations of 3G mutations. U to G changes were visualized by nuclease T1 digestions (T1 nuclease cuts RNA 3′ of Gs; Figure 2A, lanes 6, 11, 16, 21, 26 and 32). 3GI + III, 3GI + IV and 3GI + V mutations resulted in moderate reduction of cleavage at poly(A)-site positions (lanes 9, 10, 14, 15, 19 and 20). Remarkably, with 3GII + IV and 3GII + V RNAs, cleavage at the poly(A) site was observed mainly at position 61 (lanes 24 and 29) and was strongly inhibited at this position when GST–Nab4p was present (lanes 25 and 30). This effect was further enhanced when 3GII + IV + V mutations were present in the same substrate. Trace amounts of cleavage at the poly(A) site observed in the absence of GST–Nab4p were lost in its presence (lanes 34 and 35). Importantly, the 3G mutant RNAs were not impaired in cleavage at upstream sites, which thus serves as a control for processing efficiency. These data strongly suggest that U-rich sequences immediately upstream and 10–15 nt downstream of the poly(A) site contribute to recognition of the CYC1 cleavage site.
To test whether U-rich elements contribute to cleavage in other pre-mRNAs we prepared a short ADH1 RNA (sADH1; Figure 2B). When assayed for cleavage with CPF and CF IA we observed cleavage of sADH1 at positions 37, 43, 44, 62 and 63 (Figure 2B, lane 4). These sites correspond to in vivo polyadenylation sites (Heidmann et al., 1992). In the presence of GST–Nab4p, cleavage was restricted mainly to positions 62 and 63, the major polyadenylation sites used in vivo (lane 5). Minor cleavage sites at positions 67 and 68 appeared concomi tantly. Next, we mutated U-triplets to Gs immediately upstream of position 62 (3U-II; see Figure 2B) and 10–15 nt downstream of this position (3UIV+V). We found that cleavage of both 3GII and 3GIV+V substrates was strongly inhibited at positions 62 and 63 in the presence (lanes 9 and 14) and absence of GST–Nab4p (lanes 10 and 15) compared with sADH1. We conclude that U-rich elements in ADH1 also contribute to poly(A)-site cleavage.
Involvement of poly(A)-site nucleotides in cleavage site selection
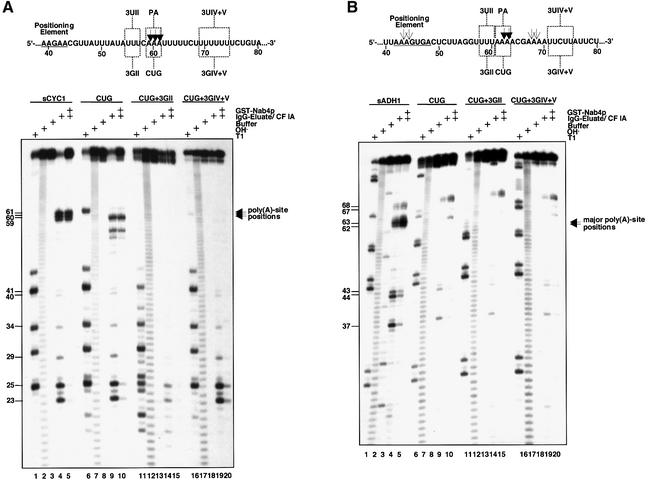
The poly(A)-site sequence Py(A)n was shown to act as polyadenylation signal (Heidmann et al., 1992). To evaluate the contribution of this element to in vitro cleavage of the short substrates, we changed triplets of adenosines at the major poly(A) sites of sCYC1 and sADH1 to CUG (Figure 3). The CUG mutation in sCYC1 changed poly(A)-site cleavage specificity but not efficiency. Cleavage occurred at position 59 (which in this case is a C) and additional cryptic sites (positions 53 and 55; lanes 9 and 10). When the CUG mutation was combined with upstream 3GII or downstream 3GIV+V mutations, cleavage at the poly(A) site was completely inhibited (lanes 14, 15, 19 and 20). In the case of sADH1, mutation of the major poly(A) site to CUG resulted in strong reduction of cleavage at positions 62 and 63 (Figure 3B, lane 9). The pronounced effect of the CUG mutation allowed only marginal enhancement of the phenotype in double mutants including 3GII or 3GIV+V mutations (lanes 14, 15, 19 and 20). These experiments show that poly(A)-site nucleotides act in concert with upstream and downstream sequences in determination of the cleavage site. Depending on the sequence context of the respective signal, multiple elements have to be destroyed to observe loss of cleavage.
Fig. 3. The poly(A) sites in CYC1 and ADH1 pre-mRNAs contribute to cleavage site selection. Sequences of sCYC1 and sADH1 poly(A)-site regions are shown in (A) and (B), respectively. Only gene-derived nucleotides are shown and positions relative to the transcription start site are indicated by numbers. Also indicated are sequences of 3UII, 3UIV+V and poly(A)-site positions (PA) as well as corresponding mutations (3GII, 3GIV+V and CUG). In vitro cleavage reactions in (A) and (B) were performed as described in the legend of Figure 1B with the 5′-32P-labelled transcripts indicated. Lane order of in vitro cleavage reactions in the absence or presence of GST–Nab4p, T1 digest, alkaline hydrolysis ladder and mock-treated reactions is as in Figure 1B. On the right of the panels filled arrowheads mark the major poly(A)-site cleavage. Length of cleavage products is indicated by numbers on the left of the panels.
