Abstract
The serum response element (SRE) is found in several immediate-early gene promoters. This DNA sequence is necessary and sufficient for rapid transcriptional induction of the human c-fos proto-oncogene in response to stimuli external to the cell. Full activation of the SRE requires the cooperative binding of a ternary complex factor (TCF) and serum response factor (SRF) to their specific DNA sites. The X-ray structure of the human SAP-1–SRF–SRE DNA ternary complex was determined (Protein Data Bank code 1hbx). It shows SAP-1 TCF bound to SRF through interactions between the SAP-1 B-box and SRF MADS domain in addition to contacts between their respective DNA-binding motifs. The SAP-1 B-box is part of a flexible linker of which 21 amino acids become ordered upon ternary complex formation. Comparison with a similar region from the yeast MATα2–MCM1–DNA complex suggests a common binding motif through which MADS-box proteins may interact with additional factors such as Fli-1.
Keywords: crystal structure/gene regulation/serum response factor/transcription complex/X-ray crystallography
Introduction
The c-fos promoter is paradigmatic for transcriptional regulation in eukaryotic cells (Treisman, 1995). Proteins known to play an important role in the regulation of c-fos expression are AP-1 (Paradis et al., 1996), C/EBP (Sealy et al., 1997), CREB (Ginty et al., 1994), HTLV-1 Tax (Fujii et al., 1992), NF-κB (Montaner et al., 1999), Phox1 (Grueneberg et al., 1992), serum response factor (SRF) (Norman et al., 1988), ternary complex factors (TCFs) (Shaw et al., 1989) and YY1 (Natesan and Gilman, 1995). The SRF homodimer and TCF monomer form a ternary complex on the serum response element (SRE) of the c-fos promoter (Schroeter et al., 1990). SRE sequences are also found in promoters of several other immediate-early genes (Hipskind et al., 1994) and viral genomes (Cahill et al., 1994). The protein complex formed at the SRE is necessary and sufficient for the rapid induction of the c-fos proto-oncogene in response to different external stimuli such as serum, growth factors and phorbol esters (Hipskind et al., 1994). Hence, the SRE DNA sequence is a point of convergence for different signal transduction cascades activated by an extensive range of agonists (Whitmarsh et al., 1995; Janknecht and Hunter, 1997). The mitogenic signal transduction pathway directly targets the TCF transcription regulators; Elk-1 (Rao et al., 1989), SAP-1a (Dalton and Treisman, 1992) and NET/SAP-2/Erp (Giovane et al., 1994; Lopez et al., 1994) are all phosphorylated in vivo and in vitro by ERK1/2 kinases (Price et al., 1995).
Human Elk-1, SAP-1a and NET are members of the ETS family of transcription factors sharing a high degree of homology in three distinct domains. The N-terminal domain encompassing 91 amino acids mediates recognition of a purine-rich DNA motif called the ETS-binding site (EBS). The structures of several ETS domains have been determined including those for Elk-1 and SAP-1 (Mo et al., 1998, 2000) and show that they belong to the winged helix–turn–helix family of transcription factors (Donaldson et al., 1996). A second region of 21 amino acids designated the B-box is required for TCF interaction with SRF (Dalton and Treisman, 1992; Shore and Sharrocks, 1994). The C-terminal region or C-box contains 56 amino acids and is the target for mitogen-activated protein kinases (MAPKs) (Janknecht et al., 1993; Marais et al., 1993). Phosphorylation most likely causes a conformational change in the protein that unmasks the DNA-binding domain and the B-box of the TCF, enabling ternary complex formation on the SRE (Masutani, 1997; Yang et al., 1999) and localization of the co-activator CBP/p300 via C-box interaction (Janknecht and Nordheim, 1996a,b). Human SRF is a founding member of the MADS domain family of transcription factors, and the X-ray structure of its core domain (amino acids 140–223) and those for the related yeast MCM1 and human MEF2A bound to DNA were determined previously (Pellegrini et al., 1995; Tan and Richmond, 1998; Huang et al., 2000; Santelli and Richmond, 2000). Although highly homologous, their structures reveal distinctive differences in DNA binding. SRF is itself a target for phosphorylation and as such the endpoint of signaling pathways separate from those activating the TCF proteins (Heidenreich et al., 1999).
