Abstract
Because of similarities in histopathology and tumor progression stages between mouse and human lung adenocarcinomas, the mouse lung tumor model with lung adenomas as the endpoint has been used extensively to evaluate the efficacy of putative lung cancer chemopreventive agents. In this study, a competitive cDNA library screening (CCLS) was employed to determine changes in the expression of mRNA in chemically induced lung adenomas compared with paired normal lung tissues. A total of 2555 clones having altered expression in tumors were observed following competitive hybridization between normal lung and lung adenomas after primary screening of over 160,000 clones from a mouse lung cDNA library. Among the 755 clones confirmed by dot blot hybridization, 240 clones were underexpressed, whereas 515 clones were overexpressed in tumors. Sixty-five clones with the most frequently altered expression in six individual tumors were confirmed by semiquantitative RT-PCR. When examining the 58 known genes, 39 clones had increased expression and 19 had decreased expression, whereas the 7 novel genes showed overexpression. A high percentage (>60%) of overexpressed or underexpressed genes was observed in at least two or three of the lesions. Reproducibly overexpressed genes included ERK-1, JAK-1, surfactant proteins A, B, and C, NFAT1, α-1 protease inhibitor, helix-loop-helix ubiquitous kinase (CHUK), α-adaptin, α-1 PI2, thioether S-methyltransferase, and CYP2C40. Reproducibly underexpressed genes included paroxanase, ALDH II, CC10, von Ebner salivary gland protein, and α- and β-globin. In addition, CCLS identified several novel genes or genes not previously associated with lung carcinogenesis, including a hypothetical protein (FLJ11240) and a guanine nucleotide exchange factor homologue. This study shows the efficacy of this methodology for identifying genes with altered expression. These genes may prove to be helpful in our understanding of the genetic basis of lung carcinogenesis and in developing biomarkers for lung cancer chemoprevention studies in mice.
Keywords: lung adenomas, CCLS, expression profile, differential changes, cancer genes
Introduction
Lung cancer is the leading cause of cancer deaths in men and women in the US [1]. Epidemiological and laboratory animal model studies have demonstrated that smoking and environmental exposure to carcinogens are closely linked to increased lung cancer risk [1–5]. Although about half of all people who had ever smoked are now former smokers, many people are unable or unwilling to stop smoking. For these reasons, chemoprevention is a potentially important approach to reduce the large number of tobacco-caused cancer deaths, especially for former smokers. The A/J mouse lung tumor model, primarily adenomas, is the most widely used preclinical model for lung cancer chemoprevention studies [3,6]. In addition to similarity between adenomas/adenocarcinomas commonly seen in mice and human lung adenocarcinomas, genetic changes found in mouse lung tumors also resemble those existing in humans [3,6]. Among the more than 50 different agents tested, several groups of chemicals have shown significant efficacy against mouse lung tumor development including glucocorticoids, green tea, nonsteroidal anti-inflammatory drugs (NSAIDs), isothiocyanates, and farnesyl transferase inhibitors [3].
Genetic changes found in mouse lung tumors include mutational activation of the K-ras gene, which is observed in 80% of both spontaneously occurring and chemically induced adenomas and adenocarcinomas of the mouse lung [3,7]. Mutation of K-ras is an early event in mouse lung tumorigenesis and persists into malignancy [3,7]. Aberrant expression of other oncogenes or tumor suppressor genes, e.g., c-myc, Rb, and p16 genes, has also been demonstrated in mouse lung tumorigenesis [8]. Allelic deletions on different chromosomes suggest the involvement of additional known and unknown genes during mouse lung tumorigenesis. Allelic loss of the p16 tumor suppressor gene occurs in approximately 50% of mouse lung adenocarcinomas [9]. Allelic loss of chromosomes 1, 4, 11, 12, and 14 are frequently associated with mouse lung tumor development [9–11]. Recently, mouse lung tumor susceptibility loci have been mapped to chromosomes 6, 9, 17, and 19. Those linked to lung tumor resistance have been mapped to chromosomes 4, 11, 12, and 18 [3].
Detection of mutations or LOH in specific oncogenes and tumor suppressor genes has been the focus in examining for genetic alterations in tumors. More global methods have recently been developed. These include CGH analysis, which allows one to examine for gene deletion or amplification, and proteomics, which allows determination of protein levels. The use of cDNA microarrays to detect altered gene expression during the neoplastic process has perhaps generated the greatest amount of effort to date. In particular, high-density oligonucleotide arrays and high-density cDNA glass slide arrays have been widely used in profiling gene expression in human and rodent tumor tissues. In the present study, we have used competitive cDNA library screening (CCLS) [12]. CCLS allows one to screen in a nonselective manner for known or unknown genes whose expression is altered between two sets of samples [12] by performing a competitive hybridization between normal lung and lung adenomas and by screening this against cDNA clones generated from normal lung. Employing this technology, we identified 65 distinct genes (58 known and 7 novel) whose expression is routinely altered in mouse lung adenomas.
Materials and Methods
Lung Adenomas
At 6 weeks of age, female A/J mice received a single intraperitoneal (i.p.) injection of N-methyl-N-nitrosourea (MNU) in acidified saline (pH 5.0) at a dose of 50 mg/kg body weight. The mice were terminated at 6 months of age and the adenomas and paired normal tissues were harvested and frozen in liquid nitrogen for RNA analysis. All lung tumors used from this lung tumor bioassay were diagnosed as lung adenomas. Lung adenomas were carefully microdissected before they were used for RNA isolation. Briefly, frozen tumor tissues were microdissected to determine the borders of tumor versus normal tissues. Tissues were embedded in Tissue Tek OCT compound (VWR Scientific Products, West Chester, PA), cryostat-sectioned, and stained with hematoxylin and eosin for microscopy. Tumor tissue sections corresponding to the microscopic sections containing only tumor cells were isolated and stored at -80°C for subsequent RNA isolation. Matching normal tissues from the same animal were also microdissected to ensure that specimens consisted of purely normal lung tissue.
Isolation of RNA
Total RNA from tumors and paired surrounding normal tissue was isolated from pulverized tissues (normal/tumor) according to an acid-guanidine-thiocyanate-phenol-chloroform method described previously [12]. The quantity and purity of the RNA were determined by spectrophotometry at wavelengths of 260/280 nm, and the RNA quality was checked by electrophoresis on a formaldehyde agarose gel.
Labeling cDNA Probes by Reverse Transcription (RT)
Two micrograms of total RNA and 2 µl of oligo (dT) 15 primer (500 µg/ml) (Promega, Madison, WI) mix were incubated at 65°C for 5 minutes, and then chilled on ice. Other reagents were added in a total of 25 µl of reaction volume containing 1x RT buffer (50 mM Tris-HCl, pH 8.3; 75 mM KCl; 3 mM MgCl2; 10 mM DTT; 0.2 mg/ml BSA); 1.0 mM dTTP, dATP, and dGTP; and either 100 µCi of [α-32P]dCTP or 1.0 mM cold dCTP; 100 U RNasin (Pro-mega); and 200 U of M-MLV reverse transcriptase (GIBCO BRL/Life Technology, Gaithersburg, MD). The reaction mixtures were incubated at 37°C for 1 hour. The probes were purified by Sephadex G-50 Columns (Boehringer Mannheim, Indianapolis, IN) and the specific labeling activity was determined by liquid scintillation counting. Specifically, for hybridization probe set 1, 2 µg of total RNA from a mouse lung adenoma was reverse-transcribed with [α-32P]dCTP for direct incorporation. The labeled tumor cDNA probe was then mixed with equal amount of unlabeled cDNA and 5 µg of mouse CotI DNA, then denatured at 95°C for 10 minutes. For hybridization probe set 2, 2 µg of total RNA from the paired normal lung was reverse-transcribed with [α-32P] dCTP for direct incorporation. The labeled normal cDNA probe was then mixed with equal amount of unlabeled cDNA and 5 µg of mouse CotI DNA and denatured.
cDNA Libraries as Targets for Competitive Hybridization
Uni-ZAP XR mouse lung cDNA library, purchased from Stratagene (La Jolla, CA), was used as the target for CCLS. Approximately 1.6x105 plaques were plated in 200 Petri dishes (100 mm) and then transferred to nitrocellulose filters following the protocol provided by the supplier. Two identical filters were made from each plate and the plaque DNA on these filters was denatured in 1.5 M NaCl/0.5 N NaOH, neutralized in 2.5 M NaCl/1 M Tris (pH 7.4), rinsed in 3x SSC, and baked at 80°C for 2 hours. After 12 hours of prehybridization with hybridization solution, one replica was hybridized to labeled tumor probe with an unlabeled competitor (set 1), whereas the other was hybridized to labeled normal probe with the same nonlabeled competitor (set 2). Hybridization solution consisted of 50% formamide, 5x SSPE, 5x Denhardt's solution, 0.1% SDS, and 100 µg/ml denatured herring sperm DNA. The hybridization was performed at 42°C for 18 hours with mild rolling or shaking. Membranes were stringently washed with agitation in 2x SSC/0.1% SDS twice (10 minutes each) at room temperature and 30 minutes in 0.1x SSC/0.1% SDS twice at 65°C. Dried filters were exposed to X-ray film for 3 to 15 days at room temperature.
