Abstract
The evolutionary history of serine proteases can be accounted for by highly conserved amino acids that form crucial structural and chemical elements of the catalytic apparatus. These residues display non- random dichotomies in either amino acid choice or serine codon usage and serve as discrete markers for tracking changes in the active site environment and supporting structures. These markers categorize serine proteases of the chymotrypsin-like, subtilisin-like and α/β-hydrolase fold clans according to phylogenetic lineages, and indicate the relative ages and order of appearance of those lineages. A common theme among these three unrelated clans of serine proteases is the development or maintenance of a catalytic tetrad, the fourth member of which is a Ser or Cys whose side chain helps stabilize other residues of the standard catalytic triad. A genetic mechanism for mutation of conserved markers, domain duplication followed by gene splitting, is suggested by analysis of evolutionary markers from newly sequenced genes with multiple protease domains.
Keywords: active site/evolution/hydrolase/serine protease/subtilisin
Introduction
Serine proteases carry out a diverse array of physiological and cellular functions, ranging from digestive and degradative processes to blood clotting, cellular and humoral immunity, fibrinolysis, fertilization, embryonic development, protein processing and tissue remodeling. Serine proteases have been classified into evolutionarily unrelated clans, which have been subdivided into families of proteases whose homology can be established statistically (Rawlings and Barrett, 1993; Barrett and Rawlings, 1995). Clans differ in terms of overall fold and the order of catalytic residues in the primary sequence. Despite these significant differences, serine proteases of clans SA (chymotrypsin-like) (Lesk and Fordham, 1996), SB (subtilisin-like) (Siezen and Leunissen, 1997) and SC (α/β-hydrolase fold) (Ollis et al., 1992) maintain a strictly conserved active site geometry among their catalytic Ser, His and Asp residues. This shared catalytic structure suggests that common architectural motifs are likely to be found in the molecular designs of active sites utilizing a Ser–His–Asp triad. Major steps in the assembly, improvement and specialization of the catalytic architecture should correspond to significant evolutionary transitions in the history of protease clans. Evolutionary markers encountered in the sequences contributing to the catalytic apparatus would thus give an account of the history of an enzyme family or clan and provide for comparative analysis with other families and clans. Therefore, the use of sequence markers associated with active site structure generates a model for protease evolution with broad applicability and potential for extension to other classes of enzymes.
The first report of a sequence marker associated with active site chemistry was the observation that both AGY and TCN codons were used to encode active site serines in a variety of enzyme families (Brenner, 1988). Since AGY→TCN interconversion is an uncommon event, it was reasoned that enzymes within the same family utilizing different active site codons belonged to different lineages. This phenomenon was shown to apply to both the enzymes of clans SA and SC. Enzymes of clan SB have been reported to use exclusively TCN for the active site serine (Rawlings, 1998a). Despite the discovery of two exceptions to that rule in this study, the paucity of AGY active site codons in clan SB suggests that codon usage by that serine provides very limited evolutionary information. Investigation of additional markers associated with catalytic function would generate a more complete story. An example of a second marker associated with catalytic function in clan SA serine proteases is residue 225 (chymotrypsinogen numbering). Residue 225 has a Pro– Tyr dichotomy; proteases with Tyr225 are Na+-activated allosteric enzymes, displaying a regulation of catalytic activity not seen in enzymes with Pro225 (Dang and Di Cera, 1996).
Several other highly conserved residues in serine protease families have been linked to catalysis, although they have not been explored as evolutionary markers. The backbone of Ser214 in chymotrypsin-like proteases contributes to the S1 binding pocket (Perona and Craik, 1995). The side chain of the same residue helps generate a polar environment for the catalytic Asp102 (McGrath et al., 1992), and both oxygens of Ser214 form hydrogen bonds with waters located in the active site cleft (Pletnev et al., 2000). Ser125 (subtilisin BPN′ numbering) in subtilisin-type proteases plays a similar role; its backbone contributes to the S1 binding pocket (Perona and Craik, 1995), whereas its side chain is directly adjacent to the catalytic residues (Siezen and Leunissen, 1997) so that the hydroxyl is within hydrogen-bonding distance of the catalytic Asp32. Several carboxypeptidases have a residue that functions similarly in the active site: the backbone nitrogen of unpaired Cys341 (yeast carboxypeptidase W numbering) hydrogen bonds to the side chain of catalytic Asp338 (Rudenko et al., 1995), while the side chain of Cys341 is proximal to the side chain of catalytic His397 (Shilton et al., 1997). Each of these residues (Ser214, Ser125 and Cys341) appears to be the fourth member of a catalytic tetrad in its respective protease clan.
We examined the above residues and found that they utilized dichotomous sequence choices. We also analyzed the sequences of proteases from clans SA, SB and SC to identify other conserved residues potentially related to active site structure or function. Absolutely conserved non-serine residues were avoided, as they were likely to yield little evolutionary information. We identified two residues (Ser190 and Ser207) in clan SB and one residue (Ser57) in clan SC that fit the conservation and dichotomy criteria for discrete evolutionary markers. Our analysis proceeded in two steps: division of the clans into families or subfamilies based on primary structure motifs surrounding the active site residues and non-active site evolutionary markers, and categorization of each family according to single-residue evolutionary markers. The results reveal the governing role of the active site in protease evolution and indicate that domain duplication followed by gene splitting may be responsible for the generation of new evolutionary lineages.
Results
Markers divide clans into families
Protease clans SA, SB and SC have been subdivided into different families (referred to as ‘subfamilies’ within the subtilisin ‘superfamily’) whose homology has been established by sequence (Rawlings and Barrett, 1993; Barrett and Rawlings, 1995). Such distinctions are based on the sequence of complete protease domains. The present study finds that the above distinctions can be recapitulated using short primary sequence elements surrounding the active site residues and other highly conserved residues that have been identified as evolutionary markers by the present study (Table I). This result indicates that alterations in active site structure accompanied the divergence of the various families within clans SB and SC. Thus, significant functional differences map to changes in narrowly defined regions of the molecule, especially within the active site.
