Abstract
Helicases not only catalyse the disruption of hydrogen bonding between complementary regions of nucleic acids, but also move along nucleic acid strands in a polar fashion. Here we show that the Rep52 and Rep40 proteins of adeno-associated virus type 2 (AAV-2) are required to translocate capsid-associated, single-stranded DNA genomes into preformed empty AAV-2 capsids, and that the DNA helicase function of Rep52/40 is essential for this process. Furthermore, DNase protection experiments suggest that insertion of AAV-2 genomes proceeds from the 3′ end, which correlates with the 3′→5′ processivity demonstrated for the Rep52/40 helicase. A model is proposed in which capsid-immobilized helicase complexes act as molecular motors to ‘pump’ single-stranded DNA across the capsid boundary.
Keywords: AAV/DNA helicase/encapsidation/virus
Introduction
Viruses package their genomes into proteinaceous capsid structures either via association of structural proteins with the viral genome or via insertion of viral genomes into pre-assembled capsids. The choice of assembly pathway appears to depend to a certain degree upon the nature of the viral genome to be encapsidated (Guo, 1994). Circular double-stranded (ds) DNA genomes seem to be encapsidated via assembly of structural proteins around the genome (Bina et al., 1983; Blasquez et al., 1983; Oppenheim et al., 1992). Viruses with ds linear genomes, including the bacteriophages, insert their genomes into preformed empty capsids (Black, 1989; Guo and Trottier, 1994), a process that involves non-structural packaging enzymes and the consumption of ATP. Studies with herpes viruses (Gibson et al., 1972; Booy et al., 1991; Newcomb et al., 1996; Trus et al., 1996) and adenoviruses (Philipson, 1984; Schmid and Hearing, 1995) suggest that their packaging pathways also involve preformed capsids and, in the case of the herpes viruses, share a number of additional features with the dsDNA bacteriophages, such as the presence of scaffold proteins (Lee et al., 1988; Newcomb and Brown, 1991), concatameric genomes (Varmuza et al., 1985; Deiss and Frenkel, 1986) and non-structural DNA packaging enzymes with a consensus ATP-binding domain (McNabb and Courtney, 1992; Yu and Weller, 1998a,b). Several viruses with single-stranded (ss) DNA or ssRNA genomes assemble their protein shell around the genome, a process driven by protein–nucleic acid interactions. This group is represented by the tobacco mosaic virus and the F1 and M13 bacteriophages. Interestingly, a number of ssDNA and ssRNA viruses, such as bacteriophages φX174 and φ6, parvoviruses and poliovirus also package their genomes into preformed capsids.
DNA helicases are enzymes that catalyse the unwinding of DNA by disrupting the hydrogen bonding between paired bases as they progress along the DNA strand in a polar fashion (either 3′→5′ or 5′→3′), and have been shown to be involved in many essential cellular processes, including replication, transcription, DNA repair and translation (for reviews see West, 1996; Bird et al., 1998; Lohman et al., 1998; Hall and Matson, 1999). The energy required for both the translocation and strand separation reactions is generated by NTP hydrolysis. A number of conserved ‘helicase motifs’ exist, which via structure–function analyses have been shown to be involved in nucleotide binding and hydrolysis. Most helicases act as oligomers—dimers or hexamers—or exist as integral parts of larger protein complexes in which they provide molecular motor functions, and are thus similar in many respects to other motor proteins such as cytoplasmic kinesin (Gilbert et al., 1995; Moore and Lohman, 1995; Lohman et al., 1998). A number of proteins that are known to be required for virus/phage genome packaging have been shown to contain helicase-like motifs (Yu and Weller, 1998b; Catalano, 2000).
Adeno-associated virus (AAV) assembly has been proposed to occur in two steps (Myers and Carter, 1980). Initially, empty capsids form and are then slowly filled by ss genomes that are generated by strand displacement synthesis from ds templates. At early stages of a productive infection, capsid assembly and viral DNA replication occur in a spatially and temporally distinct fashion, at specific sites within the cell nucleus, while at later time points, replication proteins, capsids and viral DNA seem to be co-distributed throughout the nucleoplasm (Wistuba et al., 1997). Replication of the AAV genome proceeds via a self-priming strand displacement mechanism (for review see Berns, 1990). The two large virus-encoded Rep proteins (Rep78 and Rep68) catalyse the resolution of the covalently closed AAV termini, a process that involves sequence-specific AAV DNA binding, strand-specific nicking and helicase activity under ATP consumption (Im and Muzyczka, 1990, 1992; Brister and Muzyczka, 1999, 2000). The small Rep proteins (Rep52 and Rep40) are not required for ds DNA replication (Ni et al., 1994). As a result of the Rep78/68-mediated terminal resolution, large Rep molecules remain covalently attached to the 5′ genome termini (Im and Muzyczka, 1990; Prasad and Trempe, 1995). The observation that large and small Rep proteins are able to bind to capsids as well as to one another led us to hypothesize that the interaction of large Rep-bound genomes with empty capsids is mediated by the Rep proteins (Dubielzig et al., 1999). However, the actual mechanism by which DNA is inserted into the capsids remained unknown. Interestingly, mutagenesis of the AAV genome showed that the small Rep proteins are required for the accumulation of ssDNA in the cell, which had been interpreted as representing DNA encapsidation (Chejanovsky and Carter, 1989). More recently, it has been demonstrated that the ATPase/helicase domains common to both the large and small Rep proteins are also functional in Rep52 (Smith and Kotin, 1998).
The aim of this study was to identify the mechanism by which the small Rep proteins increase the accumulation of ss AAV-2 genomes, and to determine whether or not the Rep52/40 DNA helicase activity is involved. The function of the small Rep proteins was analysed by preventing Rep52/40 expression from an infectious clone and complementation via co-expression of either wild-type (wt) Rep52/40 or Rep52/40 DNA helicase mutants. Viral genomes were isolated and analysed to determine the point at which small Rep proteins act to improve genome encapsidation. We show here that the small Rep helicase activity is required to translocate capsid-bound, full-length genomes into the capsid.
Results
Small Rep proteins are required for efficient packaging of AAV-2 DNA
The wt AAV-2 construct (Figure 1A, pTAV2-0) was mutated so as to destroy the translation initiation codon for the small Rep proteins (pTAV2-1), as has been described previously (Chejanovsky and Carter, 1989). Western blot analysis of pTAV2-1-transfected cells confirmed that Rep52 and Rep40 could no longer be detected, and that expression of the large Rep proteins and capsid proteins remained unchanged (data not shown).