We report here the X-ray structure of the ternary complex comprising the ETS domain through B-box region of human SAP-1 (SRF-associated protein 1), the homodimeric core of human SRF, and a 26 bp DNA fragment containing the consensus SRE sequence (SAP-1–SRF–DNA). The B-box is the primary region of SAP-1 contributing to the cooperativity of binding of SAP-1 to SRF, and the crystal structure shows that it adopts an unusual 310-helix/β-strand/310-helix conformation to facilitate this interaction. Comparison of the structures of the SAP-1–SRF–DNA ternary complex with that for MATα2–MCM1–DNA (Tan and Richmond, 1998) permits identification of common features in the protein sequence that may be important for the interaction of other associated transcription factors binding to MADS-box proteins. As a consequence, a B-box-like sequence could be identified in the human ETS transcription factor Fli-1 (Watson et al., 1997).
Results
Architecture of the SAP-1–SRF–DNA ternary complex
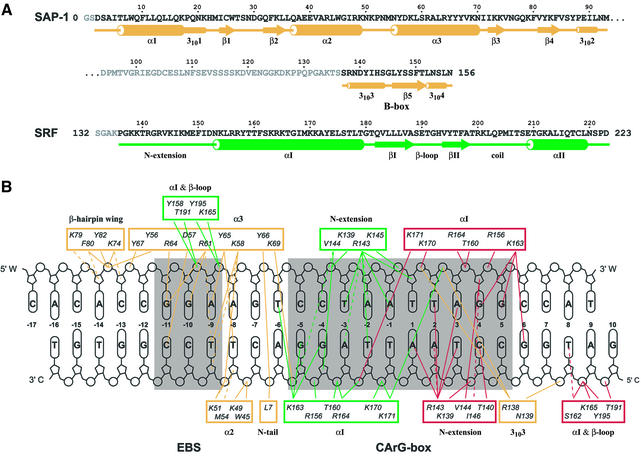
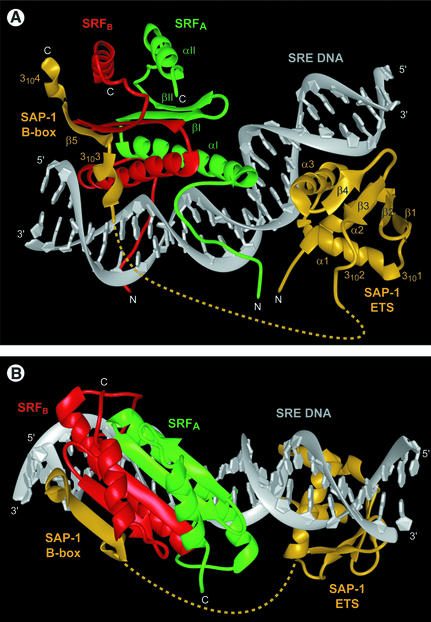
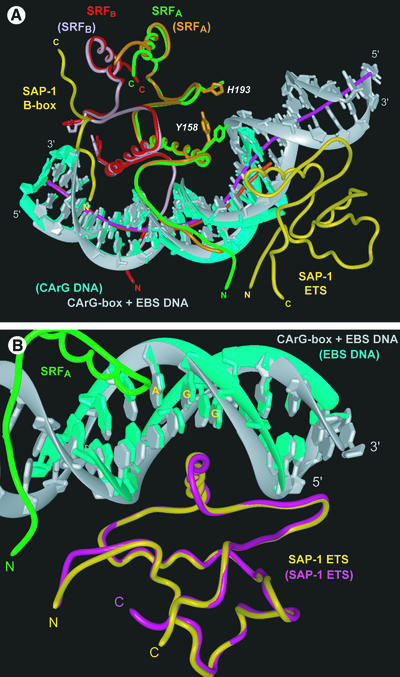
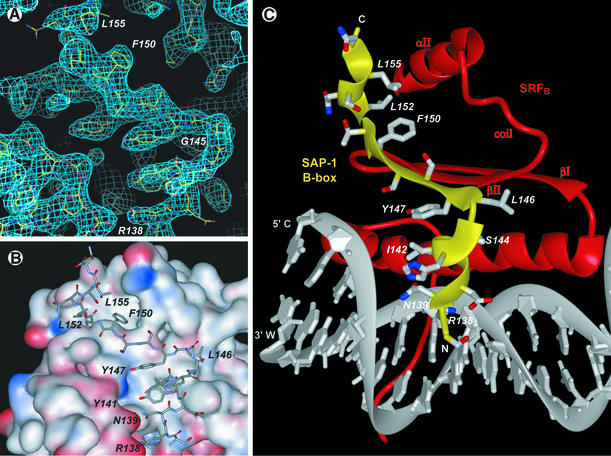
The crystal structure of a 56 kDa complex comprising the homodimeric core domain of human SRF protein (amino acids 132–223), the DNA-binding domain through B-box domain of human SAP-1 (amino acids 1–156) and a 26 bp DNA oligonucleotide was solved by X-ray crystallo graphy to 3.15 Å resolution (Figure 1; Table I). The DNA duplex is B-form, having Dy values over –1.0 for all base pairs (El Hassan and Calladine, 1997), bent overall by 77° and extensively covered with protein along its entire length. All base pairs with the exception of the two at positions –15 and –16 are involved in protein–DNA contacts through either direct hydrogen bonds to the bases, van der Waals interactions to the phosphodiester backbone or hydrophobic contacts to the deoxyribose moieties (Figure 1B). The DNA duplex contains both an EBS consensus sequence for SAP-1 and a CArG-box binding site for SRF. As the centers of these two DNA sites are separated by 9.5 bp, SAP-1 and SRF are bound essentially on opposite faces of the DNA duplex in the major and minor grooves, respectively (Figure 2). The overall orientation and binding modes of SAP-1 and SRF to