Table I. Sequence motifs surrounding evolutionary markers and active site residues for major families/subfamilies of clans SA, SB and SC.
| SA family |
His57* |
Asp102* |
Ser195* |
Ser214 |
Pro/Tyr225 |
|
| S1 |
T A A H C |
D I A L |
G D S G G P |
G I V S W |
P/Y G V Y/F |
|
| SB subfamily |
Asp32* |
His64* |
Ser125 |
Ser190 |
Ser207 |
Ser221* |
| S8A | D T G I | G H G T H | N M S L G G | F S N Y | I L S T | G T S M A T |
| S8B | D D G I | K/R H G T R | S A S W G P | Y S E X | I X T T | G T S A S A |
| S8C |
D T G I |
G H G T H |
N/S X S L G X |
F/W S S R |
I L S X |
G T S M A T/S |
| SC family |
Ser57 |
Ser146* |
Asp338* and Val/Cys341 |
His397* |
|
|
| S9 | G G P G – S X | G W S Y G G | G X X D X N V/C | G A G H | ||
| S10 | G G P G C S S | G E S Y A G | G D X D X X V/C | D E X H |
Conserved and active site residues are in bold. Active site residues are marked with an asterisk (*). ‘X’ indicates significant variation for a particular residue.
Markers divide families into lineages
Within families, sequence changes occurring at active site and marker locations are subtler. These sequence changes can be traced through residues that feature usage of two dissimilar amino acids, such as Pro/Tyr225 in clan SA and Val/Cys341 in clan SC. Similar alterations can be observed in the silent but non-conservative difference between usage of TCN or AGY codons for highly conserved serines, such as Ser195 and Ser214 in clan SA; Ser125, Ser190 and Ser207 in clan SB; and Ser57 and Ser146 in clan SC. The above sequence differences represent significant mutations, and chronicle changes in and around the active site. Such changes, all of which require double-nucleotide shifts, would be extremely rare. Accordingly, they represent the best candidates for serving as discrete evolutionary markers that subdivide protease families into distinct lineages. The protease domains analyzed in this study are categorized according to evolutionary markers in Table II.
Table II. Categories of serine proteases according to evolutionary markers.
| A |
|
|
| Ser214:TCN |
Ser214:AGY |
|
| Ser195:TCN Pro225 | Chymotrypsin (vertebrates, invertebrates); chymotrypsin-like (human); easter (fly); enterokinase (mammals); granzymes (mammals)a; kallikrein—tissue and glandular (mammals); trypsin (vertebrates, invertebrates) | elastase (mammals); factor Be (mammals); factor C2 (mammals)e; factor XI (human); factor XII (mammals); HGF (mammals)f; HGFA (human); kallikrein—plasma (mammals); Sp14D1 (mosquito); t-PA (mammals) |
| Ser195:TCN Tyr225 | MASP-1 (mammals); gastrulation defective (fly) | factor B (sea urchin); haptoglobin (mammals)g; nudel (fly) |
| Ser195:AGY Pro225 | apolipoprotein(a) (mammals)b; CG-18735 (fly); masqueradec (fly); neurotrypsin (mammals); plasmin (mammals) | acrosin (mammals); hepsin (mammals) |
| Ser195:AGY Tyr225 |
CASP (hamster); cercarial protease (schistosome)d; factor C1r (human); factor C1s (human); MASP-2 (human); thrombin (vertebrates) |
factor VII (mammals)h; factor IX (mammals); factor X (mammals, birds); protein C (mammals) |
| B |
|
|
| Ser214:TCN |
Ser214:AGY |
|
| Ser195:TCN Pro225 | degradative, development, cell-mediated immunity, kallikrein | cell-mediated immunity, clotting, degradative, fibrinolytic/HGFA, HGF, humoral immunity |
| Ser195:TCN Tyr225 | humoral immunity, development | development, humoral immunity |
| Ser195:AGY Pro225 | degradative, development, fibrinolytic | degradative |
| Ser195:AGY Tyr225 |
clotting, humoral immunity |
clotting |
| C |
|
|
| Ser190:TCN |
Ser190:AGY |
|
| Ser125:TCN Ser207:TCN | S8A: intracellular protease I (Bacillus subtilis); proteinase K (Tritirachium album); sexual differentiation serine protease (Schizosaccharomyces pombe); subtilisin 1 (B.subtilis); subtilisin-like protease III (Saccharomyces cerevisiae) | S8A: alkaline protease (Acremonium chrysogenum); alkaline protease (Trichoderma harzianum)l; serine protease (Paenibacillus polymyxa) |
| S8C: cucumisin (melon)i; cucumisin-like AF036960 (soybean)i; TagB,C (Dictyostelium discoides); TMP (tomato)i | S8C: cucumisin-like ARA12 (A.thaliana)i; cucumisin-like sbt1 (tomato)i | |
| Ser125:TCN Ser207:AGY | S8A: minor extracellular serine protease vpr (B.subtilis) S8B: blisterase 4 (C.elegans)j,k; celfur (C.elegans)k; kex2-like endoprotease 1 (fly); kexin 2 (Candida albicans)l | S8B: amon (fly)k,p; kexin 1 (S.pombe)k; kexin 2 (h.crab)k S8C: TPP-II-like F21H12.3 (C.elegans)q,r |
| Ser125:AGY Ser207:TCN | S8A: aqualysin I (Thermus aquaticus); C5a peptidase (Streptococcus pyogenes); subtilisin-like (Plasmodium berghei) | S8C: TPP-II-like SPAP8A3.12c (S.pombe)r,s; TPP-II (fly)l |
| S8C: SKI-1 (mammals); TPP-II (mammals); TPP-II-like T13K14.10 (Arabidopsis thaliana)m | ||
| Ser125:AGY Ser207:AGY | S8A: alkaline protease (B.amyloliquifaciens); cell wall protease (B.subtilis); elastase (Aspergillus flavus); epidermin (Staphylococcus epidermidis); subtilisin carlsberg (Bacillus licheniformis); subtilisin E (B.subtilis); | S8A: nisin operon protease (Lactococcus lactis)k |