Introduction of a U-rich element downstream of the GAL7 poly(A) site strongly enhances cleavage
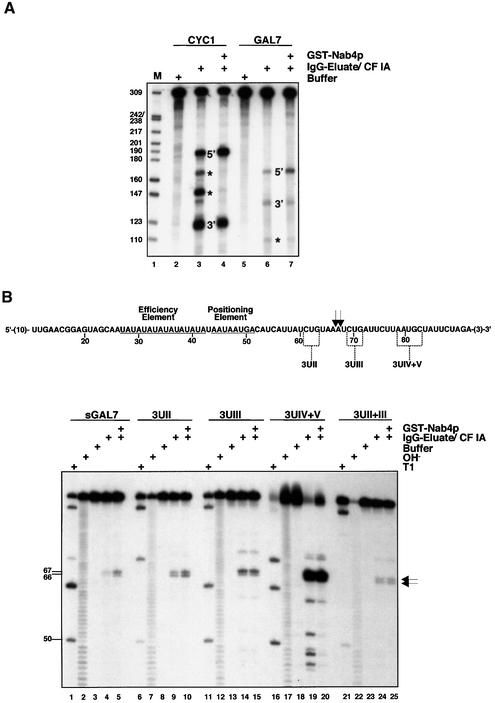
The results shown in Figures 2 and 3 argue in favour of an involvement of U-rich upstream and downstream sequences in poly(A)-site cleavage of yeast pre-mRNAs by CPF and CF IA. Such elements are, however, not always found at yeast poly(A) sites (Graber et al., 1999). GAL7 pre-mRNA displays a lower uridine content at the poly(A)-site region compared with CYC1 (see Figures 1A and 4B). When assayed in parallel, it is apparent that in vitro processing reactions on a long standard GAL7 substrate were significantly less efficient compared with a long CYC1 substrate (Figure 4A). Based on the disappearance of full-length substrate 40% of CYC1 RNA was cleaved in the absence of Nap4p and 64% when Nab4p was present (lanes 3 and 4). GAL7 pre-mRNA, in contrast, was cleaved with 7 and 19% efficiency, in the absence or presence of Nab4p/Hrp1p, respectively (lanes 6 and 7).
Fig. 4. Introduction of a U-rich downstream element strongly enhances cleavage of sGAL7 RNA by CPF and CF IA in vitro. (A) In vitro processing reactions with ‘long’ CYC1 and GAL7 pre-mRNA substrates. Internally 32P-labelled CYC1 and GAL7 RNAs were incubated either with buffer (lanes 2 and 5) or CPF and CF IA in the absence (lanes 3 and 6) or presence of GST–Nab4p (lanes 4 and 7). RNAs were resolved on a 6% polyacrylamide/8.3 M urea gel and quantified on a PhosphorImager (Molecular Dynamics). Upstream (5′) and downstream (3′) cleavage products that originate from cleavage at the poly(A) site are indicated. Cryptic cleavage products that are obtained in the absence of GST–Nab4p are indicated by an asterisk. HpaII-digested pBR322 fragments were 5′-end labelled and served as marker bands (M; lane 1); length of size marker bands is indicated by numbers on the left of the panel. (B) Sequence of the sGAL7 transcript. Only GAL7-derived nucleotides are shown and positions relative to the transcription start site are indicated by numbers. Vector-derived nucleotides are indicated by numbers in brackets. EE and PE are underlined and indicated. Also indicated are sequences of mutant 3UII, 3UIII and 3UIV+V positions. In vitro cleavage reactions were performed as described in the legend of Figure 1B with the 5′-32P-labelled transcripts indicated. Lane order of in vitro cleavage reactions in the absence or presence of GST–Nab4p, T1 digest, alkaline hydrolysis ladder and mock-treated reactions is as in Figure 1B. Arrows mark the poly(A)-site cleavage positions. Length of cleavage products is indicated by numbers on the left of the panel.
A short GAL7 substrate (sGAL7; Figure 4B) was cleaved inefficiently as well, albeit site-specifically at the in vivo poly(A) site (positions 66 and 67; Figure 4B, lanes 4 and 5). As a firm test of our hypothesis that U-rich elements contribute to cleavage we reasoned that processing of sGAL7 should improve upon introduction of such elements. Us were introduced immediately upstream of the poly(A) site (3UII) and 10–15 nt downstream of the cleavage site (3UIV+V; Figure 4B). Additionally, we introduced a U-triplet immediately downstream of the poly(A) site (3UIII) and we also produced all combinations of double mutants (Figure 4B and not shown). 3UII, 3UIII and 3UII+III mutations only marginally improved cleavage at the poly(A) site compared with wild-type sGAL7, independently of the absence (Figure 4B, lanes 9, 14 and 24) or presence (lanes 10, 15 and 25) of GST–Nab4p. Apparently, the sequence element UCUGU found immediately upstream of the GAL7 poly(A) site cannot be improved for in vitro cleavage when changed to UUUUU. In strong contrast, introduction of four uridine residues at the 3UIV+V position resulted in a substrate that was almost completely cleaved (Figure 4B, lane 19). This mutant RNA was also processed at cryptic sites that were not used in sGAL7. Inclusion of GST–Nab4p in the processing reaction inhibited cleavage at these minor sites, thus restricting cleavage to the poly(A)-site positions (lane 20). The same cleavage efficiency and pattern was observed when 3UII+IV+V and 3UIII+IV+V RNAs were tested (data not shown). These results demonstrate that GAL7 RNA carries a downstream element that is highly unfavourable for in vitro processing by CPF and CF IA, and highlight the importance of downstream elements for cleavage efficiency of yeast pre-mRNAs in general.