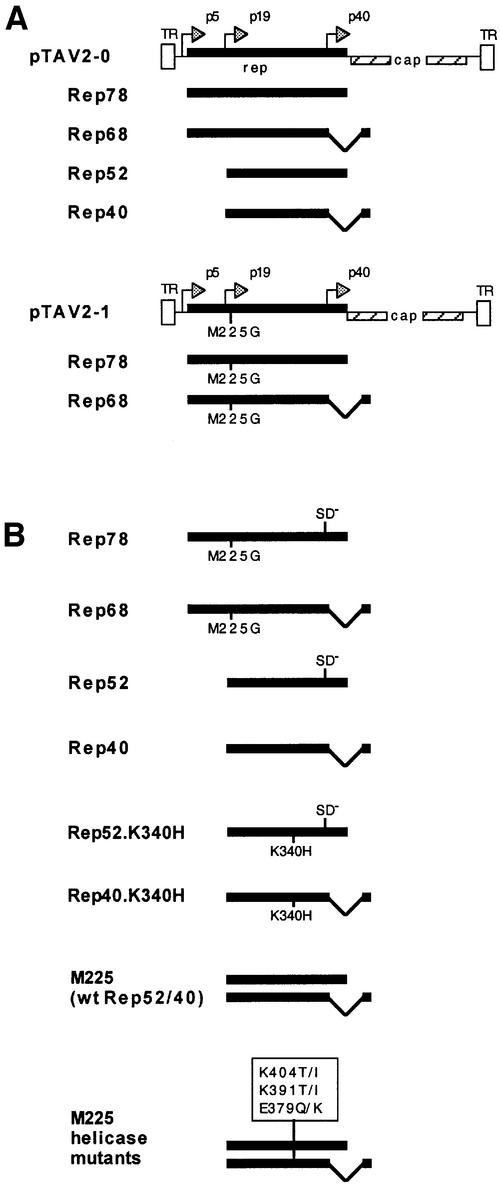
Fig. 1. AAV viral constructs and Rep expression plasmids. (A) pTAV2-0 consists of the entire AAV-2 genome, including both inverted terminal repeats (TR). pTAV2-1 is derived from pTAV2-0, but has the Rep52/40 translation start site methionine 225 mutated to glycine. Spliced and unspliced gene products expressed from the rep open reading frames (ORFs) of the two virus genomes are also shown. AAV promoters are denoted by p5, p19 and p40. (B) The Rep78, Rep68, Rep52 and Rep40 proteins are individually expressed from a CMV promoter within the pKEX-XL plasmid (Hörer et al., 1995). The K340H small Rep mutants contain a lysine to histidine mutation at amino acid 116 (corresponding to amino acids 340 of the complete Rep ORF), which mutates the ATP binding site (Chejanovsky and Carter, 1990). M225 is also based on pKEX-XL and allows expression of both Rep52 and Rep40, as the splice sites have not been altered. A series of helicase mutations (E379Q, E379K, K391I, K391T, K404I, K404T) were introduced into M225 as described in Materials and methods. SD– denotes a mutated splice donor site.
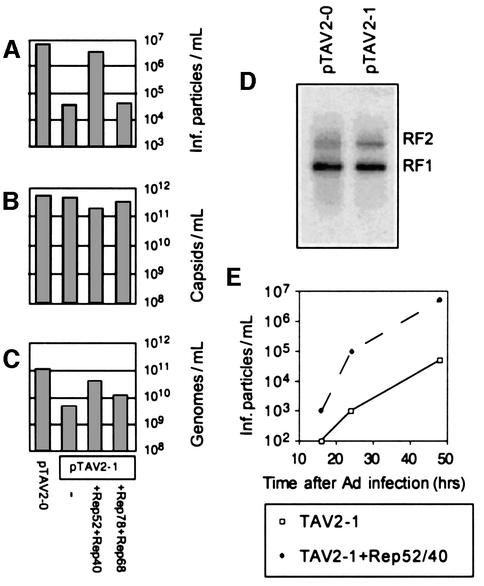
We then analysed virus production in the absence of small Rep proteins using the pTAV2-1 construct. The infectious titre was reduced 200-fold (Figure 2A), confirming previous observations (Chejanovsky and Carter, 1989; Hölscher et al., 1995). This reduction was not due to impaired capsid formation as analysed by capsid ELISA titration (Figure 2B) nor to differences in viral DNA replication (Figure 2D), but rather to a large (20-fold) reduction in the amount of encapsidated DNA (Figure 2C). Since the dot-blot quantification of encapsidated DNA also detects incompletely packaged and thus non-infectious genomes, the reduction in infectious titre is more pronounced than the decrease in encapsidated DNA. Complementation of pTAV2-1 by co-transfecting Rep52- and Rep40-expressing plasmids (Figure 1B) efficiently rescued the TAV2-1 mutation, as shown by increased production of infectious particles. Additional expression of the large Rep proteins—which contain the small Rep coding regions, but do not express them as separate small Rep proteins due to the mutated Rep52/40 translation start codon Met225—did not. The dependence of virus production upon Rep52/40 was not transient, but remained stable throughout a productive AAV-2/Ad-5 infection (Figure 2E). Taken together, these experiments suggest that the small Rep proteins are involved in the DNA packaging process.
Fig. 2. Requirement of Rep52/40 for efficient DNA encapsidation. Viral supernatants were generated from cells transfected either with the wt pTAV2-0 construct or with the mutant pTAV2-1 construct and in combination with plasmids expressing either the small or large Rep proteins. Supernatants were assayed for (A) infectious viral titre, (B) ELISA-based AAV-2 capsid titre and (C) quantity of encapsidated AAV-2 DNA. (D) Low molecular weight RF (replication form) DNAs were isolated from cells transfected with pTAV2-0 or pTAV2-1 using a modified Hirt extraction procedure, and detected by Southern blotting using an AAV rep-specific probe. (E) Virus production was measured over time in the presence and absence of Rep52/40.
Helicase activity of the small Rep proteins is required for their DNA encapsidation function
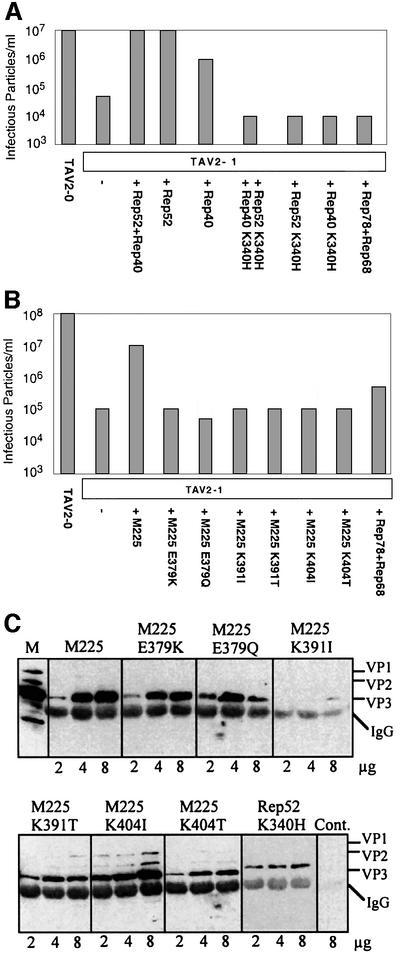
Efficient complementation of pTAV2-1 did not require expression of both small Rep proteins (Figure 3A), indicating a certain level of redundancy between Rep52 and Rep40 function. However, complementation of pTAV2-1 with Rep52- and Rep40-expressing plasmids in which the putative ATP binding site was mutated (Figure 1B, K340H; amino acid numbering relates to the large Rep proteins) failed to rescue the defect in genome packaging (Figure 3A). This mutation in Rep52 has been shown to abrogate both DNA helicase and ATPase activities (Smith and Kotin, 1998).