| S8B: furin (Aplysia californica)k,n; PACE4 (mammals)j,k; PC1 (mammals)o; PC5 (mammals, lancelet)j,k | S8B: calcium-dependent protease (A.variabilis)k; PC2 (mammals)k,p; PC3 (mammals, frog)k; PC3-like (hydra)k,l,t | |
| |
|
S8C: CG-7169 (fly) |
| D |
|
|
| Ser146:TCN |
Ser146:AGY |
|
| Ser57:TCN Val341 | S9: APEH-like (Thermoplasma acidophilum)u; APEH (mammals); APEH-like (A.thaliana); DPP-B (S.cerevisiae); ome (fly) | S9: APEH-like (C.elegans)u; DPP (S.pombe); oligopeptidase B (Escherichia coli)u |
| S10: CG-4572 (fly); carboxypeptidase I (A.thaliana, barley, rice, tomato); glucose acyltransferase (potatov, tomato); vitellogenic carboxypeptidase (human) | S10: BG: DS00365.3(a) (fly); carboxypeptidase D (barley, ricev); carboxypeptidase D-like AP002539 (rice); carboxypeptidase II (rice)v | |
| Ser57:TCN Cys341 | S10: carboxypeptidase A (C.elegans); carboxypeptidase Y (C.albicans); vitellogenic carboxypeptidase (mosquito) | S9: APEH-like orf-c01017 (Sulfolobus solfataricus)ab; APEH-like APE1547 (A.pernix)ab |
| S10: BG:DS00365.3(b) (fly)u; CG-3344 (fly); carboxypeptidase D (S.cerevisiae) | ||
| Ser57:AGY Val341 | S9: allergen (Trichophyton rubrum)w,x; APEH-like APE2441 (Aeropyrum pernix); DPP-IV (vertebrates); FAP (mammals); prolyl oligopeptidase (mammals)y | S9: alanyl DPP (Aspergillus oryzaex, Xylella fastidiosav,w); DPP-VI (mammals)v,ac |
| S10: virulence protein NF314 (Naegleria fowleri) | ||
| Ser57:AGY Cys341 | S9: APEH-like PH0863 (Pyrococcus horikoshii)z; APEH-like SC66t3.21c (Streptomyces coelicolor)aa | S9: APEH-like PAB1418 (Pyrococcus abyssi)w; APEH-like YuxL (B.subtilis)ad; APEH-like DR0165 (Deinococcus radiodurans)z |
| S10: carboxypeptidase Y (A.thaliana); carboxypeptidase-like (Matricaria chamomilla) | S10: carboxypeptidase A (mammals) |
A, clan SA category assignments. B, functional assignments of clan SA marker lineages. C, clan SB category assignments. D, clan SC category assignments. Gene accession numbers are provided for newly sequenced genes of undetermined function. Letters in parentheses following gene names indicate individual protease domains from multi-protease genes; the alphabetical order indicates the 5′→3′ sequence of the domains within the gene. Annotations are provided for those enzymes whose category title does not precisely match their amino acid or codon usage; such exceptions are placed in the categories that most closely correspond to the enzymes’ amino acid or codon usage.
aMouse granzyme F, Thr214; rat granzyme II, Ser214:AGT; brhesus monkey, Asn195; cGly195; dIle225; einsertion after residue 224; fTyr195, Val214; gAla195; hmouse, Ile225; rabbit, Val225; iAla207; jLeu190; kThr207; lThr190; mPro207; nThr125; oAla190; pAsp190; qGly207; rSer221:AGT; sCys190; tCys125; uAla57; vIle341; wGly57; xLeu341; yAsn57; zThr57; aaTrp57; abThr341; acAsp146; adMet57.
APEH, acyl-peptide hydrolase; CASP, calcium-activated serine protease; DPP, dipeptidyl peptidase; FAP, fibroblast activation protein; HGF, hepatocyte growth factor; HGFA, hepatocyte growth factor activator; MASP, mannose-associated lectin-binding serine protease; PACE, paired basic amino acid cleaving enzyme; PC, prohormone convertase; SKI, subtilisin/kexin isozyme; Sp, serine protease; Tag, tight aggregate stage; TMP, tomato meiotic proteinase; t-PA, tissue plasminogen activator; TPP, tripeptidyl peptidase. CG- and BG- prefixes refer to Drosophila genes sequenced by Celera Genomics and the Berkeley Drosophila Genome Project, respectively. This table provides a limited sample of sequences from each protease clan. A complete listing of the sequences analyzed and categorized is available as Supplementary data (at The EMBO Journal Online).
As each peptidase clan has been assigned three binary evolutionary markers, each clan is subdivided into eight groups. The segregation is not random. Enzymes that have high sequence similarity tend to group into the same lineage. Different copies of enzymes from the same organism or closely related organisms are almost always categorized within the same lineage. For example, the enzymes considered to be part of the plasminogen activator/hepatocyte growth factor activator subfamily of serine proteases (Miyazawa et al., 1998) are all part of the Ser195:TCN/Ser214:AGY/Pro225 lineage in clan SA. The mammalian enzymes of the prohormone and proprotein convertase subfamily (Seidah et al., 1994) are found exclusively within two lineages, Ser125:AGY/Ser190: TCN/Ser207:AGY and Ser125:AGY/Ser190:AGY/Ser207: AGY, of clan SB. Occasionally, proteases with low sequence similarity are found in the same group, as with the schistosomal cercarial protease and thrombin in lineage Ser195:AGY/Ser214:TCN/Tyr225. The low mutation rate of the binary markers suggests common ancestry for proteases such as the cercarial protease and thrombin, sequences that might otherwise be considered unrelated.