Ydh1p/Cft2p does not require efficiency and positioning elements for RNA binding
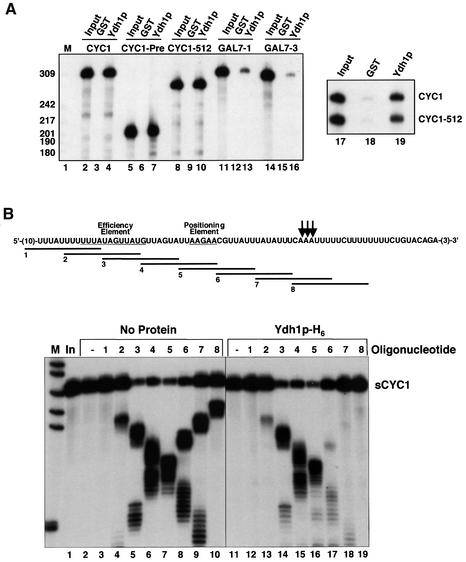
Our analysis of cis-acting signals suggested that sequences encompassing the poly(A) site are recognized by CPF, CF IA or both factors. To gain more insight into the RNA-binding properties of CPF and its components we analysed recombinant Ydh1p/Cft2p, which was expressed in Escherichia coli with a GST tag fused to the N-terminus and a His6 tag at the C-terminus (GST–Ydh1p-H6). In GST pull-down experiments (see Materials and methods) the protein bound efficiently to CYC1 RNA (Figure 5A, lane 4), CYC1-Pre RNA, which lacked sequences downstream of the poly(A) site, and CYC1-512 RNA, which has a 38 nt deletion encompassing both the EE and PE (Figure 5A, lanes 7 and 10). CYC1 was bound preferentially over CYC1-512 RNA in a ratio of ∼1.5:1.0, when both RNAs were included in the reaction (lane 19). We also tested wild-type GAL7-1 and GAL7-3 RNA, which has a deletion in the EE. Under the same conditions, binding of GAL7 RNAs was weaker compared with CYC1 RNAs (Figure 5A, lanes 13 and 16). Binding was specific for both protein and RNA since reactions contained excess unlabelled E.coli tRNA and no binding could be observed with GST alone (lanes 3, 6, 9, 12, 15 and 18). In summary, Ydh1p/Cft2p bound more efficiently to CYC1 RNAs compared with GAL7 RNAs. Sequences downstream of the CYC1 poly(A) site were not required for binding. CYC1 sequences deleted in CYC1-512 mutant RNA contributed to binding but were not essential for the interactions to occur.
Fig. 5. RNA-binding analysis of Ydh1p/Cft2p. (A) GST pull-down experiments with 0.5 µg of GST (lanes 3, 6, 9, 12 and 15) or GST–Ydh1p-H6 (Ydh1p; lanes 4, 7, 10, 13, 16 and 19) and indicated in vitro transcribed 32P-labelled RNAs. Lanes 2, 5, 8, 11, 14 and 17 show 10% of RNA included in binding reactions (Input). Length of marker bands (M; lane 1) is indicated on the left. Bands in lanes 17–19 were quantified on a PhosphorImager (Molecular Dynamics). (B) Sequence of the sCYC1 transcript. Arrows mark the poly(A) site. Positions of sequences complementary to 14mer DNA oligonucleotides (1–8) are indicated. The lower panel shows the RNase H protection profile of Ydh1p-H6 on sCYC1. In vitro transcribed 32P-labelled sCYC1 RNA was incubated with binding buffer (No Protein; lanes 2–10) or GST–Ydh1p-H6 (lanes 11–19). During binding to RNA the GST moiety was removed by cleavage with TEV protease included in the binding buffer. Protein-containing assays are therefore indicated by Ydh1p-H6. After pre-incubation the substrate was cleaved by addition of RNase H and oligonucleotides as indicated. For control, oligonucleotides were omitted in lanes 2 and 11. The RNA input is shown in lane 1. HpaII-digested pBR322 fragments were 5′-end labelled and served as marker bands (M).
To locate binding sites of Ydh1p/Cft2p on sCYC1 we carried out RNase H protection experiments. Internally labelled RNA was incubated either in the absence or presence of protein. Following pre-incubation, reaction mixtures were supplemented with antisense DNA oligonucleotides and RNase H. Eight overlapping 14mer oligonucleotides were employed, which covered most of sCYC1 (oligos 1–8; Figure 5B). Accessibility of the RNA to oligos 2–8 in the absence of protein resulted in the formation of RNA–DNA hybrids that were efficiently recognized by RNase H (Figure 5B, lanes 4–10). Appearance of distinct cleavage products showed that RNase H cleavage was specifically directed by antisense oligonucleotide sequences. Oligo 1 was not functional in this assay (lane 3). Pre-incubation of sCYC1 RNA with GST–Ydh1-H6 prevented hybrid formation with oligos 6, 7 and 8 and subsequent RNase H cleavage (lanes 17–19). Cleavage directed by oligo 2 was reduced in multiple experiments (lane 13). No significant protection could be observed when oligos 3, 4 and 5 were used (lanes 14–16). We cannot entirely exclude the possibility that binding of Ydh1/Cft2p on sCYC1 at a certain site influences RNA conformation and thus oligo accessibility at another position. This RNA-binding analysis is, however, consistent with results obtained from GST pull-down assays (see above) and previous UV-crosslinking experiments (Zhao et al., 1997). Thus, we conclude that Ydh1p/Cft2p binds sCYC1 with high affinity at sequences surrounding the poly(A) site and weaker at the EE.