Reverse Dot Blot Analysis
Positive clones found in the CCLS primary screening were verified by reverse dot blot analysis that served as a secondary screen to eliminate false positives. DNA from positive clones was isolated using the standard mini prep method described previously [12]. DNA was denatured by adding 0.1 vol of 2 N NaOH, 2 mM EDTA at 37°C for 30 minutes and neutralized with 0.1 vol of 3 M sodium acetate (pH 4.8). Two micrograms of denatured DNA was then spotted onto nylon membranes using the HYBRI.DOT Manifold Apparatus (GIBCO BRL/Life Technology). Duplicate membranes were prepared for each clone. These membranes were hybridized with probes prepared as described above for the CCLS primary screening. The washing conditions were also the same as above.
DNA Sequence Analysis
Differentially expressed clones as identified by dot blot analyses were selected for sequencing using vector-specific primers. Cycling sequencing with Taq polymerase was performed with fluorescent-labeled dideoxynucleotides (BigDye Terminator DNA Sequence Kit; PE Applied Biosystems, Forster City, CA) with phagmid DNA as template according to the manufacturer's instructions. After the reaction, the samples were resolved on an ABI fluorescent DNA sequencer. GenBank database matching was performed with BLAST sequence comparison programs at NCBI (http://www.ncbi.nlm.nih.gov/blast).
Gene-Specific Quantitative RT-PCR Analysis
Two micrograms of total RNA was used to synthesize cDNA in a total volume of 30 µl. After incubation of the RNA in 19 µl of DEPC-treated water at 65°C for 10 minutes, the following components were added: 1 µl of 45 nM oligo-dT, 5x reaction buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl), 0.6 µl of 50 U/µl RNase inhibitor, and 2 µl of 200 U/µl MMLV reverse transcriptase. The reaction mixture was incubated at 37°C for 1 hour. The reaction was then terminated at 95°C for 10 minutes. We performed comparative multiplex PCR to semiquantitatively evaluate the gene expression differences. In comparative multiplex PCR, primer pairs from the control cDNA, GAPDH, and the target gene cDNA would be included in each reaction at equivalent concentrations. The coamplification of the control cDNA and the target gene cDNA in tumor and normal tissues would provide a means to control for PCR amplification and enable the relative level of the target gene expression to be quantified. A pair of primers specific for mouse GAPDH cDNA was used as an internal control. Prior to PCR, one primer from each pair (GAPDH and target gene) was 5′ end-labeled with T4 polynucleotide kinase (United States Biochemical, Cleveland, OH). Forty picomoles of each target primer pair and GAPDH primer pair was then combined with 1 µl of aliquot of cDNA, 100 µM of each deoxyribonucleotide (dATP, dCTP dGTP, and dTTP), 1.0 U of Taq DNA polymerase (Promega, Madison, WI), 50 mM KCl, 10 mM Tris-HCl (pH 9.0), and 0.1% Triton X-100. A reaction volume of 20 µl is subjected to 18 to 24 cycles of PCR amplification. Each cycle consisted of 1 minute at 94°C, 2 minutes at 57 to 60°C, and 1 min at 72°C. To determine the linear range of each PCR reactions, a series of three to four PCR reactions using 18, 20, 22, or 24 cycles were performed for each target gene, and one of the reactions that fits into the linear range was used for further quantitation. Approximately 2.5 µl of reaction mixture from each PCR was loaded on the 8% polyacrylamide gel and run at 60 W for about 2 hours. Gels were dried and exposed to a Phosphor Image screen for 48 hours. The signals collected by the screen were analyzed by computer software Image-Quant Version1.1. The relative intensities of the target products were then normalized to the level of GAPDH control. The normalized intensities of normal lung tissues and those of tumors were compared to assess for gene expression differences. For hard copy of the image, dried gels were also exposed to X-ray films overnight.
Statistical Analysis
After normalization to the level of GAPDH cDNA amplification in the multiplex PCR reactions using cDNA from both normal lungs and lung adenomas, Student's t test was used to determine the difference in the signal intensity of phosphor imaging between normal lungs and lung adenomas.
Results
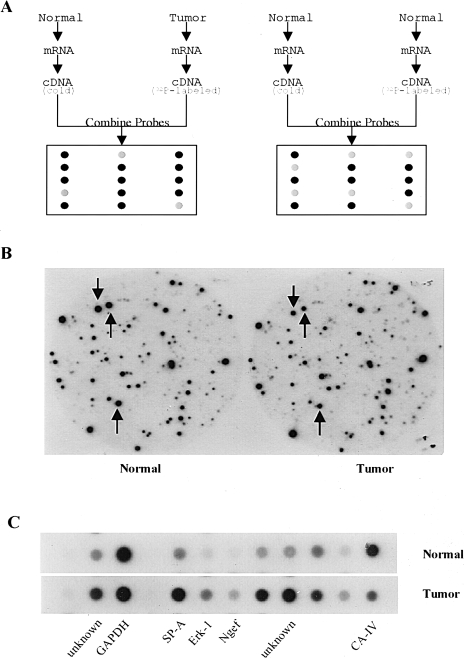
Six pairs of lung adenomas and normal lung tissues were used in the primary screening to detect differentially expressed genes. Lung adenomas were obtained from A/J mice using a standard 6-month protocol employing MNU as the carcinogen. As shown in Figure 1, lung adenomas used in the present study are characterized by a monomorphic growth pattern and are generally comprised of well-differentiated pulmonary cells. A total of 200 pairs of nitrocellulose filters, containing nearly 1.6x105 clones from a mouse lung cDNA library, were screened with two sets of labeled cDNA probes (Figure 2, A and B). One set was a mixture from a [α-32P]dCTP labeled cDNA derived from an MNU-induced lung adenoma and unlabeled DNA derived from a normal paired lung. The other set was prepared from mixture of a [α-32P]dCTP labeled cDNA from a normal paired lung and a nonlabeled cDNA prepared from an equal amount of total RNA from normal lung tissue. Approximately 2500 clones were found to have substantially altered expression in mouse lung tumors. These clones underwent secondary screening by reverse dot blot analysis to eliminate the false-positive clones (Figure 2C). Sequence analysis was performed on 755 clones confirmed by dot blot analysis, and the sequences were entered into BLAST of National Center for Biotechnology Information (NCBI) to search for matches in the GenBank database. The results of the BLAST search showed that many of the 755 clones matched to the mouse Clara cell 10-kDa (CC10) protein (197) or the surfactant protein C (371), reflecting their high expression resulting in multiple independent clones in the non-normalized mouse lung cDNA library employed for CCLS analysis.
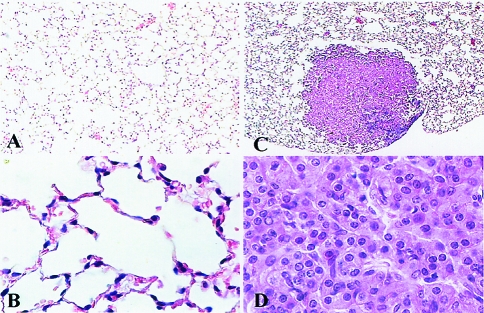
Figure 1.