Clan SA is unique among the three clans studied in that nearly all of its enzymes are extracellular. As a result, proteases of that clan have been shown to participate in a wide variety of physiological functions. Functions map roughly to the categories, with individual functions mapping to between one and four categories (Table IIB). In cases where particular functions map to multiple categories, these categories differ by one or two binary markers in the majority of cases. Categorizations that place enzymes associated with a particular function in the same category are in agreement with phylogenetic information based on extensive or complete sequence comparisons indicating a common lineage. Categorizations that place enzymes associated with a particular function (such as clotting) in significantly different categories are suggestive of different lineages giving rise to the enzymes of that function (e.g. factors X and XI in the clotting system) and also fit previous phylogenetic analyses (Krem et al., 1999). Enzymes of the complement and vitamin K-dependent clotting systems map to the same categories, but they do not map to the same categories as enzymes of fibrinolysis, cell-mediated immunity and tissue degradation (which includes digestion). This result concurs with previous studies indicating different lineages for the clotting and fibrinolytic cascades (Patthy, 1985, 1990). Thus, the separation of chymotrypsin-like proteases into discrete lineages agrees with previous partitioning of serine proteases and duplicates recognizable trends, doing so with minimal information. All three markers contribute information, as elimination of any one of the markers would place sequences that have been previously documented as having limited similarity in the same categories and impair the already limited functional resolution of the method. For example, elimination of the Pro/Tyr225 marker would merge the predominantly degradative category of Ser195:AGY/Ser214:TCN/Pro225 with the predominantly complement category of Ser195:AGY/Ser214: TCN/Tyr225. While the use of binary markers appears less suited to accurate functional prediction than sequence comparison and dendrogram construction, changes in the active site environment appear to have contributed significantly to functional radiation within clan SA.
In clan SB, the clearest example of the segregative power of a binary marker is the exclusive use of categories with Ser207:AGY (including Thr207:ACN) by subfamily S8B; no enzymes in the kexin subfamily (S8B) utilize TCN at residue 207. Thus, a codon switch at residue 207 occurred with the emergence of the kexin subfamily. The kexin subfamily appears to be further subdivided by codon usage at Ser125. With the exception of the calcium-dependent protease of Anabaena variabilis, Ser125:AGY is found exclusively in metazoans. Some metazoans, yeast and fungi use Ser125:TCN. Mammalian sequences from subfamily S8B—and also subfamily S8C—only use Ser125:AGY. In subfamily S8A, the most highly conserved codon choice is found at residue Ser190. Thirty-nine of 45 sequences surveyed utilized TCN. Five of the six sequences utilizing Ser190:AGY mapped to the same lineage. As with clan SA, all three markers chosen for clan SB contain evolutionary information. The pattern of category occupation in clan SB further suggests that the various lineages of each subfamily of the subtilisin clan diverged subsequently to the division of clan SB into subfamilies.
In clan SC, families S9 and S10 differ in category occupation, although not as dramatically as the subfamilies of clan SB. Nevertheless, it appears that the individual lineages within families radiated after the evolution of individual families within clan SC. Within family S10, plant and animal/fungal carboxypeptidases tend to occupy different lineages. Plant sequences dominate categories Ser57:TCN/Ser146:AGY/Val341 and Ser57:AGY/Ser146: TCN/Cys341; animal and fungal carboxypeptidases dom inate categories Ser57:TCN/Ser146:TCN/Cys341, Ser57: TCN/Ser146:AGY/Cys341 and Ser57:AGY/Ser146:AGY/Cys341. Within family S9, the major species divide resides within the Val/Cys341 marker. No eukaryotic sequences have Cys341; that distinction belongs to acyl peptide hydrolase-like enzymes from a variety of thermophilic bacteria. The resulting paradox is that in family S9 the category Ser57:AGY/Ser146:AGY/Cys341 is occupied solely by thermophilic bacteria, while in family S10 the very same category is occupied solely by mammalian lysosomal carboxypeptidases. The utilization of Cys341 in family S9 appears to be a development unique to thermophilic bacteria.
Construction of phylogenies based on markers
Discrete evolutionary markers lend themselves to the construction of phylogenies. Phylogenies in this study were based on the presumption that TCN, Pro and Val are primordial compared with AGY, Tyr and Cys at the residues serving as markers. Evidence for the primordiality of TCN comes from previous sequence analyses comparing conserved and non-conserved serine residues. Diaz-Lazcoz et al. (1995) examined several classes of enzymes employing serine nucleophiles, including serine proteases, and demonstrated that catalytic serine residues utilized TCN codons to a greater degree than non-conserved serine residues. For serine proteases, the greater preference for TCN at active site residues was found to be statistically significant, with P(χ2) = 0.04. Because highly conserved residues are more likely than non-conserved residues to retain their original codon usage, it was concluded that TCN codons were the primordial serine codons. Comparative analysis of mammalian and fruit fly chymotrypsin-like sequences at residue 225 also agrees with functional evidence (Guinto et al., 1999) of the primordiality of Pro at that position. The fruit fly has Pro225 91% of the time compared with 82% for mammals, with P(χ2) <0.05. The case for the primordiality of Val as opposed to Cys at residue 341 of clan SC is implied by the preponderance of Val in families S9 [76% Val, n = 45, P(χ2) <0.01] and S10 [67% Val, n = 64, P(χ2) <0.01]. Perhaps the most convincing argument for the modernity of AGY, Tyr and Cys at the positions described above is that the enzymes utilizing those choices tend to be from higher metazoans and perform functions associated with a high degree of physiological complexity, such as blood clotting and prohormone processing.