Poly(A)-site recognition and cleavage depend on binding of RNA elements by CPF
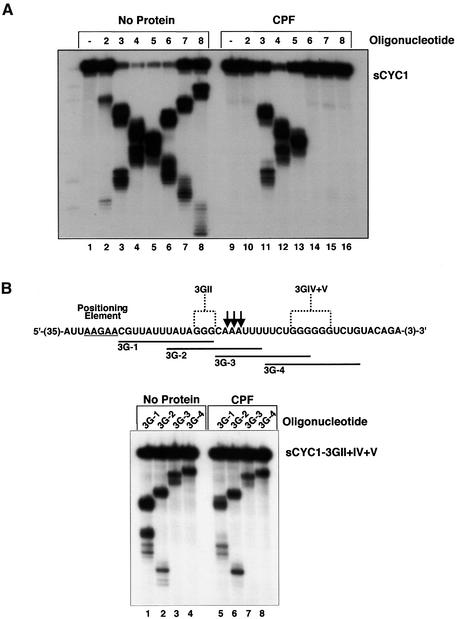
Binding sites of Ydh1p/Cft2p on sCYC1 overlapped with cleavage sites observed in vitro (see Figure 1B, lane 4), suggesting that CPF binding might determine the cleavage site. To test this hypothesis we used purified CPF and performed RNase H protection experiments on sCYC1. CPF completely protected the RNA from RNase H degradation directed by oligonucleotides 2, 6, 7 and 8 (Figure 6A, lanes 10 and 14–16). Some accumulation of full-length RNA was observed with oligonucleotides 3, 4 and 5 (lanes 11–13). This protection pattern paralleled the one found for Ydh1p/Cft2p. Consequently, high-affinity binding sites of CPF overlap with major cleavage sites in the efficiency element (oligo 2) and the poly(A) site (oligos 6, 7 and 8), respectively. Partial protection of low-affinity binding sites occurred where minor in vitro cleavage sites were observed. To test rigorously whether binding of CPF on sCYC1 determines cleavage, we performed RNase H protection experiments on a substrate that is not cleaved at the poly(A) site (3GII+IV+V RNA; see Figure 2A, lanes 34 and 35) using antisense DNA oligonucleotides covering the poly(A) site and mutated regions. As shown in Figure 6B we did not observe significant protection of RNase H degradation directed by oligonucleotides 3G-1 to 3G-4 compared with reactions without protein (lanes 1–8). These results strongly suggest that cleavage of sCYC1 at the poly(A) site depends on recognition of sequences encompassing the poly(A) site by CPF.
Fig. 6. Poly(A)-site cleavage requires binding by CPF. (A) RNase H protection profile of purified CPF on sCYC1. The assays were performed with the same substrate and DNA oligonucleotides as described in the legend of Figure 4B. Protein-containing samples are indicated by CPF (lanes 9–16). For control, oligonucleotides were omitted in lanes 1 and 9. (B) RNase H protection assay as described above with purified CPF and mutant sCYC1-3GI+IV+V substrate. Only the region of the RNA substrate containing the mutated nucleotides is shown. Arrows indicate the poly(A)-site positions within sCYC1 and mutations are indicated as 3GII and 3GIV+V, respectively. Positions of sequences complementary to 14mer DNA oligonucleotides (3G-1 to 3G-4) are indicated.
U-rich elements upstream and downstream of the poly(A) site act as polyadenylation signals in vivo
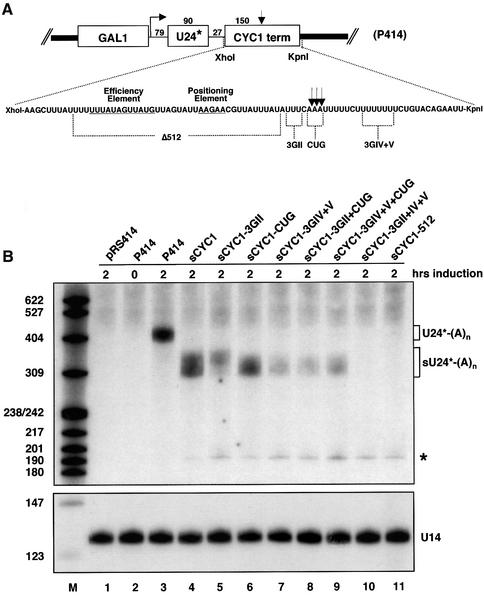
Finally, we analysed the effects of mutations within the newly identified elements on 3′-end formation in vivo. A reporter system described by Fatica et al. (2000) allows expression of U24* snoRNA under the control of an inducible GAL1 promotor (P414; Figure 7A). Following galactose induction, transcripts are detected by northern analysis with a probe complementary to a specific tag sequence. Fatica et al. (2000) showed that the U24* transcript is specifically polyadenylated when a downstream CYC1 terminator sequence is present. Consist ently, we found that a U24* RNA of >400 nt accumulated after galactose induction for 2 h (Figure 7B, lane 3), corresponding to the polyadenylated form of the U24* RNA [U24*-(A)n; Fatica et al., 2000]. This RNA cannot be detected in yeast cells carrying an empty vector (pRS414; Figure 7B, lane 1) or prior to galactose induction (lane 2). The CYC1-terminator of P414 was replaced by sequences encoding wild-type or mutant sCYC1 3′-UTR sequences. sCYC1 was sufficient for accumulation of a U24* transcript of >310 nt [Figure 7B, lane 4; referred to as sU24*-(A)n]. This RNA is shorter compared with U24*-(A)n derived from P414, due to less CYC1 sequence (∼100 nt) upstream of the cleavage site. Poly(A) sites containing 3GII and 3GIV+V single mutations showed clearly reduced accumulation of sU24*-(A)n (Figure 7B, lanes 5 and 7); the CUG mutation had a weak effect (lane 6). All double mutants tested (lanes 8–10) showed reduced levels of sU24*-(A)n. The 3GII+IV+V mutation did not allow detectable accumulation of sU24*-(A)n (Figure 7B, lane 10). The Δ512 mutation served as control and no sU24*-(A)n could be detected (lane 11), consistent with previous results (Zaret and Sherman, 1982). Interestingly, we observed minor accumulation of a 190 nt U24* RNA in all cells carrying the small CYC1 3′-UTR constructs (*; lanes 4–11). Fatica et al. (2000) identified a U24* species of the same size as U24*, which was trimmed to the 3′-end of the snoRNA body by 3′→5′ exonucleases. Thus, constructs carrying a short sCYC1 3′-UTR produce some snoRNA transcripts that are subjected to exonuclease trimming. Loading of equal amounts of total RNA is shown by comparable levels of U14 snoRNA in all lanes (Figure 7B, lanes 1–11, lower panel). In summary, U-rich elements encompassing the poly(A) site were required for 3′-end formation in vivo. Consistent with results obtained in vitro, combinations of mutations upstream and downstream of the poly(A) site were most effective.