Fig. 3. Effect of small Rep DNA helicase on AAV DNA packaging. (A) Cells were transfected either with pTAV2-0 or with pTAV2-1, either alone or together with plasmids encoding wt or small Rep proteins mutated in the helicase domain (K340H) or large Rep proteins. Extracts were prepared and their infectious particle titres determined. (B) A similar analysis was carried out using plasmids expressing either the wt or mutant forms of both small Rep proteins, in which point mutations had been introduced at additional conserved amino acid residues of the helicase domain (see Figure 1). (C) The capacity of these mutants to form complexes with AAV-2 capsid proteins was tested by co-transfecting increasing amounts (2, 4 and 8 µg) of the capsid protein-expressing plasmid (CMV-VP) with a fixed amount (3 μg) of each small Rep expression plasmid. Complexes were immunoprecipitated from cell extracts using an anti-Rep antiserum and analysed via western blotting using a monoclonal anti-VP antibody to detect capsid proteins. A control reaction was carried out using extracts of cells that had not been transfected with a Rep-expressing plasmid (Cont). M, standard showing the expression of the three capsid proteins (VP1–3). IgG indicates the position of the immunoglobulin heavy chain of the antiserum used for the immunoprecipitation.
A series of Rep52/40 mutants was then constructed, with single amino acid substitutions of conserved nucleotide-binding motif residues (Bradley et al., 1987) that are present in several viral non-structural proteins (Jindal et al., 1994) and important for large Rep ATPase activity, upon which the DNA helicase activity depends (McCarty et al., 1992; Walker et al., 1997). None of these helicase mutants was able to complement the pTAV2-1 packaging defect (Figure 3B). The helicase activity of the small Rep proteins is thus required for their packaging activity.
Large and small Rep proteins are able to form complexes with AAV-2 capsids (Dubielzig et al., 1999). The formation of such complexes is believed to be a prerequisite for DNA encapsidation. It has previously been observed that small Rep proteins with a mutated ATP binding site (Figure 1B, K340H) form Rep–capsid complexes less efficiently than the wt small Rep proteins (Dubielzig et al., 1999). We tested the ability of the helicase mutant small Rep proteins to form complexes with capsids. A fixed amount of each of the small Rep proteins was co-expressed with increasing amounts of capsid proteins. Anti-Rep antisera were used to immunoprecipitate Rep–capsid complexes, which were then analysed via western blotting using an anti-VP monoclonal antibody (Figure 3C). In accordance with previous observations, the K340H mutant bound lower levels of capsid proteins than the wt small Rep proteins. In addition, three of the new mutants (K391I, K391T and K404T) also showed reduced capsid complex formation, while another three (E379K, E379Q and K404I) formed complexes as efficiently as wt Rep52/40. The fact that the latter three helicase mutants bound to capsids efficiently argues that the missing ATPase/helicase activity and not poor capsid association was responsible for their inability to package AAV-2 genomes efficiently.
Analysis of encapsidated genomes
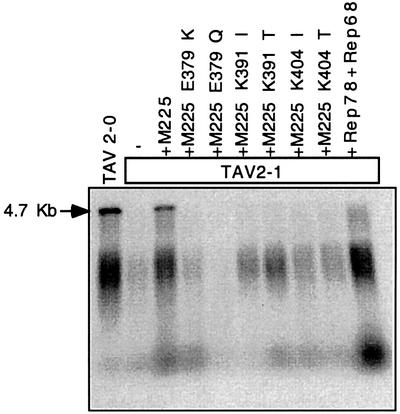
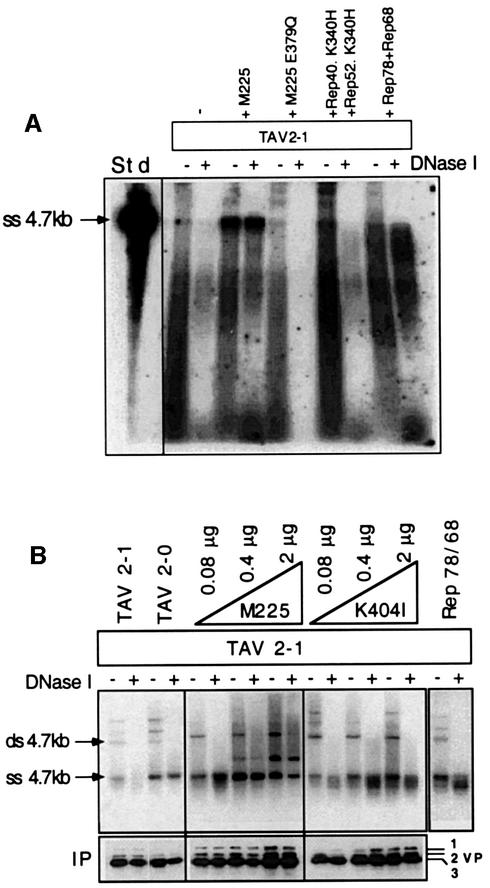
Viral genomes were extracted from virus produced in the above described experiments, following pre-treatment of viral supernatants with DNase I. This DNase I-resistant DNA was interpreted as being encapsidated, or at least protein protected. Alkaline agarose gels were then used to analyse the ss genomes (Figure 4). Three main populations of genomes were observed. A clear band at 4.7 kb represented the fully encapsidated AAV-2 genomes. A minor population of very short molecules was interpreted as representing genome fragments that had associated with encapsidation complexes or been inserted only a short way into the capsids. The third population was visible as a non-uniform smear of DNA, and taken to represent genomes that had been inserted to ∼50–75% of full length. Clear bands at 4.7 kb were only detected with wt virus (Figure 4, TAV2-0) or with the TAV2-1 virus complemented with wt small Rep proteins (Figure 4, M225). In the absence of wt small Rep proteins or in the presence of the helicase mutant small Rep proteins, only partially encapsidated genomes were detected. Additional expression of the large Rep proteins resulted in an increased number of partially inserted genomes.
Fig. 4. Genome encapsidation in the presence of Rep52/40 DNA helicase mutants. Viral genomes from virus preparations (see Figure 3B) were isolated from the virus supernatants after extensive DNase I digestion, separated by alkaline agarose gel electrophoresis and Southern blotted. AAV DNA was detected using a rep-specific probe. 4.7 kb indicates full-length encapsidated AAV DNA.