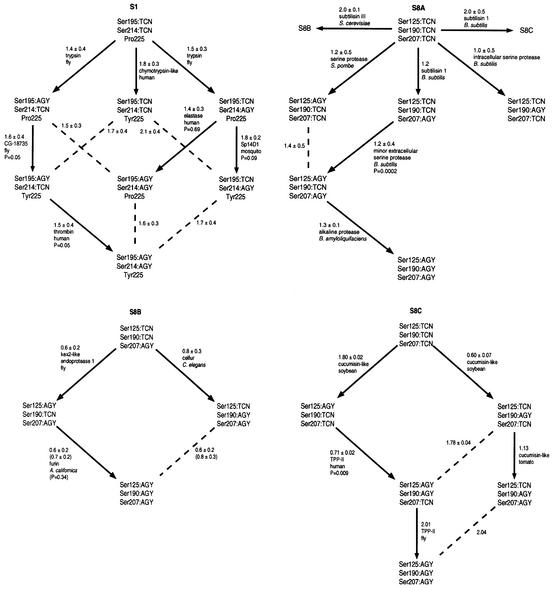
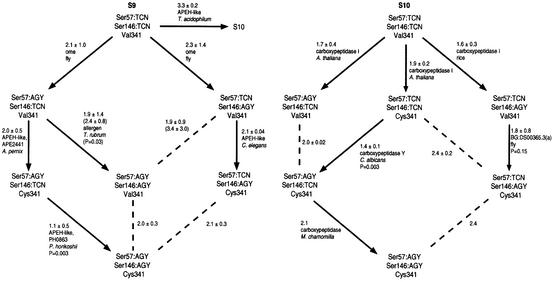
Figure 1 depicts phylogenies that are rooted based upon the primordiality of TCN, Pro and Val; despite this fact, the topology is invariant. Transitions involving two or three markers at one time were presumed to be unlikely. To identify the more likely single-marker transitions, we searched for individual enzymes that have close sequence relationships to enzymes in more modern categories. The protein–protein distances between each enzyme in a potential parent group (‘parent enzyme’) and the enzymes in the potential daughter group were averaged. Those smallest average distance values were used to choose the likely parent groups and enzymes for the daughter groups. This method allows the identification of the parent enzyme, which is similar to the ancestral enzyme that would have given rise to the daughter group (Figure 1). A small number of the pathways selected are not statistically superior to their alternatives. This raises the possibility that enzymes in some categories may have reached common evolutionary ‘destinations’ by following different pathways. Large amounts of variation within certain lineages also inflate the standard deviations of several average distances. In cases of numerical ties, the average distances between all enzymes in the parent and daughter groups were calculated and used to select a pathway.
Fig. 1. Phylogenetic category-based evolutionary pathways of serine proteases. More likely evolutionary transitions are shown with arrows; dashed lines indicate less likely transitions. Numbers indicate average distances ± standard deviations of the closest enzyme in each potential parent group to the enzymes in the potential daughter group(s). Numbers in parentheses indicate average distances ± standard deviations of all enzymes in each potential parent group to the enzymes in the potential daughter groups(s). Names of ‘parent enzymes’ and their species of origin are listed beneath average distances. Beneath names of parent enzymes are the probability values that pathway choices are the result of a random distance distribution. Probability values in parentheses correspond to average distance values in parentheses. Distances without standard deviations indicate daughter groups that contain only one enzyme.
The phylogeny of family S1 of clan SA yields a new perspective of the evolution of that family. Trypsin is the selected parent enzyme for the two largest daughter groups emerging from the Ser195:TCN/Ser214:TCN/Pro225 lineage. Those two daughter groups, Ser195:AGY/Ser214: TCN/Pro225 and Ser195:TCN/Ser214:AGY/Pro225, contain a variety of enzymes that perform both degradative and more advanced physiological functions such as fibrinolysis and embryonic development. The enzymes in the daughter categories could therefore be viewed as more specialized relatives of the degradative enzymes like trypsin. The above daughter categories give rise to their own daughter categories, which show further enzyme specialization. The lineage Ser195:TCN/Ser214:AGY/Pro225 gives rise to Ser195:TCN/Ser214:AGY/Tyr225 through the parent enzyme Sp14D1. The daughter lineage contains enzymes involved in immunity (haptoglobin and sea urchin factor B) and development (fruit fly nudel); Sp14D1 is a hemolymph protein that has been implicated in the mosquito immune response (Gorman et al., 2000). The lineage Ser195:AGY/Ser214:TCN/Pro225 gives rise to Ser195:AGY/Ser214:TCN/Tyr225 through the uncharacterized fly gene CG-18735. The daughter lineage primarily contains enzymes involved in clotting (thrombin) and complement (CASP, factor C1r, factor C1s and MASP-2). Thrombin, in turn, is the parent enzyme for the lineage Ser195:AGY/Ser214:AGY/Tyr225, which contains blood clotting factors. Thus, in the functionally diverse family S1, enzymes become more functionally specialized as one follows the evolutionary pathway from primordial to modern lineages.
The phylogeny of clan SB is constructed with subfamily S8A as the ancestral group, as subfamilies S8B and S8C are devoid of prokaryotic sequences, with the single exception of the calcium-dependent protease of A.variabilis. Thus, subfamilies S8B and S8C are depicted as daughter lineages of the Ser125:TCN/Ser190:TCN/Ser207:TCN lineage of subfamily S8A. Each subfamily displays a unique evolutionary pathway, reinforcing the concept that subfamilies diverged prior to the emergence of marker lineages. In subfamily S8B, the individual parent enzyme for the lineage Ser125:AGY/Ser190:AGY/Ser207:AGY can not be identified with statistical confidence; however, as a group, Ser125:AGY/Ser190:TCN/Ser207:AGY appears to have a closer evolutionary relationship and was selected as the parent group. This selection is consistent with previous analyses that reveal the functional and sequence similarity of the mammalian prohormone convertases (Seidah et al., 1994). In subfamily S8C, cucumisin-like enzymes from plants are the selected parent enzymes for groups containing other cucumisin-like enzymes and tripeptidyl peptidase (TPP)-like enzymes. It is difficult to establish this trend as definitive since only a limited number of TPP-like sequences are currently known.
The phylogeny of clan SC is constructed with family S9 as the ancestral group, since family S10 is absent from prokaryotes, unlike family S9 (Rawlings, 1998b). This places family S9 ancestral to family S10 based on current sequence data. As with clan SB, each family has a unique evolutionary pathway. In family S9, the parent enzyme of lineage Ser57:AGY/Ser146:AGY/Val341 can not be identified with statistical confidence; as a group, Ser57:AGY/Ser146:TCN/Val341 appears to have a closer evolutionary relationship. In family S10, plant carboxypeptidases serve as parent enzymes for animal/fungal carboxypeptidase groups, and vice versa. This result suggests that plants, animals and fungi started with roughly the same lineages of carboxypeptidases, and that each kingdom utilized different lineages of carboxypeptidases according to their varied physiological requirements.