Fig. 7. In vivo analysis of the requirement for U-rich elements in 3′-end formation. (A) The structure of the GAL1-U24*-CYC1 terminator construct (P414) is depicted schematically. Indicated are vector sequences (thick lines), the GAL1 promotor sequence, the transcription start site, the body of the U24* snoRNA and the CYC1 terminator sequence and the size of sequences in nucleotides. Wild-type and mutant short CYC1 3′-UTRs replacing the XhoI–KpnI fragment of P414 are indicated (RNA sequences are shown). Positions of Δ512, 3GII, CUG and 3GIV+V mutations are indicated. The Δ512 sequence contains 38 nt of additional 5′-CYC1 sequence. The poly(A) site is marked by arrows. (B) Northern analysis of total RNA extracted from wild-type W303 cells carrying GAL1-U24* constructs [see (A)] as indicated on top of each lane. pRS414 (lane 1; Stratagene) served as negative control. Induction with galactose is indicated in hours. Total RNA (8 µg) was resolved on 8.3 M urea/8% polyacrylamide and transferred to Hybond N+ membrane. Hybridization was performed with oligonucleotides complementary to the tag of U24* and U14 snoRNA. Indicated are positions of polyadenylated U24* RNA [U24*-(A)n, short polyadenylated U24* RNA (sU24*-(A)n] and a 3′→5′-trimmed product (*). Sizes of 5′-end labelled marker bands (HpaII-digested pBR322 fragments) are indicated on the left of the panel.
Discussion
EE, PE and poly(A)-site signals were suggested to be sufficient to direct cleavage and polyadenylation of yeast pre-mRNAs (Guo and Sherman, 1996b). CF IA, CF IB and CF II trans-acting factors were initially defined as sufficient for cleavage in vitro (Kessler et al., 1996). Subsequent work showed, however, that CF IB is dispensable for cleavage activity and instead controls cleavage-site selection (Minvielle-Sebastia et al., 1998). These observations also resulted in the surprising finding that CYC1-512 RNA is cleaved by CF IA and CF II in vitro when CF IB is absent, suggesting that both EE and PE are dispensable for cleavage.
We extended this observation by analysing cleavage of a short CYC1 substrate by CPF and CF IA in vitro. Consistent with previous results CF IB was not required for cleavage activity but restricted cleavage to the poly(A) site. While deletion of EE sequences in sCYC1 did not influence poly(A)-site cleavage, removal of the PE had a strong effect. In contrast, a mutant RNA containing deletions of both EE and PE (Δ512) was specifically and efficiently cleaved. Thus, the PE contributed to poly(A)-site cleavage only when an intact EE was present. It seems unlikely that an RNA-binding activity recognizing the PE is required in the context of the ΔPE substrate but not the Δ512 RNA. We propose that deletion of the PE generated an upstream-binding site for the processing complex. Increased use of an otherwise minor site (position 29; Figure 1B, lane 19) supports this interpretation. Thus, the PE contributed to cleavage at the poly(A) site by preventing assembly of the cleavage complex at adjacent positions. In summary, we conclude that in vitro cleavage of sCYC1 by CPF and CF IA can occur at the poly(A) site in the absence of EE and PE.
As a consequence, additional signals directing cleavage by CPF and CF IA were predicted. Mutations within U-rich elements upstream and downstream of the cleavage site resulted in inhibition of cleavage, suggesting that these elements represent cleavage signals. Furthermore, these sequences act in concert with the poly(A) site. In sCYC1, single mutations of neither U-rich element nor the poly(A) site alone completely inhibited cleavage, whereas combinations of mutations were highly effective. We interpret these observations to mean that multiple interactions occur at the poly(A)-site region and that effects of mutations within these elements depend on the strength of remaining interactions. Mutation of U-rich upstream and downstream sequences and the poly(A) site in sADH1 strongly reduced cleavage, indicating a general requirement for these elements in yeast pre-mRNA 3′-end formation. Strikingly, single mutations in sADH1 were more effective compared with sCYC1, suggesting that the sequence context of the poly(A) site or secondary structure constraints influence poly(A)-site recognition.
The missing requirement for downstream elements in yeast pre-mRNA 3′-end formation is a major difference compared with higher eukaryotes. Previous analyses in vivo and in vitro showed that many yeast pre-mRNAs can be cleaved and polyadenylated in the absence of extensive downstream sequences (reviewed in Zhao et al., 1999). We identified downstream sequences as part of a larger RNA recognition surface that is recognized by CPF. Loss of an RNA–protein interaction downstream of the poly(A) site will affect 3′-end formation only when the remaining contacts are sufficiently weak. This is likely to be the case for sADH1 cleavage in vitro and polyadenylation of ADH2 pre-mRNA in vivo, which requires a downstream sequence (Hyman et al., 1991). The intriguing observation that introduction of four uridine residues downstream of the GAL7 poly(A) site strongly enhances cleavage in vitro suggests that downstream elements are involved in 3′-end formation of yeast pre-mRNAs in general.