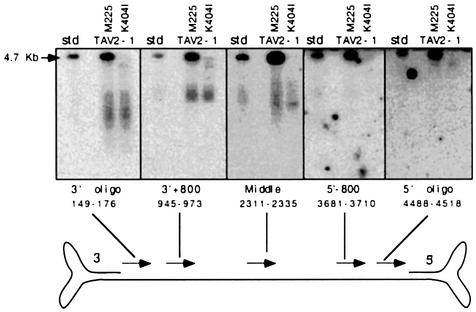
In order to determine whether specific regions of the genome were selectively protected against DNase I digestion, and thus more likely to be located inside the capsid, labelled probes specific for various genome regions were used for hybridizations (Figure 5). Probes specific for the 3′ half of the genome (3′ oligo and 3′+800) detected both full-length and partially packaged genomes, whereas probes specific for regions towards the 5′ end (5′–800 and 5′ oligo) reacted only with full-length genomes. The under-representation of 5′ ends in the incompletely packaged pool indicates that these ends were inserted at a late stage in the encapsidation process and suggests that packaging proceeds in a 3′→5′ direction.
Fig. 5. Directionality of AAV packaging. Cells were transfected with the small Rep-deleted pTAV2-1 plasmid and either the wt Rep52/40 (M225) or the K404I helicase mutant Rep52/40 construct. DNase I-protected genomes from lysates were separated on alkaline agarose gels. Following Southern blotting, partially and fully packaged genomes were detected using end-labelled oligonucleotides that hybridize to the 3′ end (3′ oligo), a region 800 bp from the 3′ end (3′+800), a central region (middle), a region 800 bp from the 5′ end (5′–800) and the 5′ end (5′ oligo) of one of the two packagable genome strands, as indicated (arrowheads point in the 5′→3′ direction).
Analysis of capsid-associated versus encapsidated AAV-2 DNA
The analyses of viral genomes shown in Figures 4 and 5 are limited to encapsidated DNA. To analyse also genomes arrested at various stages of the packaging process, a capsid-specific immunoprecipitation was performed upon extracts that had been nuclease pre-treated or not, prior to genome isolation and Southern blot analysis. Extracts were derived from transfections of pTAV2-1 alone and from co-transfections of pTAV2-1 with either wt small Rep plasmid (M225), two different helicase mutant small Reps (K340H and E379Q), or additional large Rep proteins. With wt small Rep proteins (Figure 6A, lanes M225), dominant DNase I-resistant 4.7 kb bands were present in both the DNase-treated and untreated lanes, indicating that full-length genomes had been efficiently packaged. In addition, two major populations of incompletely packaged genomes were detected, which were partially DNase I resistant as previously observed in Figure 4. In contrast, virus produced either in the absence of wt small Rep proteins or in the presence of the helicase mutant small Reps (Figure 6A, lanes M225 E379Q and Rep52.K340H +Rep40.K340H) contained predominantly partially encapsidated genomes, the majority of capsid-coprecipitated genomes being DNase I sensitive. Interest ingly, in the DNase I-untreated samples, faint bands at 4.7 kb were also detected in the absence of wt small Reps or in the presence of helicase mutant small Reps. These were interpreted as full-length, capsid-associated genomes that had not been efficiently encapsidated. Overexpression of the large Rep proteins again resulted in an increase in the number of partially packaged genomes. Remarkably, under these conditions, most of the capsid-coprecipitated genomes were DNase resistant and thus encapsidated. However, as before, no full-length encapsidated DNA was visible at 4.7 kb. AAV DNAs significantly larger than 4.7 kb were also present. These were DNase I sensitive and thus interpretated as being non-encapsidated replication intermediates. As they were also able to bind to protein A–Sepharose in the absence of A20 antibody, it is not clear whether they were directly associated with the capsid or merely non-specifically coprecipitated. A non-specific binding of ss genomes or capsids to the protein A–Sepharose was, however, not detected (data not shown).
Fig. 6. Analysis of capsid-associated and encapsidated genomes. (A) Viral supernatants obtained from cells that had been transfected with pTAV2-1 alone, or together with either both large Rep expression plasmids (Rep78/68), wt (M225) or helicase mutant (E379Q or K340H) small Rep expression plasmids, were digested (+) or not (–) with DNase I to discriminate between capsid-associated and packaged genomes. Capsids were then immunoprecipitated from each extract using equal amounts of protein A–Sepharose-bound A20 antibody. Coprecipitated genomes were isolated and electrophoresed on an alkaline agarose gel, Southern blotted and detected using a rep-specific probe. A 4.7 kb AAV genome fragment was used as a standard (Std). (B) Cells were transfected with pTAV2-0 or pTAV2-1 alone or with pTAV2-1 and both large Rep expression plasmids (Rep78/68) or increasing amounts of either the wt Rep52/40 (M225) plasmid or the M225-K404I helicase mutant. Cell extracts were then processed as described in (A). Viral genomes were extracted and electrophoresed on neutral agarose gels, Southern blotted and detected using a random-primed, rep-specific probe. A portion of each immunoprecipitate was tested for the level of capsid recovery via western analysis.
Alkaline gel electrophoresis of capsid-associated and encapsidated DNA efficiently resolved the lower molecular weight molecules, but did not allow us to distinguish ss from ds 4.7 kb molecules, as all DNAs were denatured. For this reason we performed similar experiments using neutral agarose gels. Cells were transfected with pTAV2-1 either alone or together with increasing amounts of the wt and helicase mutant Rep52/40 constructs (Figure 6B). In addition, a portion of the cells harvested was supplemented with EDTA to prevent degradation of capsid-bound ss genomes during the freeze–thaw cell lysis step. In the presence of wt small Rep proteins (Figure 6B, TAV2-0 or TAV2-1 complemented with increasing amounts of M225), clear bands were visible at the position of ss 4.7 kb DNA in both the DNase I-treated and untreated lanes, indicating that full-length genomes had been efficiently packaged. In the absence of wt small Rep (Figure 6B, lanes TAV2-1 and TAV2-1 + K404I), a 4.7 kb ssDNA was also detected in association with capsids. However, these genomes were DNase I sensitive, indicating that they had not been encapsidated. Western blot analysis confirmed that equivalent amounts of capsids were analysed. Overexpression of Rep78/68 had the same effect and, in addition, revealed an increased binding of ss genomes to the capsid, similar to that observed after overexpression of the K404I mutant. Again the high molecular weight bands corresponding to ds monomer, dimer and tetramer replication form intermediates were DNase I sensitive. The additional band present between the 4.7 kb ss and ds molecules where 0.4 or 2 µg of M225 had been co-transfected is probably due to re-annealing of ss genomes and only occurred when high concentrations of encapsidated DNA were isolated and separated on neutral gels. This experiment proves three points. (i) ss genomes are produced and associate with the capsid in the absence of small Rep proteins, clearly indicating that these proteins are not required for the generation of ss genomes during strand displacement synthesis. (ii) Small Rep proteins are required for efficient translocation of full-length ss genomes from the capsid surface to the inside. (iii) This translocation function requires a functional small Rep DNA helicase.