Differences in marker usage within multi-protease genes
Several genes in the Drosophila (Adams et al., 2000) and Caenorhabditis elegans (C. elegans Sequencing Consortium, 1998) genomes contain multiple serine protease domains. It is unclear whether these domains function as part of a single polypeptide or are cleaved into separate active units. These genes are interesting because they allow the determination of sequence markers multiple times within a single gene. In most cases, all of the sequence markers for the different protease domains are in agreement. For some Drosophila genes within clan SA, however, there are discrepancies among different domains as to codon usage for Ser214 and residue choice for Pro/Tyr225. And one protease domain of a double-carboxypeptidase (clan SC) with a dichotomy in Val/Cys341 usage from Drosophila was selected as a parent enzyme for the lineage containing the second protease domain. Thus, the fruit fly has been ‘caught in the act’ of exploring catalytic variations in clans SA and SC. This type of natural sequence experimentation may have been partly responsible for generating the diversity seen among serine proteases.
Discussion
Active site structure governs serine protease evolution
Previously, our laboratory reported that phylogenetic trees based on protease domain sequences from family S1 of clan SA segregated serine proteases into physiological functional groups; the apparent driving force behind this phenomenon was substrate recognition (Krem et al., 1999). Our current results indicate that development and maintenance of the active site structure played at least an equally important role in the evolution of proteolytic enzymes. This was not revealed by the original study because phylogenetic trees provide fundamentally different information from active site markers. Phylogenetic trees appear to be governed by surface-exposed residues that control substrate and modulatory ligand recognition. On the other hand, active site evolutionary markers are buried or invariant regardless of changes in substrates or other ligands. The correlation between family divisions and the sequence motifs immediately surrounding active site and evolutionary marker residues illustrates the principle that variations in active site structure permit enzymes to fulfill new roles. A clear example is variation around the catalytic Ser146 in clan SC: serine carboxypeptidases (family S10) have Glu145, but family S9 most often places Trp at that position. Glu145 is responsible for the pH 4.5–5.5 optimum of the serine carboxypeptidases (Remington and Breddam, 1994). Therefore, generation of enzymes with new functions depends on the two separate processes of modifying the catalytic machinery and optimizing contacts with substrates and regulatory ligands. In fact, because trees are more function oriented, convergent evolution of enzymes to recognize similar or identical substrates could make trees less reliable indicators of evolutionary lineage than active site markers.
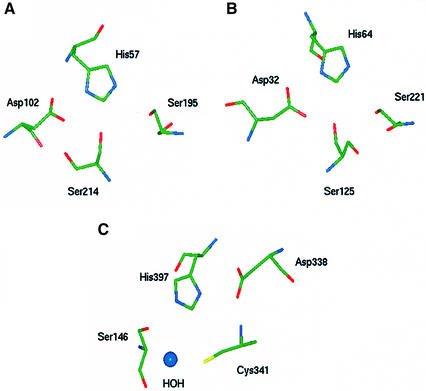
Several of the markers used to characterize serine proteases have been studied in detail regarding their contribution to the regulation of enzymatic activity. Among the chymotrypsin-like enzymes, the side chain of Ser214 hydrogen bonds with the side chain of the catalytic Asp102, as well as conserved water molecules within the active site pocket (McGrath et al., 1992; Pletnev et al., 2000) (Figure 2A). Mutation of this residue to Lys in trypsin is predicted to destabilize the transition state, implying that this residue may govern the electrostatic potential of Asp102 (McGrath et al., 1992). In the serine carboxypeptidase family of clan SC, the sulfhydryl of unpaired Cys341 plays a large role in catalytic efficiency, possibly through the correct positioning of the catalytic His397 (Jung et al., 1999). In the crystal structure of lysosomal carboxypeptidase A, the side chains of Cys341 and the catalytic Ser146 form hydrogen bonds with a shared active site water molecule. In addition, the backbone N of Cys341 forms a hydrogen bond with the carboxyl group of the catalytic Asp338 (Rudenko et al., 1995). Although not required for enzyme activity, Cys341 is likely to improve catalytic activity through direct or indirect interactions with all three residues of the charge relay system (Figure 2C). These interactions were of sufficient importance that usage of Cys341 emerged independently in both families S9 and S10. In the subtilisin clan, Ser125 is highly conserved and positioned adjacent to the catalytic triad (Siezen and Leunissen, 1997). The crystal structure of subtilisin BPN′ (Bott et al., 1988) reveals that the side chain of Ser125 is positioned such that it may form hydrogen bonds with the carboxyl group of the catalytic Asp64 (Figure 2B). In the BPN′ mutant subtiligase, additional mutation of Ser125 and Leu126 may beneficially reposition the catalytic nucleophile Cys221 (Atwell and Wells, 1999). Mutational and structural analyses therefore indicate that conserved Ser or Cys residues stabilize the charge transfer apparatus and form the fourth member of a catalytic tetrad in three unrelated enzyme clans.
Fig. 2. Three-dimensional structural representations of serine protease active sites from clans SA, SB and SC. Catalytic residues are shown in stick representation. Coordinates are from Brookhaven Protein Data Bank entries, indicated in parentheses after each enzyme name. (A) Human thrombin (1HAH) of clan SA. (B) Bacillus amyloliquefaciens subtilisin BPN′ (2ST1) of clan SB. (C) Human lysosomal carboxypeptidase A (1IVY) of clan SC.
Other markers are outside of the active site and appear to contribute indirectly to catalytic function. Pro/Tyr225 of the chymotrypsin-like proteases governs whether enzymes can take advantage of monovalent cation binding to increase their catalytic specificity, manifested by increased kcat and decreased Km (Dang and Di Cera, 1996; Guinto et al., 1999). The numerical preference for Pro225, which prevents cation binding, may be an example of a conserved proline that regulates the folding kinetics of secondary structural elements (Hardy and Nelson, 2000). In subtilisin-type proteases, mutation of residue 188 to Pro, just upstream of the conserved marker Ser190, results in enhanced thermostability (Sättler et al., 1996) and the ability to function in denaturing environments (Chen and Arnold, 1993). Ser190 is located near the active site and substrate binding pocket, implying that the loop containing the Ser190 marker is associated with proper folding of the catalytic apparatus. The role of Ser207, however, is less clear. Although Ser207 is at least 10 Å from the nearest catalytic residue, the backbone carbonyl of residue 208 hydrogen bonds with the α-helix that contains the catalytic His64 (Bott et al., 1988). Ser207 may be part of a structural motif that contributes to the stability of the catalytic triad. In clan SC proteases, Ser57 is part of a conserved sequence motif that includes Gly53, which forms part of the oxyanion hole that stabilizes the negatively charged substrate transition state (Rudenko et al., 1995). Pro54 of the same motif is positioned to engage in a ring-stacking interaction with the side chain of Tyr147, the residue that completes the oxyanion hole and is immediately downstream of the catalytic serine. Ser57 may, therefore, be a marker that reports on the stability of the oxyanion hole and the loop containing the active site Ser146.