Analysis of polyadenylation of an inducible U24* snoRNA showed that the cis-acting elements identified in this study are also required for 3′-end formation in vivo. Consistent with in vitro data, combinations of mutations within U-rich elements upstream and downstream of the poly(A) site were most effective, supporting our idea that recognition of the poly(A)-site region involves multiple RNA–protein interactions. The phenotypes of mutations within specific pre-mRNAs will therefore depend on the ‘strength’ of interactions at the respective poly(A) site. Computational analyses of yeast 3′-UTR sequences identified U-rich sequences that are statistically over-represented immediately upstream and downstream of the poly(A) site (Graber et al., 1999). Our results suggest that these U-rich sequences are genuine polyadenylation signals, which are essential for cleavage-site recognition, and that the U-content at the poly(A) site reflects the ‘strength’ of a poly(A) signal. But how is cleavage-site selection achieved in yeast pre-mRNAs with a lower uridine content, e.g. GAL7? Since this RNA is cleaved site-specifically by CPF and CF IA in vitro it seems unlikely that an alternative mode of poly(A)-site recognition exists for non-U-rich poly(A) sites. The GAL7 downstream signal is highly unfavourable for in vitro cleavage by CPF and CF IA, but 3′-end formation in vivo might involve auxiliary RNA-binding proteins that increase processing efficiency.
The functional requirement for elements encompassing the poly(A) site implies that trans-acting factors recognize these signals. We found that Ydh1/Cft2p recognized the poly(A)-site region. Furthermore, the RNA-binding profiles of the CPF subunits Yhh1p/Cft1p (B.Dichtl, unpublished data) and Yth1p (Barabino et al., 2000) overlap with that found for Ydh1p/Cft2p, suggesting that these CPF subunits form a continuous RNA interaction surface. Mapping of CPF-binding sites on sCYC1 revealed interactions with sequences encompassing the poly(A) site and the EE. Since RNA-binding sites of CPF coincided with major cleavage sites in sCYC1 we postulated that CPF interactions determine the cleavage site. That this is indeed the case could be shown by loss of CPF binding on a mutant substrate that is not cleaved at the poly(A) site.
Control of cleavage site selection by CF IB was reported for ‘longer’ pre-mRNAs (Minvielle-Sebastia et al., 1998). Consistent with CF IB binding at the EE, recombinant Nab4p/Hrp1p prevented cleavage at EE positions in sCYC1. Therefore, CF IB is most likely to exert its function in vitro by competing with CPF for binding sites on the substrate. Nab4p/Hrp1p is also likely to play an important role in the apparent discrepancy of CYC1-512 RNA processing by purified factors in vitro and the strong effect of this mutation on polyadenylation in vivo. The basis for these conflicting results remains unclear and we can only speculate about possible reasons: (i) Nab4p/Hrp1p competes with CPF for binding at the poly(A) site of the emerging transcript in vivo due to the absence of its ‘normal’ binding site (EE). (ii) Binding of Nab4p/Hrp1p to the EE is required to ‘mark’ an upcoming poly(A) signal and to enable in vivo poly(A)-site recognition by CPF. (iii) Efficient recognition of the poly(A) site by CPF in vivo requires interactions with Nab4p/Hrp1p that are non-essential in vitro. Irrespective of the question of whether these scenarios correctly describe the phenotype of the CYC1-512 mutation, poly(A)-site recognition occurs by interactions of CPF with signals encompassing the poly(A) site, as demonstrated here.
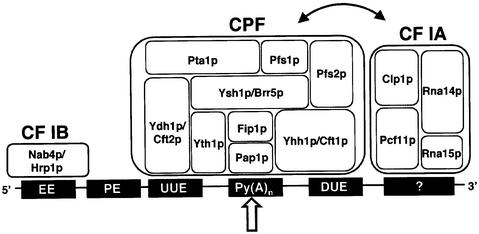
How does the 3′-end formation complex assemble at yeast poly(A) sites? We propose that CPF makes essential RNA–protein contacts with signals encompassing the cleavage site. CF IB binding to the EE prevents cleavage at this position and thus restricts CPF binding to the poly(A)-site sequence. An active processing complex is formed through association of CF IA with CPF mediated by protein–protein interactions (Ohnacker et al., 2000). The CF IA subunit Rna15p might contribute to recognition of U-rich elements, since SELEX experiments identified U-rich consensus sequences as high-affinity ligands (Takagaki and Manley, 1997). A scenario where CPF and CF IA sub-complexes assemble across the poly(A) site seems attractive (Figure 8). Such an organization of 3′-end formation factors occurs at poly(A) sites of higher eukaryotes where CPSF binds the hexanucleotide AAUAAA upstream of the poly(A) site and CstF makes contact to the downstream element. Moreover, CPF contains four proteins (Yhh1p/Cft1p, Ydh1p/Cft2p, Ysh1p/Brr5p and Yth1p) that are homologous to mammalian CPSF subunits (160, 100, 73 and 30 kDa), and CF IA contains two proteins (Rna15p and Rna14p) that are homologous to CstF subunits (77 and 64 kDa). Conservation of 3′-end formation factors and their topological arrangement at the site of catalysis suggests equivalent mechanisms for the production of poly(A) tails in both organisms.
Fig. 8. Schematic model for poly(A)-site recognition in yeast pre-mRNAs. Shown are functionally important cis-acting elements found at yeast polyadenylation sites and their interactions with trans-acting factors. The EE is the binding site of CF IB. The PE, which often occurs 3′ to the EE, could so far not be shown to interact with a polyadenylation factor. The Py(A)n sequence at the polyadenylation site, upstream U-rich element (UUE) and the downstream U-rich element (DUE) sequences identified in this work are bound by CPF and its RNA-binding subunits. Since several RNA–protein interactions are predicted to occur simultaneously we cannot assign exact binding sites of Ydh1p/Cft2p, Yth1p and Yhh1p/Cft1p. It is unclear which RNA element is bound by the RNA-binding components of CF IA, as indicated by a bar with a question mark. Protein–protein interactions between CPF and CF IA sub-complexes are indicated by a curved arrow. The arrangement of polypeptide subunits within the sub-complexes ignores known protein–protein interactions. The broad arrow marks the site of cleavage and polyadenylation.