DNA in the SAP-1–SRF–DNA ternary complex structure are identical to those observed in the crystals of the individual subcomplexes of SAP-1–DNA (Mo et al., 1998) and SRF–DNA (Pellegrini et al., 1995) (Figure 3), having root mean square deviations (r.m.s.ds) between structures of 0.75 and 0.98 Å2, respectively, based on 88 protein Cα atoms and 20 DNA phosphorus atoms for SAP-1–DNA, and 164 protein Cα atoms and 34 DNA phosphorus atoms for SRF–DNA. The 21 amino acids (136–156) defining the B-box essential for cooperative binding of SAP-1 to SRF are clearly visible in near entirety (amino acids 137–156 and 138–154) for both independent copies of the complex in the asymmetric unit (Figure 4A). The 44 amino acids of SAP-1 spanning between positions 93 and 136 or 137 could not be traced unambiguously in the electron density map, indicating that they constitute a flexible linker connecting the ETS domain and the B-box (Figures 1A and 2). The electron density of each of the four copies of the SRF polypeptide in the asymmetric unit begins variably between amino acids 136 and 140, and continues to the C-terminus.
Fig. 1. Sequences of the protein and DNA components used for the human SAP-1–SRF–DNA crystals. (A) SAP-1 ETS domain through the B-box (amino acids 1–156) and SRF core domain (MADS-box plus SAM domains, amino acids 132–223). The terminal amino acids and the SAP-1 linker region disordered in the structure are printed in gray. The N-terminal methionine of the wild-type SAP-1 sequence was replaced by glycine followed by serine (amino acids 0–1). Secondary structure elements are indicated below the sequences where the larger tubes represent α-helices, smaller tubes 310-helices and arrows β-strands. (B) SRE consensus sequence with demarcation of protein–DNA interactions. The EBS (GGA core) and the CArG sequence are on a gray background. The base pair numbering starts at the 2-fold symmetric sequence center of the CArG-box with the two different strands labeled W and C. Amino acids contacting DNA are grouped by secondary structure elements with monomer SRFA in green, monomer SRFB in red and SAP-1 in orange. Hydrogen bond interactions are shown as solid lines and hydrophobic contacts as dotted lines.
Table I. Crystallographic data and refinement statistics.