Histology of a normal lung and a lung adenoma used in this study. Light photomicrographs of a normal lung (A and B) and a lung adenoma (C and D) at x4 and x40 magnifications, respectively.
Figure 2.
Analysis of differentially expressed genes in mouse lung adenomas using CCLS. (A) Schematic illustration of CCLS. Equal amounts of total RNA from lung tumors and normal tissues were converted to cDNA probes with incorporation of 32P into the cDNA strands during RT. Two competitors were also generated from normal lung tissues using the same procedure except for 32P incorporation. Probes 1 and 2 were used to perform differential hybridization against a mouse lung cDNA library. (B) An example of data from CCLS. Two identical filters were differentially hybridized with the cDNA probes. The left one represents hybridization with the probe generated from normal tissue, whereas the right one represents hybridization with the probe derived from a lung tumor. The three spots indicated by arrows show three differentially expressed clones that were identified by CCLS. (C) Results of dot blot analysis. Dot blot analysis was conducted as one confirmation step. Differentially expressed clones selected from CCLS were confirmed by dot blot analysis. The GAPDH was used as an internal control.
In order to confirm differential mRNA expression, 83 distinctive clones were further examined by quantitative RT-PCR analysis in duplicate to avoid false positives (Figure 3). Detected genes were divided into two categories: overexpressed and underexpressed in tumors compared with their paired normal tissues (Table 1). We confirmed 65 of 83 selected clones by quantitative RT-PCR (Table 1). Secondary confirmation of differential gene expression in lung adenomas by Northern blot analysis could not be performed due to the limited amount of RNA from the individual adenomas employed in the present study.
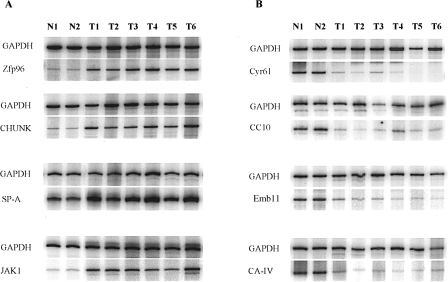
Figure 3.
RT-PCR verification of differentially expressed genes detected using CCLS. “N” represents the normal mouse lung tissue; “T” represents the MNU-induced mouse lung adenomas. GAPDH was applied as an internal control to determine the amount of template in each reaction. (A) RT-PCR confirmation of upregulated genes. Zfp96, zinc finger protein 96; CHUK, conserved helix-loop-helix ubiquitous kinase; SP-A, surfactant protein A; and JAK1 protein. (B) RT-PCR confirmation of downregulated genes. Cry61, growth factor-inducible immediate early gene; CC10, Clara cell protein 10; Emb11, 11-day embryo cDNA; and CA IV, carbonic anhydrase IV.
Table 1.
RT-PCR Confirmed Genes with Differential Expression in Mouse Lung Adenomas Using CCLS.
| Gene† | Upregulation | Gene† | Downregulation | ||||||
| Incidence‡ | Changes (fold)§ | Normal (mean±SD) | Tumor (mean±SD)¶ | Incidence‡ | Changes (fold)§ | Normal (mean±SD) | Tumor (mean±SD)¶ | ||
| mRNA for translational controlled 40-kDa polypeptide p40 | 2/6 | 2–3 | 0.732±0.002 | 1.584±0.013** | 10-Day-old male pancreas cDNA | 3/6 | 2–4 | 3.322±0.506 | 1.260±0.443* |
| Homo putative transcription factor CA150 | 4/6 | 3–5 | 0.046±0.002 | 0.194±0.055* | 11-Day embryo cDNA | 5/6 | 2–4 | 4.768±0.410 | 1.693±0.546** |
| 45S pre-rRNA | 5/6 | 2–7 | 23.31±3.225 | 79.67±34.85* | 13-Day embryo liver cDNA | 3/6 | 2–5 | 10.66±1.721 | 3.399±1.227* |
| mRNA for cysteinyl-tRNA synthetase | 6/6 | 2–6 | 0.017±0.001 | 0.071±0.027** | α-Globin mRNA | 4/6 | 2–3 | 5.731±0.011 | 2.643±0.355*** |
| mRNA for pancortin-1 and -3 | 6/6 | 2–3 | 0.014±0.003 | 0.036±0.010** | β-Globin major gene | 6/6 | 2–4 | 6.623±0.186 | 2.317±0.602*** |
| Serine hydrolase-like (Serhl) mRNA | 4/6 | 2–3 | 0.025±0.004 | 0.065±0.018* | CA IV gene | 3/6 | 3–4 | 0.127±0.007 | 0.043±0.006*** |
| Human hypothetical protein FLJ11240 | 3/6 | 2 | 0.026±0.007 | 0.057±0.020* | ALDH II mRNA | 5/6 | 2–5 | 0.575±0.076 | 0.183±0.063* |
| Gene for fibrinogen A-α-chain | 3/6 | 2–8 | 0.040±0.002 | 0.179±0.002** | Growth factor-inducible immediate-early gene, cyr61 | 5/6 | 3–10 | 0.379±0.093 | 0.055±0.018* |
| mRNA for erk-1 | 4/6 | 2–3 | 0.286±0.038 | 0.566±0.054*** | Paroxanase (PON-1) mRNA | 6/6 | 4–10 | 0.379±0.093 | 0.046±0.027* |
| JAK-1 protein | 6/6 | 3–10 | 0.029±0.010 | 0.153±0.019** | Homo sapiens glucose-regulated protein | 2/6 | 2 | 0.254±0.012 | 0.130±0.007* |
| Neuronal guanine nucleotide exchange factor | 6/6 | 3–9 | 0.016±0.002 | 0.075±0.037** | Hybridoma 12A1 immunoglobulin heavy-chain mRNA | 6/6 | 4–15 | 0.363±0.131 | 0.047±0.026* |
| Zinc finger protein 96 (Zfp96) mRNA | 6/6 | 3–7 | 0.020±0.0001 | 0.098±0.038** | Rat Ras GTPase-activating protein | 3/6 | 2–3 | 0.252±0.030 | 0.121±0.013** |
| BALB/c conserved CHUK mRNA | 5/6 | 2–3 | 0.209±0.004 | 0.431±0.104** | H. sapiens TNFa-stimulated ABC protein | 3/6 | 2 | 2.268±0.073 | 1.090±0.063** |
| mRNA for α-adaptin (C) | 5/6 | 2–6 | 0.077±0.010 | 0.264±0.132* | Mitochondrial DNA | 3/6 | 2 | 8.239±0.592 | 3.228±0.241* |
| T-cell transcription factor NFAT1 isoform A mRNA | 3/6 | 2–3 | 0.034±0.006 | 0.101±0.024* | Rat mRNA for ribosomal protein L18a | 3/6 | 2 | 0.932±0.056 | 0.474±0.041* |
| MCH class I heavy-chain precursor (H-2D(k)) mRNA | 3/6 | 2–3 | 2.691±0.561 | 7.093±1.909* | Lsp-s mRNA for lysozyme P | 2/6 | 2 | 6.114±0.122 | 3.160±0.116** |