Discussion
Packaging into preformed capsids versus capsid assembly around genomes
Ever since the pioneering work of Myers and Carter (1980), the accepted idea has been that single-stranded AAV-2 genomes are packaged into preformed, empty capsids. Although there has been no additional proof, data from experiments carried out over the last 20 years do not contradict this concept. The alternative assembly pathway, in which genomes are ‘surrounded’ by structural proteins, is usually primed by binding of structural proteins to specific genome sequences (Guo, 1994). Based on Rep–Cap interaction experiments (Dubielzig et al., 1999), it is also possible that AAV-2 viral genomes, when bound by Rep78/68, are able to interact with viral capsid proteins during the capsid assembly process, which could theoretically result in encapsidation via assembly around genomes. However, the presence of large excesses of empty capsids in AAV stocks (Grimm et al., 1999), and the fact that VP expression alone is sufficient for capsid assembly (Ruffing et al., 1992; Wistuba et al., 1997), prove that viral DNA is not required for capsid formation. In addition, the observation that capsid-associated genomes are encapsidated when a capsid-bound helicase is present strongly suggests that the genomes are translocated into preformed capsids. Finally, the experiments showing that 3′ termini are more readily detected in partially encapsidated genome populations than 5′ termini (Figure 5) further support the theory that the main AAV packaging mechanism utilizes preformed empty capsids as substrates in which the DNA is introduced in a preferred orientation. Therefore, we have adopted the concept described by Myers and Carter, and base our findings accordingly.
Rep52/40 point of action
There are three stages in the virus assembly process at which small Rep proteins could exert their function to result in increased ssDNA accumulation. Initially, they could enhance the replication process required to yield RF (replication form) DNA and ss genomes. Alternatively, they could direct the targeting of ss genomes to preformed, empty capsids. Finally, they could be involved in inserting genomes into capsids. It has been shown previously (Chejanovsky and Carter, 1989) and confirmed in this study that small Rep proteins are not required for the generation of RF DNA. This is in accordance with in vitro experiments, which showed that small Rep proteins do not stimulate ds DNA replication (Ni et al., 1994). Here we demonstrate that small Rep proteins are also not required for synthesis of ssDNA. Detection of incompletely encapsidated ss genomes was made possible using high EDTA concentrations to prevent ssDNA degradation and an immunoprecipitation technique that allowed the purification of capsid-associated molecules. Recently, in vitro experiments have also shown that ss genomes are produced in the absence of small Rep proteins, and that they accumulate when ssDNA binding proteins are present (Ward and Linden, 2000). The small Rep proteins also appear not to be neccesary for association of ss genomes with the capsid, although it is possible that their expression results in an increased level of genome–capsid association. When increasing amounts of the helicase mutant Rep52/40 proteins were expressed, the number of capsid-associated genomes increased (Figure 6B). However, as the number of capsids precipitated also increased slightly, it is difficult to assess the degree to which the small Rep proteins may contribute to genome–capsid association. Genome–capsid association in the absence of small Reps is presumably mediated directly by genome-bound large Rep molecules. A direct interaction of the genome with its capsid, as demonstrated for the MVM (minute virus of mice) parvovirus (Willwand and Hirt, 1993), was not detected for AAV-2 (our unpublished data).
The most striking observation in this study was that the small Rep proteins, and in particular their helicase activity, are required for translocation of capsid-associated genomes into the capsid. It was of utmost interest to determine whether they are involved in the entire ss genome translocation reaction or rather at a specific step (e.g. initiation and/or completion). The partial DNase I resistance of capsid-associated AAV DNA in the presence of small Rep helicase mutants or following overexpression of the large Rep proteins could be interpreted as representing genomes that have become encapsidated up until a point at which the small Rep DNA helicase activity becomes essential for completion of the encapsidation process. However, as absence of a functional small Rep helicase resulted not only in poorer full-length DNA insertion, but also in a 20-fold reduction in total DNA encapsidation (Figure 2C), a more general involvement of the small Rep helicase activity in genome encapsidation has to be proposed. In any case, an active Rep52/40 DNA helicase is required to translocate full-length genomes from the capsid surface to the inside.
Similarities to phage DNA packaging systems
The mechanism by which viruses translocate their genomes into empty capsids is still not understood. Even for the most comprehensively studied viruses such as the bacteriophages, the ‘motor’ mechanism responsible for the actual translocation reaction remains elusive. The detailed biochemical analysis of the bacteriophage packaging mechanism (Fujisawa and Morita, 1997; Catalano, 2000) using in vitro packaging systems has shown that the bacteriophage packaging complex consists of two gene products which, when bound to the genome, display a spectrum of activities very similar to the AAV-2 large and small Rep proteins, namely sequence-specific DNA binding, ATPase, endonuclease and helicase activities. Following binding of the packaging proteins to the genome terminus to be inserted, this nucleoprotein complex must dock with the portal vertex complex on the procapsid surface, through which the genome is ‘pumped’ inside. A similar mechanism can now be proposed for the AAV Rep proteins, which specifically bind to replicated AAV genomes and form complexes with AAV capsids (Dubielzig et al., 1999). However, no portal vertex-like structures have so far been detected for AAV. Several models have been proposed for phage DNA packaging, including rotation of the portal vertex structure (Guo et al., 1998; Hendrix, 1998), the ratchet model (Fujisawa and Morita, 1997) or the formation of a doughnut-shaped DNA packaging complex, similar to the structures proposed for hexameric helicase complexes (Baker and Bell, 1998).
A model for the helicase-driven DNA encapsidation of AAV genomes
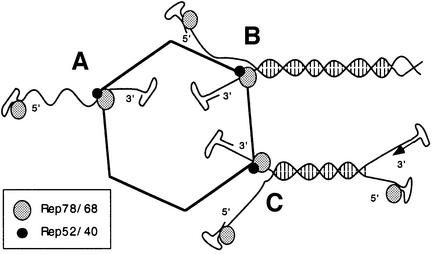
Helicases are able to move along the DNA they are unwinding under the consumption of ATP. Based on findings described here and the previously reported complex formation involving large and small Rep proteins with capsids during the capsid assembly process (Dubielzig et al., 1999), we propose that the helicase activity of capsid-immobilized small Rep proteins present in encapsidation complexes drives ss AAV genome translocation into the preformed capsids, for which three scenarios are depicted (Figure 7). In scenario A, a free ss genome is translocated by the helicase complex in a ‘scanning’-like fashion, as it would only require an unwinding activity to insert the partially ds terminal repeats. The existence of a preferred polarity for helicase movement on ssDNA is, however, still a matter for discussion (Lohman and Bjornson, 1996). In the second scenario (Figure 7B), the packaging helicase unwinds a ds concatameric genome on the capsid surface and simultaneously inserts the 3′ end of one of the single strands into the capsid. The hypothesis that dimer and multimer genomes can act as substrates for packaging would also require that the packaging reaction is terminated, most probably by the nicking activity of the large Rep proteins. The third scenario (Figure 7C) describes a similar situation in which the palindrome-containing termini of a ds monomer not engaged in the packaging reaction open up to allow a further round of strand displacement replication to occur during encapsidation. This situation would result in ‘collision’ of the packaging and replication complexes and premature strand displacement, which could either promote or inhibit encapsidation depending on whether ss or ds genomes are the preferred substrates for the packaging complex. Indeed, alkaline agarose gel analysis of packaged genomes did reveal a population of molecules ∼50–75% of the full length (Figures 4, 5 and 6A), whose presence could be due to a premature packaging stop or a transient slow down in the packaging rate, followed by a fast packaging completion step.