| Crystallographic data | Native | CH3HgNO3 derivative |
|---|---|---|
| Resolution (Å) | 51–3.00 | 60–3.4 |
| Completeness | ||
| overall (last shell) | 98.5 (96.7) | 94.5 (96.5) |
| last shell (range Å) | 3.05–3.00 | 3.52–3.4 |
| Unique reflections | 31747 | 20936 |
| Average redundancy | 5.2 | 3.9 |
| Rmerge (%) | ||
| overall (last shell) | 4.2 (58.3) | 7.8 (56.5) |
| last shell (range Å) | (3.05–3.00) | (3.52–3.4) |
| <I/σ> | 30.4 | 11.8 |
| Phasing power | 1.14 | |
| RCullis | 0.82 | |
| Unit cell | P21212 | |
| a (Å) | 142.679 | 143.172 |
| b (Å) | 144.389 | 144.100 |
| c (Å) |
75.932 |
75.319 |
| Structure refinement | ||
| Resolution (Å) | 51–3.15 | |
| Rworka 51–3.15 (3.18–3.15) (%) | 25.6 (42.5) | |
| Rfreeb 51–3.15 (3.18–3.15) (%) | 28.5 (46.3) | |
| Reflections work/free | 24785 / 2735 | |
| Atoms (two copies/ASU) | ||
| total | 6716 | |
| protein | 4596 | |
| DNA | 2120 | |
| R.m.s. deviations | ||
| bond length (Å) | 0.007 | |
| bond angle (°) | 1.42 | |
| Ramachandran plotc | ||
| favored (%) | 77.9 | |
| allowed (%) | 20.5 | |
| generously allowed (%) | 1.2 | |
| disallowed (%) | 0.4 | |
| Average B-values (Å2) | 81.9 | |
aRefined with anisotropic B-factor correction and bulk solvent correction.
bRfree was calculated using 10% of the data chosen randomly and omitted from the refinement and Rwork.
cCategories as defined by the PROCHECK analysis.
Fig. 2. Structure of the SAP-1–SRF–DNA ternary complex. (A) View of the SAP-1 monomer and SRF dimer unobscured by the SRE DNA. The B-box of SAP-1 (yellow) connected by a flexible linker (dashed line, unobserved) to the ETS domain is bound to the SRFB monomer (red) and adds an antiparallel β-strand flanked by short 310-helix segments to the four-stranded, antiparallel β-sheet of the SRF MADS-box. The SRFA monomer (green) directly contacts the SAP-1 ETS domain. The entire DNA oligonucleotide (gray) is shown and the protein secondary structure elements are labeled. (B) View showing SAP-1 and SRF bound to opposite DNA faces. The SAP-1 ETS domain is bound in the EBS major groove and SRF is centered on the CArG-box minor groove.
Fig. 3. Structural alignments of the SAP-1–SRF–DNA ternary complex with the SRF–DNA and SAP-1–DNA binary complexes. The ternary complex colors are as in Figure 2. The binary complex components have colors corresponding to their labels shown in parentheses. (A) SRF–DNA (Pellegrini et al., 1995) and SAP-1–SRF–DNA (r.m.s.d. 0.66 Å for 164 Cα atoms). The amino acids Y158 and H193 involved in binding the extremities of the DNA site have different side chain conformations in the two structures. DNA helical axes for the ternary (pink) and binary (orange) complexes were calculated with CURVES (Lavery and Sklenar, 1988). (B) SAP-1–DNA (Mo et al., 1998) and SAP-1–SRF–DNA (r.m.s.d. 0.90 Å for 88 Cα atoms). The overlap of the two protein chains is nearly exact except at the N- and C-termini. In contrast, the conformations of the DNA differ significantly outside of the central GGA sequence.
Fig. 4. Structure of the SAP-1 B-box sequence element bound to SRF. (A) Model of the SAP-1 B-box superimposed in the experimental electron density. The 2Fo – Fc electron density map calculated at 3.15 Å resolution is contoured at 1.5σ. The amino acid F150 is labeled to indicate the B-box polypeptide chain. The density at the lower left corresponds to DNA and at the upper right to SRF. (B) SAP-1 polypeptide chain bound to SRF shown as an electrostatic potential surface (red negative and blue positive charge). With the exception of the interactions with the DNA phosphate groups, the interaction surface is predominantly hydrophobic. The three aromatic amino acids Y141, Y147 and F150 contribute substantially to the SAP-1–SRF interaction interface, and the analogous amino acids in Elk-1 were identified as the most important for its addition to the ternary complex (Ling et al., 1997). (C) Side chains of the SAP-1 B-box stabilizing its bound conformation. R138 and N139 make hydrogen bonds to DNA phosphate groups. Y141, I142 and Y147 make a hydrophobic cluster by which the 3103 helix and β-strand interact. L146 extends along the surface of SRF, and F150 protrudes into the hydrophobic cleft formed by the SRF βII strand, coil region and αII helix. L152 and L155 bind the 3104 helix to SRF.