Mechanisms for changes in active site markers
The phylogenies of Figure 1 utilize TCN→AGY transitions. Such transitions may occur through different mechanisms. A number of conserved Ser positions display the occasional use of Thr or Cys residues (Table II). Thr and Cys are each a single-nucleotide change away from TCN and AGY, and most likely served as intermediates in stepwise TCN→AGY transitions. Residues that could withstand the substitution of amino acids with slightly altered steric or electronegativity profiles, such as Ser214 in clan SA and Ser125 in clan SB, seem to have employed Thr and Cys as evolutionary intermediates. Active site nucleophile Ser residues would be less tolerant to mutation. Among the catalytically active members of the enzyme families studied here, such residues are exclusively Ser. However, several Cys nucleophile variants of serine proteases are known. Picornains, cysteine proteases of positive strand RNA viruses, share secondary structure and organization of the catalytic triad with chymotrypsin-like serine proteases, strongly suggesting that picornains and chymotrypsin-like serine proteases share common ancestry (Gorbalenya et al., 1989). Dienelactone hydrolase of Pseudomonas, also with a cysteine nucleophile, is not a protease but has the α/β-hydrolase fold (Ollis et al., 1992) like the enzymes of families S9 and S10. The existence of the above cysteine nucleophiles suggests that single-nucleotide transitions involving Cys are possible in active site Ser TCN→AGY transitions. It is difficult to gauge the evolutionary role of picornains, as viruses are intracellular parasites that obtain many of their genes through lateral transfer from host organisms. Furthermore, the origin of the picornains is ambiguous, as they do not maintain Ser214 and Pro/Tyr225 as evolutionary markers, and Cys195 could be derived from either Ser195:AGY or Ser195:TCN lineages. On the other hand, serine proteases might avoid the risk of less active Thr and Cys intermediates altogether through the use of direct TCN→AGY transitions occurring by double-nucleotide substitutions, which have recently been shown to occur in both coding and non-coding sequences at the appreciable rate of 0.1 per site per billion years (Averof et al., 2000).
The identification of several newly sequenced genes with multiple protease domains suggests yet another mechanism by which codon and amino acid usage at conserved residues can change. Rather than mutation of a functional gene or gene duplication resulting in an entire ‘experimental’ copy that would probably be lost upon trial of a deleterious mutation, duplication of a protease domain within a gene would allow both the retention of the original protease domain and the experimental copy. The experimental copy could explore a range of mutations in key regions as long as the original maintained its function. Experimental copies of domains displaying advantageous changes in function would subsequently be split into their own separate genes. Domain duplication is hypothesized to have occurred in several protein families (Heringa and Taylor, 1997), but examples of it in action, producing new enzymes, have been lacking. This study finds evidence that domain duplication fosters the exploration of alterations in active site function. One example of a gene exploring variation in a catalytic marker is the clan SA gene CG-8215 from Drosophila, which displays Ile, Pro, Val and Ser at residue 225. Ser has been hypothesized to be an intermediate in the Pro→Tyr transition at residue 225 of clan SA proteases (Guinto et al., 1999).
Conclusions
Analysis of amino acid and codon usage by highly conserved residues linked to active site function in three unrelated clans of serine proteases reveals non-random sequence dichotomies that place enzymes within phylogenetically distinct lineages. The identities of the lineage-defining markers indicate that the maintenance or development of a catalytic tetrad is an evolutionary feature common to all three protease clans examined. Similar methods can be used to trace lineages and establish evolutionary timelines for other protein families, especially those with highly conserved serine residues. The advantage of such methods is that they are based on limited sequence elements shared by nearly all members of a given family. The use of nearly absolutely conserved residues allows the identification of common ancestries when extended protein sequences have diverged extensively and detailed structural information is not available. In addition, evolutionary trends resulting from changes in active site structure can be dissociated from trends that are due to similarities or differences in substrate recognition. Finally, examination of changes in active site-related residues in multi-protease genes leads to the postulation of a mechanism by which functional diversity develops within an enzyme family without the generation of evolutionary intermediates that are functionally compromised and unlikely to be retained.
Materials and methods
Amino acid and nucleotide sequences of serine proteases and homologous proteins of clans SA, SB and SC were culled from DDBJ/EMBL/GenBank at http://www.ncbi.nlm.nih.gov/Entrez. Genome sequences were translated and then aligned using CLUSTAL-W (Thompson et al., 1994) in order to identify the codons and amino acid choices for residues of interest. For calculation of protein distance matrices, nearly identical sequences were eliminated from alignments to minimize duplication. Non-protease sequences were also eliminated. Protein distances were calculated using PROTDIST from the PHYLIP package (Felsenstein, 1999). Statistical comparisons of amino acid preferences were performed with the χ2 test. Statistical comparisons of average evolutionary distances between single enzymes and groups of enzymes were carried out using two-factor ANOVA without replication; comparisons of average distances between two groups of enzymes were carried out using two-sample t-tests assuming unequal variances.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
This work was supported in part by NIH research grants HL49413 and HL58141.