Fig. 7. Model for the involvement of Rep helicases as genome packaging motors. Most helicases bind to ssDNA adjacent to duplex regions and then proceed to unwind the two strands, moving along the bound single strand, under the consumption of ATP. The AAV packaging complex represents an immobilized helicase complex, composed of large and small Rep proteins, on the capsid surface. Both ss and ds genomes are able to bind to the packaging complex via interaction with the large Rep proteins, which bind sequence specifically to both ends and covalently to the 5′ end. The genome is then translocated through the packaging complex and into the capsid either (A) as a single-stranded molecule using the initial ‘scanning’ function before the first duplexed base pairs are encountered or (B) by unwinding a ds dimer or multimer genome on the capsid surface at the same time. (C) Simultaneous replication (arrow) of a ds monomer genome being packaged would result in premature strand displacement. Large Rep molecules at 5′ ends are covalently bound.
The polarity of DNA translocation by AAV Rep helicases fits with the observed directionality of the packaging process. However, this also implies that strand displacement synthesis at the capsid surface is not the driving force for DNA packaging, because if it were it would promote 5′→3′ packaging of the displaced strand. Thus, DNA replication and packaging are unlikely to be directly connected mechanistically. Indeed, as previously mentioned, it is possible that the replication process interferes with full-length encapsidation of monomer genomes. The involvement of the large Rep helicase activity in the packaging complexes could not be assessed directly via mutagenesis analyses, as it is also essential for replication of the genome termini. However, it is interesting to note that Rep78 and Rep68 catalyse the packaging reaction up to 1000-fold less efficiently, although they possess the entire Rep52 and Rep40 coding regions, respectively. It is conceivable that the strong, sequence-specific or covalent genome binding exhibited by the large Rep proteins may prevent them from moving along the genome in a processive manner. In addition, overexpression of the large Rep proteins could lead to the increased production of ds RF monomers, which may be more sensitive to interference by strand displacement synthesis during the packaging reaction. Nevertheless, large Rep proteins are included in the packaging model as they are very likely to be required for additional steps in the encapsidation process, such as targeting of genomes to the capsid, or for termination of packaging once a ds monomer genome has been unwound and inserted. In addition, all other members of the parvovirus family require only the large Rep protein functional equivalent (NS1 protein) to package their genomes. Further refinement of AAV-2 in vitro packaging systems (Ding et al., 1997; Zhou and Muzyczka, 1998) using packaging mutants (Wu et al., 2000) or supplementing empty capsid preparations should allow the working hypothesis to be tested further.
Materials and methods
AAV and Rep expression constructs and oligonucleotides
The pTAV2-0 construct (Heilbronn et al., 1990) consists of the entire AAV-2 genome from pAV-2 (Laughlin et al., 1983), including both inverted terminal repeats cloned into the BamHI site of pBluescript II. pTAV2-1 is based on pTAV2-0, but contains a mutated Rep52/40 translation start site (methionine to glycine at amino acid 225) to prevent expression of the small Rep proteins. A 502 bp SfiI–BamHI fragment was removed from the pKEX-Rep78 plasmid, which contained the ATG to GGG mutation, and inserted into pTAV2-0 to recreate the mutation previously described by Chejanovsky and Carter (1989). The CMV-Rep78, 68, 52, 40 and Rep52/40 (M225) expression constructs are based on the pKEX-XL plasmid and have been described previously (Hörer et al., 1995). The K340H mutation destroying the Rep ATP binding site has been previously described for the large Rep proteins (Hörer et al., 1995; Walker et al., 1997) and was also used in the context of Rep52 and Rep40. Rep/Cap constructs containing specific point mutations (E379K, E379Q, K391I, K391T, K404I, K404T) in the Rep ATPase domain (McCarty et al., 1992) were kindly supplied by N.Muzyczka. SalI–SwaI fragments were removed from these constructs and cloned into the corresponding sites of the CMV-Rep52/40 construct, allowing expression of both small Reps carrying the various ATPase mutations.
The following oligonucleotides were used to analyse the directionality of AAV-2 packaging: 3′ oligo (nucleotides 149–176), GGGGTGGAGTCGTGACGTGAATTACGTC; 3′+800 oligo (nucleotides 945–973), CCCAATTCTGATGCGCCGGTGATCAGATC; Middle oligo (nucleotides 2311–2335), CGGCATAAGGACGACAGCAGGGGTC; 5′–800 oligo (nucleotides 3681–3710), GCGGATAACAACAACAGTGAATACTCGTGGAC; 5′ oligo (4488–4518), CGCCCCATTGGCACCAGATACCTGACTCGT.
Transfection of 293T cells and preparation of virus supernatants
Cells were plated out at 4 × 105 in 4 ml of Dulbecco’s modified Eagle’s medium (DMEM) [10% fetal calf serum (FCS)] per 6 cm Petri dish 1 day prior to transfection. Transfections were carried out using a total of 6–7 µg of DNA per dish according to the method of Chen and Okayama (1988). Sixteen hours post-transfection, the medium was removed and replaced with 4 ml of adenovirus 5-containing medium [m.o.i. (multiplicity of infection) of 20] and incubated at 37°C for a further 2 days. Cells were lysed by three rounds of freeze–thawing (–80 and 37°C). Cell debris was then removed by centrifugation at 400 g for 10 min. A portion of the harvested cells was removed prior to cell lysis, washed once with phosphate-buffered saline and processed for western analysis. Monoclonal antibodies 303.9 and B1, specific for the Rep and VP proteins, respectively, were used as described previously (Wistuba et al., 1995).
Estimation of infectious viral titre
Virus supernatants were serially diluted (×10) and used to infect HeLa cells that had been plated out at 5000 cells/well in 100 µl of DMEM (10% FCS) in a 96-well plate the day before. Two to three hours post-infection, 100 µl of DMEM (0% FCS) containing adenovirus 5 (m.o.i. = 10) were added per well. Three days later, cells were lysed by three rounds of freeze–thawing (–80 and 37°C). DNA was denatured upon addition of 0.4 vols of 1.5 M NaOH and transferred to a Genescreen Plus Nylon membrane (NEN, Cologne, Germany) using a vacuum blotter (Life Technologies, Karlsruhe, Germany). The membrane was then processed further as a Southern blot and hybridized using a random-primed 32P-labelled rep-specific probe.
Preparation of capsids and viral DNA
Virus supernatants were first incubated with DNase I (500 µg/ml), in the presence of 5 mM MgCl2, for 2 h at 37°C to digest non-packaged DNAs. The DNase was stopped by the addition of 16 mM EDTA and 1.5% SDS. Capsid proteins were then digested using proteinase K (320 µg/ml) at 37°C for 3 h and removed via phenol:chloroform:isoamylalcohol (24:24:1) extraction. The DNA was then ethanol precipitated, washed and dried under vacuum. DNA pellets were dissolved slowly in dH2O at 4°C and then for 10 min at room temperature with shaking. These samples were either run on a neutral TAE gel or mixed with 10 µl of 100 mM NaOH, 2 mM EDTA, 60 min prior to loading on an alkaline agarose gel.