Additional amino acids are ordered in the N-extensions of SRF in the ternary complex as compared with the SRF–DNA structure (Figures 1 and 3A). The main chain atoms of SRF amino acids 137–139 are clearly visible in the ternary complex, with the amide group of K139 making a hydrogen bond to the phosphate of base T±3 (Figure 1B). The conformation of the C-terminal amino acids 219–223 of SRF emanating from the αII helix deviates significantly in the SAP-1–SRF–DNA and SRF–DNA complexes. For the SAP-1 ETS domain, significant conformational differences are found only for the N-terminal arm amino acids 2–8 and for the C-terminal amino acids 90–93 (Figure 3B). The different conformation of the N-terminal arm in the ternary complex can be explained by its close proximity to the N-extension of the adjacent SRF subunit.
SAP-1 B-box region
The B-box sequence of SAP-1 adopts an unusual 310-helix/β-strand/310-helix motif, which contacts all three secondary structure layers of a single SRF subunit as well as DNA (Figure 4). The N-terminus of the B-box sequence is positioned above the DNA major groove, with the side chains of R138 making a direct hydrogen bond with the phosphate oxygen of T2pA3 and N139 contacting T8pG7 (Figure 4B). The main interaction of the 3103 helix with SRF is made by its Y141 side chain lying flat on M169 of the SRF αI helix. Together with I142 in the 3103 helix and Y147 in the β-strand, Y141 provides hydrophobic interactions stabilizing the orientation of 3103 with respect to the adjacent β-strand (Figure 4B and C). G145 facilitates the right angle turn in the chain going from the 3103 helix to the β-strand by adopting the Φ–Ψ parameters corresponding to a left-handed α-helix. Mutation of Elk-1 G157 to alanine (corresponding to SAP-1 G145) indicates the importance of the observed conformation as the Elk-1 mutant binds with ∼2-fold lower affinity in the ternary complex (i.e. Figure 2B; Ling et al., 1997). Amino acids 145–151 extend the four-stranded, antiparallel β-sheet of SRF by adding one further antiparallel β-strand. F150 fits into the hydrophobic pocket of SRF formed by the βII strand (V194), the coil region leading to the αII helix (I206), and the αII helix (I215) (Figures 4B, C and 6B). The chain turns again at L152 roughly at a right angle to the B-box β-strand and contributes the side chains of L152 and L155 to a hydrophobic core region below the SRF αII helix (I215 and L219). The entire SAP-1 B-box domain spanning amino acids 137–156 forms an extensive interaction surface with SRF, accounting for a loss of solvent-accessible surface area of 1646 Å2. The primary SAP-1 amino acids contributing to this interface are Y141, L146, F150 and L155 (Figure 4B).
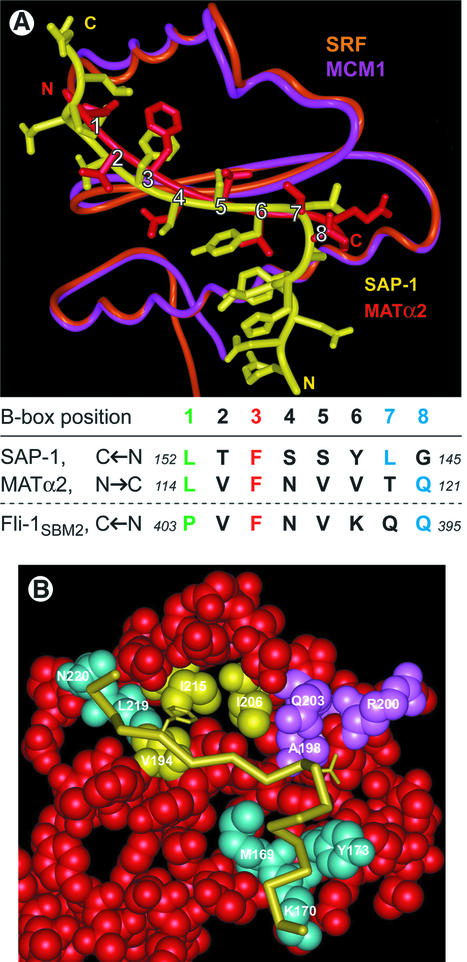
Fig. 6. SAP-1 B-box and MATα2 MBM interactions with SRF and MCM1. (A) Structural alignment of human SAP-1–SRF–DNA and yeast MATα2–MCM1–DNA ternary complexes. Cα atoms of human SRF (amino acids 143–222, orange) and yeast MCM1 (amino acids 18–97, magenta) superimpose with an r.m.s.d. of 0.5 Å. The positions of amino acids in common for human SAP-1 (yellow) and yeast MATα2 (red) and interacting with the MADS-box are labeled from 1 to 8. The identity of the amino acids is given in the B-box position table in which the direction of chains is indicated. Positions 3, 1 and 7–8 in that order appear to be the most important for specificity of binding. Reversal of the MATα2 MBM sequence to match the SAP-1 B-box allows identification of a plausible MADS-box inter action sequence in the Fli-1 SBM2 sequence as shown in the table. (B) MADS-box amino acids important for B-box binding and specificity. The main chain path and side chains F150 and L146 are shown for the SAP-1 B-box (yellow). The van der Waals surface of the SRFB monomer (red) is shown with amino acids important for specificity highlighted. The SRF cluster of V194, I206 and I215 (yellow) contacts SAP-1 F150. SRF M169, K170 and Y173 (cyan) bind helix 3103, and SRF L219 and N220 (cyan) bind helix 3104. The SRF amino acids A198, R200, L202 and Q203 (violet) were found to be important for the discrimination of binding by accessory proteins (Mueller and Nordheim, 1991).