| MCH class I heavy-chain precursor (H-2K(k)) mRNA | 3/6 | 2–6 | 0.275±0.056 | 1.122±0.528* | CC10 protein | 6/6 | 2–4 | 8.576±1.122 | 2.903±0.413* |
| Complement component C3 gene, 5′ end | 4/6 | 3–10 | 0.141±0.006 | 0.887±0.394* | Mitochondrial genes for transfer RNA | 4/6 | 2–3 | 46.64±6.237 | 17.38±4.441* |
| Mouse surfactant protein-A (SP-A) | 3/6 | 2 | 0.930±0.079 | 1.890±0.118** | von Ebner minor salivary gland protein | 6/6 | 4–14 | 0.182±0.030 | 0.027±0.030* |
| Rat mRNA for surfactant protein-B | 6/6 | 2–4 | 1.464±0.401 | 3.488±0.661*** | |||||
| Pulmonary surfactant protein SP-C | 6/6 | 2–3 | 0.314±0.014 | 0.851±0.106*** | |||||
| mRNA for sulfated glycoprotein-2 | 4/6 | 2–3 | 0.259±0.058 | 0.546±0.168* | |||||
| Serine proteinase inhibitor 6 (SPI6) | 6/6 | 2–9 | 0.026±0.005 | 0.110±0.071* | |||||
| α-1 Protease inhibitor 2 mRNA | 4/6 | 6–13 | 0.008±0.004 | 0.099±0.053* | |||||
| Human mRNA for KIAA0183 | 5/6 | 2–5 | 0.050±0.004 | 0.163±0.066* | |||||
| Human mRNA for KIAA0187 | 6/6 | 2–5 | 0.126±0.006 | 0.404±0.100*** | |||||
| Homolog of D. melanogaster flightless I | 6/6 | 2–19 | 0.054±0.011 | 0.388±0.286* | |||||
| CYP2C40 | 4/6 | 3–9 | 0.003±0.001 | 0.016±0.008* | |||||
| RIKEN cDNA 2500002L14 gene | 5/6 | 2–4 | 0.090±0.011 | 0.258±0.068** | |||||
| RIKEN cDNA 5730403B10 gene | 3/6 | 2 | 0.183±0.003 | 0.386±0.066* | |||||
| Brain cDNA, clone MNCb-5704 | 6/6 | 2–10 | 0.283±0.133 | 1.553±1.040* | |||||
| Adult male testis cDNA | 4/6 | 2 | 0.050±0.007 | 0.112±0.016** | |||||
| 0-Day neonate skin cDNA | 6/6 | 5–16 | 0.013±0.001 | 0.130±0.054** | |||||
| 10-Day embryo cDNA | 6/6 | 2–4 | 0.641±0.094 | 1.641±0.527** | |||||
| Mus musculus proline 4-hydrosylase α-1 polypeptide (P4ha1) | 6/6 | 2–4 | 0.100±0.017 | 0.307±0.094* | |||||
| M. musculus similar to KIAA1711 | 5/6 | 2–3 | 0.200±0.029 | 0.562±0.105*** | |||||
| Mouse DNA sequence from clone RP23-39409 on chromosome 11 | 5/6 | 2–4 | 1.276±0.384 | 3.562±0.936** | |||||
| M. musculus hypothetical protein MGC25836 | 5/6 | 2–4 | 0.021±0.004 | 0.071±0.019** | |||||
| H. sapiens chromosome 18, clone RP11-749G1 | 3/6 | 2 | 0.615±0.210 | 1.835±0.522* | |||||
| LRG1# | 6/6 | 2–4 | 0.022±0.005 | 0.062±0.014*** | |||||
| LRG2# | 5/6 | 3–7 | 0.072±0.007 | 0.370±0.104** | |||||
| LRG3# | 6/6 | 2–4 | 0.013±0.003 | 0.037±0.013* | |||||
| LRG4# | 6/6 | 2–7 | 0.018±0.012 | 0.082±0.032** | |||||
| LRG5# | 6/6 | 3–16 | 0.028±0.0002 | 0.391±0.154*** | |||||
| LRG6# | 6/6 | 2–8 | 0.082±0.048 | 0.492±0.165** | |||||
| LRG7# | 6/6 | 5–13 | 0.006±0.002 | 0.065±0.023*** | |||||
P< .05.
P<.01.
P<.001.
Putative identity based on high degrees of homology with known genes (see Results).
Incidence is the number of tumors had changes out of number of tumors tested.
Range of fold increase or reduction is calculated in individual tumors to control tissues.
Statistically different from normal controls.
The genes have no matches in NCBI GenBank. We named the genes as Lung tumor-Related Genes (LRG).
Nineteen genes were underexpressed, whereas 46 genes were overexpressed in lung tumors. Fifty-eight genes showed high homology (greater than 90%) to known genes, and seven genes, which did not match any known genes in the NCBI sequence database, were considered to be novel (see Table 2). Three genes, which showed lower homology to known genes, were human TNFα-stimulated ABC protein, human KIAA0187, and a human homolog of Drosophila melanogaster flightless I, which had 81%, 85%, and 87% homology, respectively.
Table 2.
Partial Sequences of the Unknown Genes Confirmed with Differential Expression in Mouse Lung Adenomas.
| Gene | bp | Sequence |
| LRG1 | 566 | GTTTTGCCTAAGTGCTGACTTCTTTATTGACTCCTTTAGAGAAAGTTATTATGACCAATACATACTCCATGTTAATCCTGTCCCTCAAGTGTTTCTTTAGGCATTGATATTATGGTACATTCTCTACATAATACTTCTAATCTGACTTTCCTTCAGGATGATGGAGTTATGAATTTTCCCCTTTATTAAAACGAGGCAAGGTGGTGCATGCCTGTAATCATAGCACCTAGTAGGCAAGGGTAGAAAGATGATGAATTTAAGCCAGTTTGGCCTTATATAATAAAGCCTCATGTGGAAAAACCGTGCAGGCAAGGCTGAGGATATAGTTCAGTGGTAACGCACTTGCTTAAGTGTGCCCCAGGTCTAATACGAGAAAACAAAGCATTTTGTTACTACTGGTTGTTGGGCAGGCGAGGTGACTTGGTGGATGTGAGTCTTGCTTTCAAACCTGATCACTCAGTTCAATATCTTGGATCCATATAGCAGAAAGTGCAAGTTGTTCTATGATTCGCATTCTGGGCGTGTGTGCTCTCTCATGCACACACAGGTGTAAATAAATGGA |
| LRG2 | 319 | GACATATGACAAGACAGAAAGCCAGAGACAAGAGATTCAGGCTTGTTTTCATTATGTTGCCTTAGTCTCCCTAGCACTAATAAAGGCCCTGAGAGAACTACTTTAATCTCCTCCAGGGGTGGCACTCCAAT GACCTAATCACCTTCATTAAGCCACGCCTCTTAAAAGCCCTGCACAATCCTGGAACCCAGCCTTCTAGGTCATGACAATTTGATGGATCCCCTCGTACACCAGACAGNGGTGTACGACCACCATCTGGCCCCTTCCATGGAGAACATACAGACTTCAGAAAACACTTGGGCAGCGGACAGGACCCAC |
| LRG3 | 378 | TTTATGGTGCCACCTTTTCCAGAGAGGAGTCCGGCTAGGGTGTCTGGAGCCTAAACAACCAGGCTGCTGGGAGAGGCAGCAAGGGGAGAGAAGGCGAAGGGCAGGAAGTTGTCAGCCTGAGTCACAGGGTGC AGGAGGGGTCTTGGAGGGCTGGCTGCCCTCCCTTCTCAGGGCAGCTGTGCCCATTTCACCGTCTGTGTCCCACCCCTACCCAAGTCCTGTGTTTGACAGCAGTTTCCACTTGTCATGGAAAAGCCTGTCTCGGCTTTGGGTGTGTGCAGATTCGGTGTCCCTGAGACTGAGCTGCAGCCCATCAAGGGCAGAGCCACTGGACTTCCAACCTCTGGATGGACCTGGGGACTAAACTATGTGCCATGA |
| LRG4 | 579 | ACCATCCAAGTGTAAGCCATTTTTGATAACTACACATTTGTTTACATCCTAAGAACTTTACATCAATTATCTTACTTCATGCTAACATCACTGGCTGGGATATATGCTTTTATTAATTCCATTCCAACATGAGAAAACTTAGCTACGGAGTCCATTTTGTATAAGGTCACACGGACAGGTATAAGCTGTATCAATGTTTAATGTCTATCTTGAGGCAGCATCCCCACTTGGCTCAGATGCCCACCTCAGGAGCTCGTGAGCAGATGTGGGCCTGTGCTGGGCCCTCACAGAGTCTCTTGGAGCAGTAGGCACTGGCCATAGTTTGCCCTCTTTTTAGCATTTACAACTGCTATTATGGCGTCTAGTGGCAGGGTATCAGCTCTAAGATATATTTCTTATTAAATCCTTAAATGATAGAGCTTTTATGATCTTGTTTATAAGTTGAGGATTAAATAGAACTCCCCTCCCCTTTTTTTTCAGCCCANGGTTGTTTGATAGAGGTGTGTGTAGGTGTTTTAATGTAAATTACCCATTTTCTTTTTTATACAAATCTGGACCTCCTTTTTCCCTCCCTT |