Purification of capsid-associated DNA as well packaged DNA was carried out in a similar fashion following immunoprecipitation of capsids from the freeze–thaw lysate. In order to prevent degradation of ss genomes in viral supernatants that were capsid associated, EDTA was supplemented to 30 mM at the freeze–thaw lysis stage. For the immunoprecipitation of capsids, protein A–Sepharose (2 mg per sample; Amersham Pharmacia, Freiburg, Germany) was allowed to swell in NET-N buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40 pH 7.5) for 15 min at room temperature. This was then washed once with NET-N buffer and added to 600 µl of A20 hybridoma (Wistuba et al., 1997) medium in the presence of 1% bovine serum albumin, 100 µg/ml herring sperm DNA, and incubated with gentle inversion for 30 min at 4°C. Following this incubation, the protein A–Sepharose-A20 was washed twice with NET-N to remove unbound antibody. Aliquots in NET-N were then added to 200–400 µl of DNase I-treated (or not) freeze–thaw supernatants, which were then incubated for 2 h at 4°C with gentle inversion mixing. The protein A–Sepharose-A20–capsid complexes were then washed twice with 900 µl of NET-N and resuspended in proteinase K digestion buffer (10 mM Tris, 1 mM EDTA, 0.5% SDS pH 7.5). Proteinase K was added to 320 µg/ml and the mixture incubated at 37°C for 3 h. Every 30 min, the Sepharose-containing mixture was briefly vortexed. Subsequent steps were identical to those used for the preparation of genomes directly from the crude virus supernatant. The efficiency of capsid immunoprecipitation was tested by performing a western blot analysis on portions of the NET-N-washed protein A– Sepharose-A20, removed prior to proteinase K digestion.
Agarose gel electrophoresis
Viral DNAs were separated using either standard non-denaturing agarose gels (TAE buffered) or alkaline agarose gels as described by Sambrook et al. (1989).
Southern blotting
DNAs were transferred to a Genescreen Plus Nylon membrane in 0.4 M NaOH overnight. The membrane was soaked in denaturation solution (0.5 M NaOH, 1.5 M NaCl) for 5 min and then in neutralizing buffer (0.5 M Tris, 0.3 M sodium citrate, 3 M NaCl pH 7), again for 5 min. The dried membrane was then UV cross-linked and incubated in hybridization buffer (7% SDS, 0.25 sodium phosphate buffer, 0.25 M NaCl, 1 mM EDTA, 45% formamide pH 7.2) for at least 60 min prior to probe hybridization. 32P-labelled, random-primed probes were generated using the Random Primed Labelling Kit (Roche Diagnostics, Mannheim, Germany) and purified over a Sephadex G50 column. The rep-specific probe was made using the 1.7 kb SacII rep gene fragment as template. Oligonucleotides were end-labelled using polynucleotide kinase and [γ-32P]ATP, and purified over a Sephadex G25 column. Labelled, random-primed probes were added to 5 ml of hybridization buffer and incubated at 42°C overnight. Membranes were then washed first with 2× SSC, 0.1% SDS at 42°C for 10 min and then twice with 0.2× SSC, 0.1% SDS at 68°C, for 20 min. Labelled oligonucleotide probes were hybridized at 60°C overnight, and then washed as above, but at 60°C instead of 42 and 68°C. Autoradiography was then carried out using Kodak Biomax MS X-ray films.
Quantitation of AAV-2 capsids
A20 capsid ELISAs were carried out according to Grimm et al. (1999) using pre-coated plates supplied by Progen GmbH, Heidelberg, Germany.
Quantitation of packaged DNA
The amount of DNA packaged was determined by comparing serial dilutions of the viral DNA spotted on to a Genescreen nylon membrane with serial dilutions of a 4.7 kb AAV sequence of known concentration. DNAs were diluted in 50 mM NaOH and spotted onto the membrane using a vacuum blotter. Membranes were treated with denaturation and neutralization solutions as described for Southern blotting, and probed using a random-primed rep-specific 32P-labelled probe (1.7 kb SacII fragment). Quantitation was carried out using a phosphorimaging device (Molecular Dynamics, Krefeld, Germany) together with ImageQuant software.
Rep/VP immunoprecipitations
Analysis of Rep–capsid complex formation by co-immunoprecipitation was performed as described by Dubielzig et al. (1999).
Acknowledgments
Acknowledgements
We thank Nicholas Muzyczka for the AAV constructs containing mutations in the rep ATPase region, Kristin Schmidt for excellent technical assistance and Michael Pawlita for his critical reading of the manuscript. This work was supported by grant No. 01KV9805/4 from the Bundesministerium für Forschung und Technik.
References
- Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- Berns K.I. (1990) Parvovirus replication. Microbiol. Rev., 54, 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina M., Ng,S.C. and Blasquez,V. (1983) Simian virus 40 chromatin interaction with the capsid proteins. J. Biomol. Struct. Dyn., 1, 689–704. [DOI] [PubMed] [Google Scholar]
- Bird L.E., Subramanya,H.S. and Wigley,D.B. (1998) Helicases: a unifying structural theme? Curr. Opin. Struct. Biol., 8, 14–18. [DOI] [PubMed] [Google Scholar]
- Black L.W. (1989) DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol., 43, 267–292. [DOI] [PubMed] [Google Scholar]
- Blasquez V., Beecher,S. and Bina,M. (1983) Simian virus 40 morphogenetic pathway. An analysis of assembly-defective tsB201 DNA protein complexes. J. Biol. Chem., 258, 8477–8484. [PubMed] [Google Scholar]
- Booy F.P., Newcomb,W.W., Trus,B.L., Brown,J.C., Baker,T.S. and Steven,A.C. (1991) Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell, 64, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M.K., Smith,T.F., Lathrop,R.H., Livingston,D.M. and Webster,T.A. (1987) Consensus topography in the ATP binding site of the simian virus 40 and polyomavirus large tumor antigens. Proc. Natl Acad. Sci. USA, 84, 4026–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister J.R. and Muzyczka,N. (1999) Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol., 73, 9325–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister J.R. and Muzyczka,N. (2000) Mechanism of Rep-mediated adeno-associated virus origin nicking. J. Virol., 74, 7762–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano C.E. (2000) The terminase enzyme from bacteriophage λ: a DNA-packaging machine. Cell. Mol. Life Sci., 57, 128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chejanovsky N. and Carter,B.J. (1989) Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology, 173, 120–128. [DOI] [PubMed] [Google Scholar]
- Chejanovsky N. and Carter,B.J. (1990) Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J. Virol., 64, 1764–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.A. and Okayama,H. (1988) Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques, 6, 632–638. [PubMed] [Google Scholar]
- Deiss L.P. and Frenkel,N. (1986) Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol., 57, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Lu,S. and Munshi,N.C. (1997) In vitro packaging of an infectious recombinant adeno-associated virus 2. Gene Ther., 4, 1167–1172. [DOI] [PubMed] [Google Scholar]
- Dubielzig R., King,J.A., Weger,S., Kern,A. and Kleinschmidt,J.A. (1999) Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J. Virol., 73, 8989–8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H. and Morita,M. (1997) Phage DNA packaging. Genes Cells, 2, 537–545. [DOI] [PubMed] [Google Scholar]