Materials and methods
Yeast procedures
Yeast strains were transformed according to Gietz and St Jean (1992). Induction experiments were performed as described by Fatica et al. (2000) with wild-type W303 cells (Mata, ura3-1, trp1-1, ade2-1, leu2-3 112, his3-11,15) carrying plasmids as indicated in Figure 7 and Table I.
Table I. Plasmids used for run-off transcription of short RNA substrates and in vivo analysis.
| Template gene | 3′-UTR sequencea | Plasmid for run-off transcriptionb | Plasmid for in vivo analysisc |
|---|---|---|---|
| CYC1 | sCYC1 (wild type) | pBD101 | pBD256 |
| CYC1 | ΔEE5′ | pBD140 | |
| CYC1 | ΔEE3′ | pBD141 | |
| CYC1 | ΔPE | pBD139 | |
| CYC1 | Δ512 | pBD138 | pBD263 |
| CYC1 | 3GI | pBD165 | |
| CYC1 | 3GII | pBD166 | pBD257 |
| CYC1 | 3GIII | pBD167 | |
| CYC1 | 3GIV | pBD168 | |
| CYC1 | 3GV | pBD169 | |
| CYC1 | 3GIV+V | pBD259 | |
| CYC1 | 3GI+III | pBD170 | |
| CYC1 | 3GI+IV | pBD171 | |
| CYC1 | 3GI+V | pBD172 | |
| CYC1 | 3GII+IV | pBD173 | |
| CYC1 | 3GII+V | pBD174 | |
| CYC1 | 3GII+IV+V | pBD176 | pBD262 |
| CYC1 | CUG | pBD208 | pBD258 |
| CYC1 | 3GII+CUG | pBD209 | pBD260 |
| CYC1 | 3GIV+V+CUG | pBD210 | pBD261 |
| ADH1 | sADH1 (wild type) | pBD159 | |
| ADH1 | 3GII | pBD192 | |
| ADH1 | 3GIV+V | pBD194 | |
| ADH1 | CUG | pBD205 | |
| ADH1 | 3GII+CUG | pBD206 | |
| ADH1 | 3GIV+V+CUG | pBD207 | |
| GAL7 | sGAL7 (wild type) | pBD142 | |
| GAL7 | 3UII | pBD198 | |
| GAL7 | 3UIII | pBD199 | |
| GAL7 | 3UIV+V | pBD200 | |
| GAL7 | 3UII+III | pBD201 | |
| GAL7 | 3UII+IV+V | pBD202 | |
| GAL7 | 3UIII+IV+V | pBD203 |
a3′-UTR sequences and corresponding mutant RNAs are indicated in the figures.
bSequences were cloned in the HindIII–EcoRI sites of pGEM3 (Promega).
c3′-UTR sequences were PCR amplified using run-off transcription plasmids as templates and cloned into the XhoI–KpnI sites of P414 (GAL1-U24*-CYC1-terminator; Fatica et al., 2000).
Plasmids
pGDV1 is a derivative of pGEX-4T-3 (Pharmacia). The multiple cloning site (MCS) of pGEX-4T-3 was replaced by the MCS of pRS315-LEU2-ProtA–TEV (Senger et al., 1998), introducing a recognition sequence for TEV protease 3′ of the GST coding region. pBD71 (GST–Ydh1p-H6) was produced by PCR amplification of the YDH1/CFT2 coding region with oligonucleotides YDH1 (AATGGCCATGGAGATGACTTATAAATACAATTGC) and YDHCH6 (TTTTTGGATCCTCAGTGGTGGTGGTGGTGGTGGATTTTTGC) and cloning of the NcoI–BamHI (blunt) fragment into the NcoI–XhoI (blunt) sites of pGDV1. DNA templates of short transcripts were obtained by PCR and are summarized in Table I. Deletions or base-changes as well as HindIII and EcoRI restriction sites were primer encoded. These sites were used to clone fragments into pGEM3 (Promega). Constructs were verified by sequencing on a Perkin-Elmer capillary sequencer. Constructs used for in vivo analysis were obtained by PCR amplification of 3′-UTR sequences using plasmids for run-off transcription as templates (see Table I). The respective sequences were cloned in the XhoI–KpnI sites of P414 (Fatica et al., 2000) with oligonucleotide-derived restriction sites.
DNA templates and in vitro transcription
Internally 32P-labelled CYC1, CYC1-512, CYC1-Pre, GAL7-1 and GAL7-3 RNAs were produced as described (Minvielle-Sebastia et al., 1998). Templates for short transcripts (Table I) were linearized with EcoRI, transcribed by SP6 RNA polymerase in the presence of [α-32P]UTP and purified on 8.3 M urea/12% polyacrylamide gels. For 5′-end labelling, RNAs were obtained by cold transcription, treated with alkaline phosphatase and labelled with [γ-32P]ATP and T4 polynucleotide kinase.
In vitro cleavage assays
CPF factor was obtained from MO20 yeast extracts as previously described (Ohnacker et al., 2000). A 1 µl aliquot of CPF was supplemented with 0.4 µl of a purified CF IA fraction (MonoQ; Minvielle-Sebastia et al., 1998) and incubated with 20 fmol of labelled RNA under standard processing conditions for 1 h at 30°C (Minvielle-Sebastia et al., 1994). To allow cleavage only, MgAc was replaced by EDTA and ATP was replaced by CTP. One hundred nanograms of GST–Nab4p (Minvielle-Sebastia et al., 1998) were added to the reactions as indicated in the figures.