| LRG5 | 454 | TTTGCCCAATGTTCCAAGCCGAAAAGTGTGTCATCATGAAGAGATGGCACTTATTTTCAGAGCTGAGGTGCCCGGGAGAACTGAACATTCTCTGACAACCAGGATGCCTCGGGGCAATGGATTCTGATGGATAGCTGGCCACAGGCTGCGAAAACCATGCAGGAAAATCTCAGGGGGGCTTGCTATGTGGTCTATGGTTATCTTGGCAGGAAACCAGGACCTGGGGCTGAACACAGTAGAAATGGGGTGTTTTCATAATTATCCCTGTTGGGTGCCATTGAGCACATACAAGTCTTTAGATGCCACACGCACTGAACATACACCTCTCTGGTCTCCAGTGCACCGTATCAGGCTTGGGACTCACTGTAGGCAGACATTAGCGATCGCGTCCTTCAATGCTTGGCCTCGTGTCTATGCAGTACTTAGCAGTGGTTAGACACTGCCAAGAGTCT |
| LRG6 | 654 | GGACTAGTCCGAGTTTTTTTTTCTTTTTTTTTTTTGAGAGTTGTGGTTCTCCTGGCGATAATTTCTCTCACACTTATACTTCCTTTGAAGATGTCTGCATTTTCACCTCATTTCTCCGAGGACACTGTTTACAGGACACAGAACTACAGGGTGGCATTCCTTCCCTCCCCTTTCAGCACCTTTGGGATTTCACCCCATCCTTCATTGGCTACCACTGCGACCATCCGACCCTGAGGTCGTCAGCTCGTCTCACTGTTCCCACTGACCAGCCCTTCTTGTCTTCCTTTGTGCTGTTTTCCAGACTCCATCAGACTCTCACCCCCTTCCCAGTCTTCACTGGTTTGGATTGATGTGTCCAGGAGTAGGTCTTTGTATAAGGAACCGCCGTTCTGCTTGGACTCCGCTGTTTCTTGGATCTGGGGGTTGGTATTTTCATTAGCTGTAAAAGCTTCTGGTCTTTATGCATTCATGTCTACATGTAGCACCCCACCCCAAACAAGTCCTCTGCATCTCTTTATGAATGCTAAAACACATTTTTTAAGATTTTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGTGGGATGCACCCTGAAAGTCTAAAAGGAGTCTGATCCTCTGGGGCTGGCTTAAAGTTTACCCCCCC |
| LRG7 | 617 | TCTTTTCTTTTCTTTTCTTTTCTTTTCTTTTCTTTTCTTCTTTTTTCTCTTCTCTTCTCTTCTCTTCTCTTTTCTTTTCTTTTCTTTCTCCCCATCTAGGATACCATCCTCTCCGTGTTCTCCATGCGCTCAGTGGGCACTTGGCATTTCCTTCAGCCCCGTTCTGTCCTTCCTGATATATGGCCGAGACAATCCACAATTATTTGCTCCCATGACCTCTGTTCAGAACTAAATTATAATAAACCCCTCCATTTTATATATTGAGCCCAAATTTCCAGTTTCTCAGAATATAAATGTAAATGCAAGTAGGACCAAAAGAGAGAGATTTAAATCGAGTTTATTAAGGGGGAACTAATTCAGCCTGGCCGGTGATCTTATGAAGAGACTGGTAATGCAGAGAGGAGATGAGCTCCAGGGAAACGACAGAGGAAAGACGAGCCTGAGGTGAGGTGGTCAAGGAAACAGCCTTGCCAGTGCCACGCCCATGCACAGCTCCCTCCTGGAACTATAAGAAAACGTGTTTTTGTATTCGGTTCTTGGAGAGGCCTTTGCAAACTAACAGTCTTCCATGGGAATGGGTTACCTTTACCCTTACCCTTTTCCTTTTCCTTTTC |
In examining the genes with altered expression, we found consistency in the alterations. Thus, of the 46 genes that were overexpressed, 35 were overexpressed in at least two or three of tumors and 30 were overexpressed in more than 80% of tumors. The degree of overexpression, even among consistently overexpressed genes, varied from gene to gene. For example, ERK-1 was increased in four of six tumors but by an average of only two-fold, whereas JAK-1 was increased in six of six tumors by an average of roughly eight-fold. Similarly, the TNFα-stimulated ABC protein was decreased in three of six tumors by roughly two-fold, whereas the paroxanase gene (PON-1) was decreased roughly six-fold in six of six tumors.
Genes that were overexpressed in lung adenomas are also shown in Table 1. Thirty-nine overexpressed genes had high homology to known genes, whereas seven showed limited homology and were designated as unknown genes. Six of the unknown genes (LRG1, LRG3, LRG4, LRG5, LRG6, and LRG7) were highly overexpressed in 100% of lung tumors (Tables 1 and 2). This consistent and high overexpression makes them potentially interesting candidates as tumor cell markers for diagnosis and early detection as well as potential targets for chemoprevention or chemotherapy studies using mouse lung tumor models. In addition, 19 genes were found to be underexpressed in lung adenomas, with six showing underexpression in at least 80% of adenomas. Other underexpressed genes include RasGAP, α-globin, β-globin, paroxanase (PON-1), carbonic anhydrase (CA) IV, Clara cell 10-kDa protein (CC10), ALDH II, growth factor-inducible immediate-early gene (cyr61), and human TNFα-stimulated ABC protein were absent or down-regulated in mouse lung tumors. Specific genes that were overexpressed in lung tumors include ERK-1, JAK-1, SPI6, fibrinogen A α-chain, surfactants A, B, and C, MCH class I heavy chain, NFAT1 isoform A, sulfated glycoprotein-2, zinc finger protein, helix-loop-helix ubiquitous kinase (CHUK), α-1PI-2, α-adaptin, thioether S-methyltransferase, human putative transcription factor CA150, C3, CYP2C40, and so forth (Table 1).
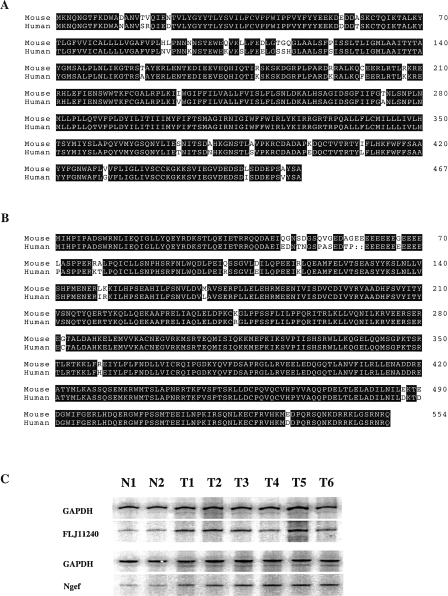
Two of the genes were further characterized through extensive sequencing and comparison of both mouse and human cDNA: a hypothetical protein (FLJ11240) and a guanine nucleotide exchange factor (GEF) homologue (Figure 4). FLJ11240 hypothetical protein was found to have limited homology with a peptidase, E1-E2 ATPase, His kinase, and Ppx/GppA phosphatase when searching the Protein-BLAST database (NCBI, NIH). The GEF gene was found to be highly homologous (>98%) to mouse neuronal GEF.
Figure 4.
Characterizations of FLJ11240 hypothetical protein, and neuronal guanine nucleotide exchange factor. (A) The sequence alignments of FLJ11240 hypothetical protein and neuronal guanine nucleotide exchange factor (Ngef) with human counterpart proteins. (B) RT-PCR verification of differential expressions of FLJ11240 hypothetical protein and neuronal guanine nucleotide exchange factor. “N” represents the normal mouse lung tissue; “T” represents the MNU-induced mouse lung adenomas. GAPDH was applied as an internal control to determine the amount of template in each reaction.