SAP-1 ETS domain interaction with SRF
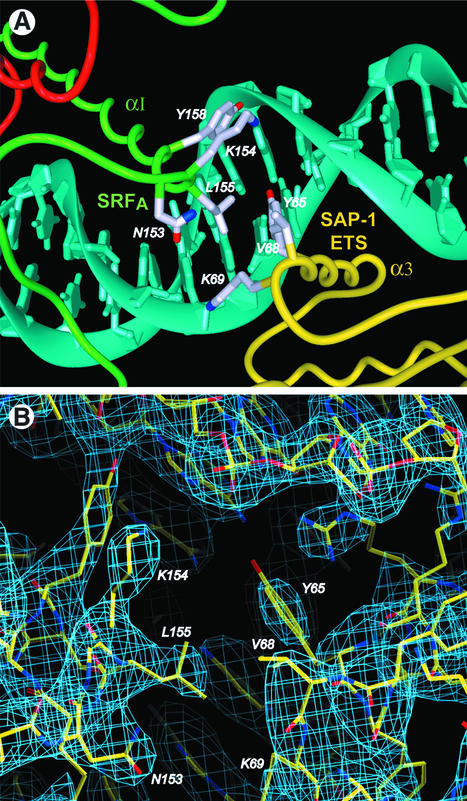
The GGA core of the EBS for SAP-1 and the CArG- box-binding site for SRF are separated by 3 bp in the DNA sequence used for crystallization. This spacing, which is based on the human c-fos promoter, enables the direct interaction of the SAP-1 and SRF DNA-binding domains through juxtaposition of the N-terminus of the SRF αI helix and the C-terminus of the SAP-1 α3 helix. SRF L155 fits into the center of a triad formed by SAP-1 side chains Y65, V68 and K69 (Figure 5). The loss of solvent-accessible surface area for this hydrophobic interface is 325 Å2, <20% of that for the B-box–SRF interaction.
Fig. 5. SAP-1 ETS domain interaction with the SRF MADS-box. (A) The amino acids in the hydrophobic interface between the SAP-1 α3 helix and the SRF αI helix are shown. (B) Electron density of the interaction interface. The simulated-annealing, 2Fo – Fc composite omit map is contoured at 1.0σ.
These interactions would be lost with a separation of DNA-binding sites of >3 bp, as can occur for natural SRE sequences, while a smaller spacing would inhibit the simultaneous binding of both proteins.
SAP-1–SRF DNA recognition
The effects of SAP-1 binding on the DNA interactions and conformation associated with SRF are not large but are significant. Comparisons of the three complete half sites seen in the SAP-1–SRF–DNA and SRF–DNA complexes show that the DNA is slightly less bent overall in the half site adjacent to the bound SAP-1. The phosphate groups of A–9 and A+9 are the first and last to be directly hydrogen bonded to the SRF β-loop. Superposition of the two half sites from SAP-1–SRF–DNA aligning the αII helices and β-strands of the MADS domain indicate that the A–9 phosphate on the side of the EBS is displaced 2.9 Å farther away from SRF than that for A+9. SRF accommodates this difference through the rotation of the χ1 torsion angle of T191, allowing the side chain hydroxyl group to make a hydrogen bond with the phosphate oxygen in both cases. The A–9 phosphate in the half site lacking the EBS has only a 0.7 Å positional difference from that for SRF–DNA. The interactions are nearly identical to SRF–DNA, with the exception that the hydrogen bond between the A–9 phosphate and H193 is broken and the imidazole ring is rotated by 90°. The additional interaction of the B-box amino acid N139 with the phosphate group of G+7 may compensate.