RNA isolation and northern analyses
Total RNA was isolated from 10.0 OD600nm of yeast cultures by a hot-phenol extraction procedure (Collart and Oliviero, 1994). RNAs were resolved on 8.3 M urea/8% polyacrylamide gels, and northern hybridization was performed according to Tollervey (1987) with oligonucleotide anti-U24-tag (TGCGGACTGCCTGGATGCCGGTT) and anti-U14 (TCACTCAGACATCCTAGG) labelled with [γ-32P]ATP and T4 polynucleotide kinase.
Recombinant protein expression and purification
Escherichia coli BL21 pLysS (Studier, 1991) transformed with plasmid pBD71 (GST–Ydh1p-H6) was grown at 25°C to OD600 2.0–4.0. After induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside, incubation was continued for 6 h. Cell lysis and purification by binding to Ni-NTA (nickel-nitrilotriacetic acid-agarose; Qiagen) were performed according to the manufacturer’s instructions. Fractions were supplemented with 100 mM KCl and further purified on glutathione Sepharose 4B as recommended (Pharmacia); however, all steps were performed at 4°C or on ice. Proteins were eluted with buffer G (10 mM reduced glutathione, 50 mM Tris-HCl pH 8.0, 0.01% NP-40, 0.1 mg/ml bovine serum albumin, 10% glycerol).
RNA–protein interactions
GST pull-down adapated from Murthy and Manley (1995): 500 ng of GST or GST–Ydh-H6 protein were bound to 15 µl of glutathione Sepharose 4B at 4°C for 60 min and the resin washed with 1 ml of RNA-binding buffer [RBB; 13 mM HEPES–KOH pH 7.9, 28 mM (NH4)2SO4, 33 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 0.2 mM dithiothreitol (DTT), 0.01% NP-40]. A 50 µl aliquot of RBB containing 50 fmol of labelled RNA, 0.5 U of RNAguard (Pharmacia) and 1 mg/ml E.coli tRNA was added, followed by rotation at 4°C for 120 min. The resin was washed three times (1 ml of RBB) for 5–15 min by rotation at 4°C and treated with 50 µl of proteinase K-mix [1 µl proteinase K (20 mg/ml), 0.5 µl glycogen (20 mg/ml), 100 mM Tris pH 7.5, 12.5 mM EDTA, 1% (w/v) SDS, 150 mM NaCl] for 30 min at 42°C. After extraction with one volume of phenol/chloroform/isoamylalcohol (25:24:1), RNA was ethanol precipitated and resolved on 8.3 M urea/6% polyacrylamide gels.
RNase H mapping: for each assay 0.5–1.0 µg of protein was bound to 50 fmol of labelled RNA in RNase H buffer [100 mM KCl, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 2 U TEV protease (Gibco-BRL), 2 U RNAguard] for 15 min at 30°C. Aliquots (10 µl) of this mix were then added to 2 µl of a pre-mix (0.5 U RNase H and 20 pmol oligonucleotide) and incubation was continued for 60 min at 30°C. After proteinase K treatment (see above), RNA was ethanol precipitated and resolved on 8.3 M urea/12% polyacrylamide gels.
Acknowledgments
Acknowledgements
We thank I.Bozzoni for suggesting the GAL1-U24* in vivo experiment and for plasmid P414. We are grateful to L.Minvielle-Sebastia for CF IA and GST–Nab4p, and to M.Ohnacker for CPF used in RNase H protection experiments. We thank S.Dettwiler and A.Gerber for critically reading the manuscript. This work was supported by the University of Basel, the Swiss National Science Foundation, the European Community (via the Bundesamt für Bildung und Wissenschaft, Bern) and the Louis-Jeantet-Foundation for Medicine. B.D. was the recipient of an EMBO long-term fellowship.
References
- Barabino S.L.M., Ohnacker,M. and Keller,W. (2000) Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.S. and Platt,T. (1988) RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science, 242, 1270–1274. [DOI] [PubMed] [Google Scholar]
- Carrington J.C. and Dougherty,W.G. (1988) A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc. Natl Acad. Sci. USA, 85, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. and Moore,C. (1992) Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell. Biol., 12, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. and Hyman,L.E. (1998) A specific RNA–protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res., 26, 4965–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Oliviero,S. (1994) Preparation of yeast RNA. In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology (New York). Current Protocols, 13.12.1–13.12.2.
- Fatica A., Morlando,M. and Bozzoni,I. (2000) Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J., 19, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1996a) 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–481. [DOI] [PubMed] [Google Scholar]
- Guo Z. and Sherman,F. (1996b) Signals sufficient for 3′-end formation of yeast mRNA. Mol. Cell. Biol., 16, 2772–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann S., Obermeier,B., Vogel,K. and Domdey,H. (1992) Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 4215–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L.E., Seiler,S.H., Whoriskey,J. and Moore,C.L. (1991) Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3′-end formation. Mol. Cell. Biol., 11, 2004–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M., Zhao,J. and Moore,C.L. (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J. Biol. Chem., 271, 27167–27175. [DOI] [PubMed] [Google Scholar]
- Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C.C., Wilusz,J. and Shenk,T. (1994) The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor,B., Bonneaud,N. and Lacroute,F. (1991) Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol., 11, 3075–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′-end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K.G.K. and Manley,J.L. (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Ohnacker M., Barabino,S.M., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Simos,G., Bischoff,F.R., Podtelejnikov,A., Mann,M. and Hurt,E. (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J., 17, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- Takagaki Y. and Manley,J.L. (1994) A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature, 372, 471–474. [DOI] [PubMed] [Google Scholar]
- Takagaki Y. and Manley,J.L. (1997) RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol., 17, 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini S.R., Weiss,V.H. and Silver,P.A. (1999) Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA, 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S. and Sherman,F. (1982) DNA sequence required for efficient transcription termination in yeast. Cell, 28, 563–573. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kessler,M.M. and Moore,C.L. (1997) Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem., 272, 10831–10838. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]