Discussion
In the present study, we used CCLS to identify differentially expressed genes in mouse lung adenomas. The CCLS method allows one to define differentially expressed genes, based on competitive hybridization between normal lung RNA and RNA derived from lung adenomas, in a nonselective manner and allows one to readily clone differentially expressed genes. There are many advantages in the use of CCLS to determine gene expression changes in cancer. For example, expression differences for both known and novel genes can be detected. Because the cDNA library is not normalized to ensure approximately equal representation of polyA+ RNA sequences, detection frequencies of differentially expressed genes can be determined, indicating the relative frequency of mRNA expression in the normal lung tissue. Additionally, in-depth sampling of gene expression changes for more than 100,000 clones is possible. Although CCLS is laborious and time-consuming, screening data are extensive and allow for the further characterization and functional analysis of unknown genes and examination of the potential roles of known genes in lung tumorigenesis. Some disadvantages of CCLS also exist. The use of CCLS methodology that employs a normal mouse lung cDNA library yields a number of implications. First, if a gene that is found in tumor tissue were not expressed at all in the normal lung, it would not be detected in this study. Secondly, genes that are expressed at very low levels in normal lung were probably missed using this method. Thirdly, because the cDNA library is not normalized, clones associated with significantly overexpressed genes may be repeatedly selected. Thus, there were hundreds of independent clones identified that proved by sequencing to be either surfactants or CC10.
Sixty-five genes were found to be differentially expressed in lung adenomas when compared to normal lung. Nineteen genes were underexpressed and 46 were overexpressed. Seven clones do not match any of the known genes in the NCBI sequence database, whereas 58 had high homology to known genes. For most of the genes, the changes were highly reproducible; thus, 37 of 49 genes displaying overexpression in tumors demonstrated such overexpression in at least two or three of the adenomas. Moreover, 24 of 37 genes were overexpressed at least three-fold and 12 of 37 at least five-fold. Similarly, 10 of 19 underexpressed genes were underexpressed in at least two or three of the adenomas and 6 of 19 were underexpressed at least three-fold in adenomas. Although some of the genes appear to be mechanistically more relevant to the cancer process or are more obvious candidate targets for therapy (see Discussion below), any of the defined genes may be candidate markers for early detection of lesions and as potential endpoint biomarkers. Known genes found to be differentially expressed in lung adenomas including 45S pre-rRNA, pancotin, α-globin, β-globin, fibrinogen A α-chain, paroxanase, cysteinyl-tRNA synthetase, homolog of D. melanogaster flightless I gene, von Ebner minor salivary gland protein, and TNFα-stimulated ABC protein. Although the role of these genes in mouse lung tumorigenesis is still unknown, they are candidate biomarkers for lung tumorigenesis and potential targets for chemoprevention studies. Many of the differentially expressed genes were detected reproducibly and were highly altered in tumors versus normal parenchyma including JAK-1, zinc finger 96, α-1 protease inhibitor, and homolog of D. melanogaster flightless I gene.
Three particularly intriguing overexpressed genes code for the kinases: ERK-1, JAK-1, and CHUK. All three genes are overexpressed in at least 67% of adenomas, and JAK-1 levels are overexpressed almost five-fold in adenomas. Members of the various kinase families are particularly appealing for chemotherapy studies using mouse lung tumor model because small molecule inhibitors have been developed against this family of enzymes. ERK-1 belongs to the MAK kinase family and is a component of signaling pathways that influences cellular proliferation and differentiation, while JAK-1 is a member of the intracellular tyrosine kinase family (Janus kinases), and activation of JAKs is the initial step in cytokine signaling. Studies have shown that ERK activation increased 15-fold, whereas ERK expression levels were only 1.3-fold higher in prostate cancer [19]. CHUK contains a serine-threonine kinase catalytic domain and may be targeted to a helix-loop-helix and/or a leucine zipper transcription factor [20]. CHUK links kinase cascades to NF-κB activation [21].
RasGAP was downregulated and both ERK-1 and JAK-1 were upregulated in mouse lung tumors. RasGAP, which is downregulated in adenomas, is a ubiquitous 120-kDa protein that hydrolyzes GTP bound to p21Ras [22,23]. Underexpression of RasGAP, which should increase the levels of Ras proteins in the activated state, and overexpression of ERK-1 appear to be crucial to Ras-RasGAP cycling-Raf-1-MAPK kinase signal transduction in mouse lung tumor development. Studies show that α-adaptin interacts with GHR and mediates endocytosis of GHR [24] upon hormone stimulation. The interaction of Shc with α-adaptin is also involved in receptor endocytosis [25]. Our data showed that α-adaptin was overexpressed in 80% of mouse lung adenomas (five of six).
NFAT1, CA150, CHUK, and zinc finger protein 96 were overexpressed in 50%, 67%, 83%, and 100% of lung tumors, respectively. These four genes play crucial roles in signal transduction and gene expression. NFAT1 orients the two subunits of AP-1, c-Jun, and c-Fos on DNA through direct protein-protein interaction to regulate transcription [26,27]. Evidence suggests that CA150, a nuclear protein associated with the human RNA polymerase II holoenzyme, plays a role in the regulation of cellular transcriptional processes [28]. The functions of CA150 in the mouse have not as yet been reported. CHUK contains a serine-threonine kinase catalytic domain and may be targeted to helix-loop-helix and/or leucine zipper transcription factors [20]. CHUK links kinase cascades to NF-κB activation [21]. The zinc finger motif is generally present in most transcription factors that regulate gene expression. Overexpression of NFAT1, CA150, CHUK, and the zinc finger protein 96 in mouse lung tumor cells is likely to facilitate DNA transcription upon growth stimulation during tumor development.
The result showing a decrease in cyr61 appears to contradict the explanation that it is a factor that will stimulate tumor growth. Cyr61 is a secreted, cysteine-rich, heparin-binding protein encoded by a growth factor-inducible early gene, which acts as an extracellular, matrix-associated signaling molecule promoting the adhesion of endothelial cells through interaction with integrin αVβ3 [29–31]. Studies suggest that cyr61 is an angiogenic inducer that promotes tumor growth and vascularization through integrin αVβ3-dependent pathways [32]. We found that cyr61 was underexpressed in five of six lung tumors.
Three genes that encode metabolizing enzymes were differentially expressed in mouse lung tumors. CA IV and ALDH II were downregulated in 50% and 83% of tumors, respectively. CYP2C40 was overexpressed in 67% of tumors. CA IV is a glycoprotein associated with cell membranes in lung and kidney [33,34]. Altered expression of various CA isozymes has been observed in a variety of tumor types. CA, an NADPH-dependent enzyme, has many functions: elimination of CO2 and metabolites, pH regulation, and participation in membrane transport events during active cell growth [35]. ALDH II is a member of the ALDH family and plays a role in ethanol detoxification [36]. Similar to other p450 enzymes, CYP2C40 plays an important role in bioactivation and detoxification of certain hepatoxins.
In this study, sulfated glycoprotein-2 (clusterin) was overexpressed in more than 67% of lung tumors. Clusterin is a widely expressed, well-conserved, secreted glycoprotein that inhibits apoptosis. Secreted proteins such as clusterin become particularly attractive candidate proteins as biomarkers of cancer in serum. Recent studies indicate that overexpression of clusterin confers cellular protection against heat shock and oxidative stress [37] and exogenous clusterin reduces the sensitivity of cells to TNF [38].
MHC class I, immunoglobulin, and complement components are involved in immune surveillance [39]. In this study, both the MHC class I heavy-chain precursors, H-2D(k) and H-2K(k), are upregulated in tumors. Alterations in expression of these genes may reflect differences in the numbers of lymphoid cells observed in tumors as contrasted with normal lung parenchyma. Complement C3 is also upregulated, whereas 12A1 immunoglobulin heavy chain is downregulated.