- Gibson W. and Roizman,B. (1972) Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol., 10, 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.P., Webb,M.R., Brune,M. and Johnson,K.A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature, 373, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Kern,A., Pawlita,M., Ferrari,F., Samulski,R. and Kleinschmidt,J.A. (1999) Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther., 6, 1322–1330. [DOI] [PubMed] [Google Scholar]
- Guo P. (1994) Principles, perspectives and potential applications in viral assembly. Semin. Virol., 5, 1–3. [Google Scholar]
- Guo P. and Trottier,M. (1994) Biological and biochemical properties of the small viral RNA (pRNA) essential for the packaging of the dsDNA of phage F29. Semin. Virol., 5, 27–37. [Google Scholar]
- Guo P., Zhang,C., Chen,C., Garver,K. and Trottier,M. (1998) Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. [DOI] [PubMed] [Google Scholar]
- Heilbronn R., Burkle,A., Stephan,S. and zur Hausen,H. (1990) The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol., 64, 3012–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R.W. (1998) Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell, 94, 147–150. [DOI] [PubMed] [Google Scholar]
- Hölscher C., Kleinschmidt,J.A. and Burkle,A. (1995) High-level expression of adeno-associated virus (AAV) Rep78 or Rep68 protein is sufficient for infectious-particle formation by a rep-negative AAV mutant. J. Virol., 69, 6880–6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörer M., Weger,S., Butz,K., Hoppe,S.F., Geisen,C. and Kleinschmidt,J.A. (1995) Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J. Virol., 69, 5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im D.S. and Muzyczka,N. (1990) The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell, 61, 447–457. [DOI] [PubMed] [Google Scholar]
- Im D.S. and Muzyczka,N. (1992) Partial purification of adeno-associated virus Rep78, Rep52 and Rep40 and their biochemical characterization. J. Virol., 66, 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal H.K., Yong,C.B., Wilson,G.M., Tam,P. and Astell,C.R. (1994) Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem., 269, 3283–3289. [PubMed] [Google Scholar]
- Laughlin C.A., Tratschin,J.D., Coon,H. and Carter,B.J. (1983) Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene, 23, 65–73. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Irmiere,A. and Gibson,W. (1988) Primate cytomegalovirus assembly: evidence that DNA packaging occurs subsequent to B capsid assembly. Virology, 167, 87–96. [DOI] [PubMed] [Google Scholar]
- Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- Lohman T.M., Thorn,K. and Vale,R.D. (1998) Staying on track: common features of DNA helicases and microtubule motors. Cell, 93, 9–12. [DOI] [PubMed] [Google Scholar]
- McCarty D.M., Ni,T.H. and Muzyczka,N. (1992) Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J. Virol., 66, 4050–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D.S. and Courtney,R.J. (1992) Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol., 66, 7581–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.J. and Lohman,T.M. (1995) Helicase-catalyzed DNA unwinding: energy coupling by DNA motor proteins. Biophys. J., 68, 180S–185S. [PMC free article] [PubMed] [Google Scholar]
- Myers M.W. and Carter,B.J. (1980) Assembly of adeno-associated virus. Virology, 102, 71–82. [DOI] [PubMed] [Google Scholar]
- Newcomb W.W. and Brown,J.C. (1991) Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol., 65, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb W.W., Homa,F.L., Thomsen,D.R., Booy,F.P., Trus,B.L., Steven,A.C., Spencer,J.V. and Brown,J.C. (1996) Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol., 263, 432–446. [DOI] [PubMed] [Google Scholar]
- Ni T.H., Zhou,X., McCarty,D.M., Zolotukhin,I. and Muzyczka,N. (1994) In vitro replication of adeno-associated virus DNA. J. Virol., 68, 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim A., Sandalon,Z., Peleg,A., Shaul,O., Nicolis,S. and Ottolenghi,S. (1992) A cis-acting DNA signal for encapsidation of simian virus 40. J. Virol., 66, 5320–5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson L. (1984) Structure and assembly of adenoviruses. Curr. Top. Microbiol. Immunol., 109, 1–52. [DOI] [PubMed] [Google Scholar]
- Prasad K.M. and Trempe,J.P. (1995) The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology, 214, 360–370. [DOI] [PubMed] [Google Scholar]
- Ruffing M., Zentgraf,H. and Kleinschmidt,J.A. (1992) Assembly of virus-like particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J. Virol., 66, 6922–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmid S.I. and Hearing,P. (1995) Selective encapsidation of adenovirus DNA. Curr. Top. Microbiol. Immunol., 199, 67–80. [DOI] [PubMed] [Google Scholar]
- Smith R.H. and Kotin,R.M. (1998) The rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J. Virol., 72, 4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus B.L., Booy,F.P., Newcomb,W.W., Brown,J.C., Homa,F.L., Thomsen,D.R. and Steven,A.C. (1996) The herpes simplex virus procapsid: structure, conformational changes upon maturation and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol., 263, 447–462. [DOI] [PubMed] [Google Scholar]
- Varmuza S.L. and Smiley,J.R. (1985) Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell, 41, 793–802. [DOI] [PubMed] [Google Scholar]
- Walker S.L., Wonderling,R.S. and Owens,R.A. (1997) Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol., 71, 6996–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. and Linden,R.M. (2000) A role for single-stranded templates in cell-free adeno-associated virus DNA replication. J. Virol., 74, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. (1996) DNA helicases: new breeds of translocating motors and molecular pumps. Cell, 86, 177–180. [DOI] [PubMed] [Google Scholar]
- Willwand K. and Hirt,B. (1993) The major capsid protein VP2 of minute virus of mice (MVM) can form particles which bind to the 3′-terminal hairpin of MVM replicative-form DNA and package single-stranded viral progeny DNA. J. Virol., 67, 5660–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba A., Weger,S., Kern,A. and Kleinschmidt,J.A. (1995) Inter mediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol., 69, 5311–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistuba A., Kern,A., Weger,S., Grimm,D. and Kleinschmidt,J.A. (1997) Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol., 71, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Xiao,W., Conlon,T., Hughes,J., Agbandje-McKenna,M., Ferkol,T., Flotte,T. and Muzyczka,N. (2000) Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol., 74, 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D. and Weller,S.K. (1998a) Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol., 72, 7428–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D. and Weller,S.K. (1998b) Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology, 243, 32–44. [DOI] [PubMed] [Google Scholar]
- Zhou X. and Muzyczka,N. (1998) In vitro packaging of adeno-associated virus DNA. J. Virol., 72, 3241–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]