References
- Adams M.D. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Atwell S. and Wells,J.A. (1999) Selection for improved subtiligases by phage display. Proc. Natl Acad. Sci. USA, 96, 9497–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof M., Rokas,A., Wolfe,K.H. and Sharp,P.M. (2000) Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science, 287, 1283–1286. [DOI] [PubMed] [Google Scholar]
- Barrett A.J. and Rawlings,R.D. (1995) Families and clans of serine peptidases. Arch. Biochem. Biophys., 318, 247–250. [DOI] [PubMed] [Google Scholar]
- Bott R., Ultsch,M., Kossiakoff,A., Graycar,T., Katz,B. and Power,S. (1988) The three-dimensional structure of Bacillus amyloliquefaciens subtilisin at 1.8 Å and an analysis of the structural consequences of peroxide inactivation. J. Biol. Chem., 263, 7895–7906. [PubMed] [Google Scholar]
- Brenner S. (1988) The molecular evolution of genes and proteins: a tale of two serines. Nature, 334, 528–530. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chen K. and Arnold,F.H. (1993) Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl Acad. Sci. USA, 90, 5618–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Q.D. and Di Cera,E. (1996) Residue 225 determines the Na+-induced allosteric regulation of catalytic activity in serine proteases. Proc. Natl Acad. Sci. USA, 93, 10653–10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lazcoz Y., Hénaut,A., Vigier,P. and Risler,J.-L. (1995) Differential codon usage for conserved amino acids: evidence that the serine codons TCN were primordial. J. Mol. Biol., 250, 123–127. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1999) PHYLIP (phylogeny inference package), version 3.5c, distributed by the author, Department of Genetics, University of Washington, Seattle, WA.
- Gorbalenya A.E., Donchenko,A.P., Blinov,V.M. and Koonin,E.V. (1989) Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases: a distinct protein superfamily with a common structural fold. FEBS Lett., 243, 103–114. [DOI] [PubMed] [Google Scholar]
- Gorman M.J., Andreeva,O. and Paskewitz,S.M. (2000) Molecular characterization of five serine protease genes cloned from Anopheles gambiae hemolymph. Insect Biochem. Mol. Biol., 30, 35–46. [DOI] [PubMed] [Google Scholar]
- Guinto E.R., Caccia,S., Rose,T., Fütterer,K., Waksman,G. and Di Cera,E. (1999) Unexpected crucial role of residue 225 in serine proteases. Proc. Natl Acad. Sci. USA, 96, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J.A. and Nelson,H.C.M. (2000) Proline in α-helical kink is required for folding kinetics but not for kinked structure, function, or stability of heat shock transcription factor. Protein Sci., 9, 2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heringa J. and Taylor,W.R. (1997) Three-dimensional domain duplication, swapping and stealing. Curr. Opin. Struct. Biol., 7, 416–421. [DOI] [PubMed] [Google Scholar]
- Jung G., Ueno,H. and Hayashi,R. (1999) Carboxypeptidase Y: structural basis for protein sorting and catalytic triad. J. Biochem., 126, 1–6. [DOI] [PubMed] [Google Scholar]
- Krem M.M., Rose,T. and Di Cera,E. (1999) The C-terminal sequence encodes function in serine proteases. J. Biol. Chem., 274, 28063–28066. [DOI] [PubMed] [Google Scholar]
- Lesk A.M. and Fordham,W.D. (1996) Conservation and variability in the structures of serine proteinases of the chymotrypsin family. J. Mol. Biol., 258, 501–537. [DOI] [PubMed] [Google Scholar]
- McGrath M.E., Vasquez,J.R., Craik,C.S., Yang,A.S., Honig,B. and Fletterick,R.J. (1992) Perturbing the polar environment of Asp102 in trypsin: consequences of replacing conserved Ser214. Biochemistry, 31, 3059–3064. [DOI] [PubMed] [Google Scholar]
- Miyazawa K., Wang,Y., Minoshima,S., Shimizu,N. and Kitamura,N. (1998) Structural organization and chromosomal location of the human hepatocyte growth factor activator gene. Eur. J. Biochem., 258, 355–361. [DOI] [PubMed] [Google Scholar]
- Ollis D.L. et al. (1992) The α/β hydrolase fold. Protein Eng., 5, 197–211. [DOI] [PubMed] [Google Scholar]
- Patthy L. (1985) Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell, 41, 657–663. [DOI] [PubMed] [Google Scholar]
- Patthy L. (1990) Evolution of blood coagulation and fibrinolysis. Blood Coagul. Fibrinolysis, 1, 153–166. [PubMed] [Google Scholar]
- Perona J.J. and Craik,C.S. (1995) Structural basis of substrate specificity in the serine proteases. Protein Sci., 4, 337–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev V.Z., Zamolodchikova,T.S., Pangborn,W.A. and Duax,W.L. (2000) Crystal structure of bovine duodenase, a serine protease, with dual trypsin and chymotrypsin-like specificities. Proteins, 41, 8–16. [PubMed] [Google Scholar]
- Rawlings N.D. (1998a) Introduction: clan SB containing the subtilisin family. In Barrett,A.J., Rawlings,N.D. and Woessner,J.F. (eds), Handbook of Proteolytic Enzymes. Academic Press, San Diego, CA, pp. 284–288.
- Rawlings N.D. (1998b) Introduction: clan SC containing peptidases with the α/β hydrolase fold. In Barrett,A.J., Rawlings,N.D. and Woessner,J.F. (eds), Handbook of Proteolytic Enzymes. Academic Press, San Diego, CA, pp. 369–372.
- Rawlings N.D. and Barrett,A.J. (1993) Evolutionary families of peptidases. Biochem. J., 290, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington S.J. and Breddam,K. (1994) Carboxypeptidases C and D. Methods Enzymol., 244, 231–248. [DOI] [PubMed] [Google Scholar]
- Rudenko G., Bonten,E., d’Azzo,A. and Hol,W.G.J. (1995) Three-dimensional structure of the human protective protein: structure of the precursor form suggests a complex activation mechanism. Structure, 3, 1249–1259. [DOI] [PubMed] [Google Scholar]
- Sättler A., Kanka,S., Maurer,K.-H. and Riesner,D. (1996) Thermostable variants of subtilisin selected by temperature-gradient gel electro phoresis. Electrophoresis, 17, 784–792. [DOI] [PubMed] [Google Scholar]
- Seidah N.G., Chrétien,M. and Day,R. (1994) The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie, 76, 197–209. [DOI] [PubMed] [Google Scholar]
- Shilton B.H., Thomas,D.Y. and Cygler,M. (1997) Crystal structure of Kex1Δp. Biochemistry, 36, 9002–9012. [DOI] [PubMed] [Google Scholar]
- Siezen R.J. and Leunissen,J.A.M. (1997) Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci., 6, 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal_W: improving the sensitivity of progressive multiple sequence align ment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]