A number of genes that are commonly expressed in normal lung parenchyma were overexpressed or underexpressed in lung adenomas. We have also shown that surfactant-associated proteins (SPs) A, B, and C are upregulated in three of six (50%), six of six (100%), and six of six (100%), whereas CC10 was downregulated in all lung adenomas examined. These results suggest that most of the lung adenoma cells were derived from Type II cells instead of Clara cells. Alternatively, altered expression of these genes may have functional implications. For example, CC10 may function to bind to calcium, proteins, or other ligands and may be an important immunomodulatory and anti-inflammatory protein [40,41]. Overexpression of CC10 cDNA in the NSCLC cell line A549 markedly reduces its invasiveness. CC10-transfected cell lines also exhibit decreased adhesiveness to fibronectin [42]. These results support the conclusion that loss of CC10 may contribute to carcinogenesis. SP-A, SP-B, and SP-C, are known to be required for optimal surfactant function [43]. The functional significance of these proteins is unknown. Studies have shown that SP mRNA are present in all lung tumors, with SP-A, SP-B, and SP-C being coexpressed in 10 of 12 (83%) adenomas and four of five (80%) carcinomas [44].
There is a possibility that some of the differentially expressed genes are due to differences in the density of mouse Type II cells or Clara cells in normal lungs versus and in lung adenomas with respect to amounts of relative to stromal and other contaminating cells. We have not been successful in isolating pure Type II cells or Clara cells from normal surrounding lungs of the animals bearing adenomas. Furthermore, numerous steps and treatments required for the current methodology available for isolating these cells would not make the isolated cells suitable control cells for lung adenomas that did not undergo such a process for gene expression profiling studies.
As was mentioned in the Introduction, one of the strengths of the CCLS technique is its ability to allow the identification and cloning of unknown or minimally described genes. Thus, in addition to the better known genes described above, an additional seven unknown genes were described as being overexpressed in the majority of lung adenomas. In fact, most of these genes were overexpressed at least three-fold. Two of the genes were further characterized by extensive sequencing and comparison of both mouse and human cDNA: a hypothetical protein (FLJ11240) and a GEF homologue (Figure 4). FLJ11240 hypothetical protein was found to have partial homology with a peptidase, E1–E2 ATPase, His kinase, and Ppx/GppA phosphatase when searching the Protein-BLAST database (NCBI, NIH). The GEF gene was found to be highly homologous (>98%) to mouse neuronal GEF. GEFs have been shown to play important roles in the Ras signaling pathway, which is frequently activated by the binding of Ras to Raf protein kinases, Type I phosphatidylinositol-3 (PI3) kinases, or Ral-specific guanine nucleotide exchange factors (RalGEFs) [13]. RalGEFs interact with Ras to form the GTP-bound state of the Ral family GTPases, leading to enhanced transcription of c-fos, cyclin D1, and genes containing the TATA-binding protein promoter [14–17]. Recently, activation of the RalGEF pathway has been shown to promote tumor metastasis [18]. This result suggests that overexpression of the neuronal GEF gene is associated with lung carcinogenesis in mice. These results show the strength of this approach for identifying unknown or minimally characterized genes with altered expression.
Finally, several genes differentially expressed, observed in this study using the CCLS method, are also found to be differentially expressed in mouse lung tumors through immunohistochemistry. For example, Mason et al. [45] reported increased expression of SP-A and SP-C in mouse lung adenomas and the lack of expression of CC10 in mouse lung adenomas regardless of morphology (solid or papillary) using immunohistochemistry. Another report by Ramakrishna et al. [46] found that expression of Erk1/2 was increased in mouse lung tumors using immunoblotting method. These reports provide further confirmation of our results using a different methodology.
The genes identified in this study can be employed in a variety of ways: 1) for use as early detection markers for lung lesions in the A/J model; 2) to compare the gene expression changes observed in the A/J model compared with human adenocarcinomas; 3) for basic understanding of the cancer process; 4) to help define potential molecular targets, which can be tested in this highly reproducible lung tumor model; and 5) to serve as potential modulatable biomarkers, which can be employed in screening for potential agents or in determining the efficacy of those agents.
Acknowledgements
We are grateful to G. Stoner for critical reading of this manuscript and helpful discussions. We thank E. Wiley for secretarial assistance.
Abbreviations
- CCLS
competitive cDNA library screening
- MNU
N-methyl-N-nitrosourea
- PCR
polymerase chain reaction
- RT
reverse transcription
Footnotes
Supported by Public Health Service grants CN05113, CA58554, CA78797, and CA16058 from the National Cancer Institute, the National Institutes of Health, and the Department of Health and Human Services.
References
- 1.Beckett WS. Epidemiology and etiology of lung cancer. Clin Chest Med. 1993;14:1–15. [PubMed] [Google Scholar]
- 2.Fielding JE. Smoking: health effects and control (1) N Engl J Med. 1985;313:491–498. doi: 10.1056/NEJM198508223130807. [DOI] [PubMed] [Google Scholar]
- 3.Herzog CR, Lubet RA, You M. Genetic alterations in mouse lung tumors: implications for cancer chemoprevention. J Cell Biochem Suppl. 1997;28–29:49–63. [PubMed] [Google Scholar]
- 4.Witschi H, Espiritu I, Maronpot RR, Pinkerton KE, Jones AD. The carcinogenic potential of the gas phase of environmental tobacco smoke. Carcinogenesis. 1997;18:2035–2042. doi: 10.1093/carcin/18.11.2035. [DOI] [PubMed] [Google Scholar]
- 5.Witschi H, Espiritu I, Peake JL, Wu K, Maronpot RR, Pinkerton KE. The carcinogenicity of environmental tobacco smoke. Carcinogenesis. 1997;18:575–586. doi: 10.1093/carcin/18.3.575. [DOI] [PubMed] [Google Scholar]
- 6.Malkinson AM. Primary lung tumors in mice: an experimentally manipulable model of human adenocarcinoma. Cancer Res. 1992;52:2670s–2676s. [PubMed] [Google Scholar]
- 7.You M, Candrian U, Maronpot RR, Stoner GD, Anderson MW. Activation of the Ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci USA. 1989;86:3070–3074. doi: 10.1073/pnas.86.9.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Re FC, Manenti G, Borrello MG, Colombo MP, Fisher JH, Pierotti MA, Della Port G, Dragani TA. Multiple molecular alterations in mouse lung tumors. Mol Carcinog. 1992;5:155–160. doi: 10.1002/mc.2940050211. [DOI] [PubMed] [Google Scholar]
- 9.Herzog CR, Wang Y, You M. Allelic loss of distal chromosome 4 in mouse lung tumors localize a putative tumor suppressor gene to a region homologous with human chromosome 1p36. Oncogene. 1995;11:1811–1815. [PubMed] [Google Scholar]
- 10.Herzog CR, Chen B, Wang Y, Schut HA, You M. Loss of heterozygosity on chromosomes 1, 11, 12, and 14 in hybrid mouse lung adenocarcinomas. Mol Carcinog. 1996;16:83–90. doi: 10.1002/(SICI)1098-2744(199606)16:2<83::AID-MC4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 11.Hegi ME, Devereux TR, Dietrich WF, Cochran CJ, Lander ES, Foley JF, Maronpot RR, Anderson MW, Wiseman RW. Allelotype analysis of mouse lung carcinomas reveals frequent allelic losses on chromosome 4 and an association between allelic imbalances on chromosome 6 and K-ras activation. Cancer Res. 1994;54:6257–6264. [PubMed] [Google Scholar]
- 12.Wang Y, Hu L, Yao R, Wang M, Crist KA, Grubbs CJ, Johanning GL, Lubet RA, You M. Altered gene expression profile in chemically induced rat mammary adenocarcinomas and its modulation by an aromatase inhibitor. Oncogene. 2001;20:7710–7721. doi: 10.1038/sj.onc.1204941. [DOI] [PubMed] [Google Scholar]
- 13.Joneson T, Bar-Sagi D. Ras effectors and their role in mitogenesis and oncogenesis. J Mol Med. 1997;75:587–593. doi: 10.1007/s001090050143. [DOI] [PubMed] [Google Scholar]
- 14.Wolthuis RM, Bos JL. Ras caught in another affair: the exchange factors for Ral. Curr Opin Genet Dev. 1999;9:112–117. doi: 10.1016/s0959-437x(99)80016-1. [DOI] [PubMed] [Google Scholar]
- 15.Okazaki M, Kishida S, Hinoi T, Hasegawa T, Tamada M, Kataoka T, Kikuchi A. Synergistic activation of c-fos promoter activity by Raf and Ral GDP dissociation stimulator. Oncogene. 1997;14:515–521. doi: 10.1038/sj.onc.1200860. [DOI] [PubMed] [Google Scholar]
- 16.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 17.Johnson SA, Mandavia N, Wang HD, Johnson DL. Transcriptional regulation of the TATA-binding protein by Ras cellular signaling. Mol Cell Biol. 2000;20:5000–5009. doi: 10.1128/mcb.20.14.5000-5009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward Y, Wang W, Woodhouse E, Linnoila I, Liotta L, Kelly K. Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol Cell Biol. 2001;21:5958–5969. doi: 10.1128/MCB.21.17.5958-5969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price DT, Rocca GD, Guo C, Ballo MS, Schwinn DA, Luttrell LM. Activation of extracellular signal-regulated kinase in human prostate cancer. J Urol. 1999;162:1537–1542. [PubMed] [Google Scholar]
- 20.Connelly MA, Marcu KB. CHUK, a new member of the helix-loop-helix and leucine zipper families of interacting proteins, contains a serine-threonine kinase catalytic domain. Cell Mol Biol Res. 1995;41:537–549. [PubMed] [Google Scholar]
- 21.Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 22.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 23.Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, Long CM, Crosier WJ, Watt K, Koths K, McCormick F. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1998;242:1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- 24.Vleurick L, Pezet A, Kuhn ER, Decuypere E, Edery M. A beta-turn endocytic code is required for optimal internalization of the growth hormone receptor but not for alpha-adaptin association. Mol Endocrinol. 1999;13:1823–1831. doi: 10.1210/mend.13.11.0371. [DOI] [PubMed] [Google Scholar]
- 25.Okabayashi Y, Sugimoto Y, Tolty NF, Hsuan J, Kido Y, Sakaguchi K, Gout I, Waterfield MD, Kasuga M. Interaction of Shc with adaptor protein adaptins. J Biol Chem. 1996;271:5265–5269. doi: 10.1074/jbc.271.9.5265. [DOI] [PubMed] [Google Scholar]
- 26.Viola JP, Rao A. Role of the cyclosporin-sensitive transcription factor NFAT1 in the allergic response. Mem Inst Oswaldo Cruz. 1997;92(Suppl 2):147–155. doi: 10.1590/s0074-02761997000800020. [DOI] [PubMed] [Google Scholar]
- 27.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT.AP-1.DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 28.Sune C, Garcia-Blanco MA. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol Cell Biol. 1999;19:4719–4728. doi: 10.1128/mcb.19.7.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kireeva ML, Latinkic BV, Kolesnikova TV, Chen CC, Yang GP, Abler AS, Lau LF. Cyr61 and Fisp12 are both ECM-associated signaling molecules: activities, metabolism, and localization during development. Exp Cell Res. 1997;233:63–77. doi: 10.1006/excr.1997.3548. [DOI] [PubMed] [Google Scholar]
- 30.Kolesnikova TV, Lau LF. Human CYR61-mediated enhancement of bFGF-induced DNA synthesis in human umbilical vein endothelial cells. Oncogene. 1998;16:747–754. doi: 10.1038/sj.onc.1201572. [DOI] [PubMed] [Google Scholar]
- 31.Kireeva ML, Lam SC, Lau LF. Adhesion of human umbilical vein endothelial cells to the immediate-early gene product Cyr61 is mediated through integrin alphavbeta3. J Biol Chem. 1998;273:3090–3096. doi: 10.1074/jbc.273.5.3090. [DOI] [PubMed] [Google Scholar]
- 32.Babic AM, Kireeva ML, Kolesnikova TV, Lau LF. CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci USA. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waheed A, Zhu XL, Sly WS. Membrane-associated carbonic anhydrase from rat lung. Purification, characterization, tissue distribution, and comparison with carbonic anhydrase IVs of other mammals. J Biol Chem. 1992;267:3308–3311. [PubMed] [Google Scholar]
- 34.Lonnerholm G, Wistrand PJ. Membrane-bound carbonic anhydrase CA IV in the human kidney. Acta Physiol Scand. 1991;141:231–234. doi: 10.1111/j.1748-1716.1991.tb09072.x. [DOI] [PubMed] [Google Scholar]
- 35.Christie KN, Thomson C, Xue L, Lucocq JM, Hopwood D. Carbonic anhydrase isoenzymes I, II, III, and IV are present in human esophageal epithelium. J Histochem Cytochem. 1997;45:35–40. doi: 10.1177/002215549704500105. [DOI] [PubMed] [Google Scholar]
- 36.Dockham PA, Lee MO, Sladek NE. Identification of human liver aldehyde dehydrogenases that catalyze the oxidation of aldophosphamide and retinaldehyde. Biochem Pharmacol. 1992;43:2453–2469. doi: 10.1016/0006-2952(92)90326-e. [DOI] [PubMed] [Google Scholar]
- 37.Viard I, Wehrli P, Jornot L, Bullani R, Vechietti JL, Schifferli JA, Tschopp J, French LE. Clusterin gene expression mediates resistance to apoptotic cell death induced by heat shock and oxidative stress. J Invest Dermatol. 1999;112:290–296. doi: 10.1046/j.1523-1747.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 38.Sintich SM, Steinberg J, Kozlowski JM, Lee C, Pruden S, Sayeed S, Sensibar JA. Cytotoxic sensitivity to tumor necrosis factor-alpha in PC3 and LNCaP prostatic cancer cells is regulated by extracellular levels of SGP-2 (clusterin) Prostate. 1999;39:87–93. doi: 10.1002/(sici)1097-0045(19990501)39:2<87::aid-pros2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 39.Lodish H, Baltimore D, Berk A, Zipurski SL, Matsudaira P, Darnell J. Molecular Cell Biology. 3rd ed. New York: W.H. Freeman; 1995. [Google Scholar]
- 40.Singh G, Singh J, Katyal SL, Brown WE, Kramps JA, Paradis IL, Dauber JH, Macpherson TA, Squeglia N. Identification, cellular localization, isolation, and characterization of human Clara cell-specific 10 KD protein. J Histochem Cytochem. 1988;36:73–80. doi: 10.1177/36.1.3275712. [DOI] [PubMed] [Google Scholar]
- 41.Andersson O, Nordlund-Moller L, Barnes HJ, Lund J. Heterologous expression of human uteroglobin/polychlorinated biphenyl-binding protein. Determination of ligand binding parameters and mechanism of phospholipase A2 inhibition in vitro. J Biol Chem. 1994;269:19081–19087. [PubMed] [Google Scholar]
- 42.Szabo E, Goheer A, Witschi H, Linnoila RI. Overexpression of CC10 modifies neoplastic potential in lung cancer cells. Cell Growth Differ. 1998;9:475–485. [PubMed] [Google Scholar]
- 43.Weaver TE, Whitsett JA. Function and regulation of expression of pulmonary surfactant-associated proteins. Biochem J. 1991;273(Part 2):249–264. doi: 10.1042/bj2730249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilling AM, Mifsud NA, Jones SA, Endersby-Wood HJ, Turton JA. Expression of surfactant protein mRNA in normal and neoplastic lung of B6C3F1 mice as demonstrated by in situ hybridization. Vet Pathol. 1999;36:57–63. doi: 10.1354/vp.36-1-57. [DOI] [PubMed] [Google Scholar]
- 45.Mason RJ, Kalina M, Nielsen LD, Malkinson AM, Shannon JM. Surfactant protein C expression in urethane-induced murine pulmonary tumors. Am J Pathol. 2000;156:175–182. doi: 10.1016/S0002-9440(10)64717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramakrishna G, Perella C, Birely L, Diwan BA, Fornwald LW, Anderson LM. Decrease in K-ras p21 and increase in Raf1 and activated Erk 1 and 2 in murine lung tumors initiated by N-nitrosodimethylamine and promoted by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2002;179:21–34. doi: 10.1006/taap.2001.9344. [DOI] [PubMed] [Google Scholar]