The DNA is bent overall to 77° in the ternary complex as compared with 72° for SRF–DNA, which contains one incomplete half site extending only to base pair –8 and is therefore not fully bound. In the partial half site, the β-loop and αI helix contact phosphate groups at positions –8, –9 and –10 are not possible (Figure 3A). Both SRF subunits in the ternary complex bend the two DNA half sites to the same extent, resulting in bending of 31° into the major groove over the CC and CT base pair steps beginning and ending the CArG-box, and 15° into the minor groove over the five central TA, AA and AT base pair steps. DNA bending by SRF is essential to facilitate the contact of the N-termini of the SRF αI helix and SAP-1 α3 helix. Nevertheless, the SAP-1 ETS domain bends the DNA by 11° in roughly the opposite direction to the curvature induced by SRF. The displacement of the A–9 phosphate group away from SRF and towards SAP-1 is thus explained, as base pairs –9 and –10 are part of both the SRF- and SAP-1-binding sites. The SAP-1–SRF–DNA and SAP-1–DNA complexes show no significant deviations in their DNA conformation or protein interactions (Mo et al., 1998) (Figure 3B).
Discussion
Induction of the B-box conformation by SRF binding
The cooperative binding of TCF proteins such as SAP-1 to SRF is mediated primarily by the B-box sequence (Treisman et al., 1992). The B-box structure seen in the SAP-1–SRF–DNA complex comprises a six-amino-acid β-strand flanked by two short 310 helices. It is not α-helical as suggested previously (Ling et al., 1997). These secondary structure elements are not known for stability in isolation and do not fold together to yield a hydrophobic core region of significant size in the ternary complex structure. Substantially more interaction occurs with SRF than within the B-box itself, such that its conformation is most probably induced by its fit to the SRF surface and its contacts to DNA. Circular dichroism measurements of a B-box polypeptide in aqueous solution free of α-helix-inducing trifluoroethanol showed that the B-box was lacking defined secondary structure (Ling et al., 1997). As an element of an intact TCF protein, the B-box sequence may take on an ordered structure through interaction with the ETS domain, thereby inhibiting transcription activation by blocking DNA binding or by binding the C-terminal C-box and preventing it from association with coactivator (Yang et al., 1999). Release of this autoinhibition may occur by specific phosphorylation of the C-box via signal transduction (Cruzalegui et al., 1999).
Flexibility of the B-box–ETS domain linker
The 43 amino acids (94–136) linking the B-box to the ETS domain of SAP-1 are unobserved in the electron density map and are therefore likely to be highly disordered. This finding is consistent with the biochemical and genetic data indicating that this linker region must be flexible as it accommodates cooperative binding of SAP-1 and SRF on sites disposed with virtually any spacing of up to 10 bp as well as direction reversal of the EBS with respect to the CArG-box (Treisman et al., 1992). Furthermore, natural SRE sequences exhibit several different arrangements of CArG-box and EBS sequences (Treisman, 1990; Hipskind et al., 1994; Latinkic et al., 1996). The ternary complex structure indicates that both SRF monomers could be bound to a B-box sequence from different SAP-1 molecules simultaneously. Indeed, a quaternary complex of Elk-1 and SRF bound to the c-fos SRE DNA has been reported in which the second TCF protein is recruited only via protein–protein interactions (Gille et al., 1996). Moreover, the Egr-1 promoter containing two EBSs on either side of the CArG-box is capable of forming a quaternary complex with SRF and Elk-1 (Watson et al., 1997).
Despite the apparent flexibility of the ETS–B-box linker, the SAP-1 B-box binds exclusively to one monomer of the SRF dimer in the crystal structure (Figure 2). For both copies of the complex in the crystallographic asymmetric unit, the linkage is to the most distal SRF subunit (SRFB) with respect to SAP-1. The distance between the last C-terminal amino acid of the ETS domain observed and the first N-terminal amino acid of the B-box observed is ∼50 Å (Figure 2). The analogous distance to a hypothetical B-box element bound to the SAP-1 proximal SRF monomer (SRFA) would be ∼55 Å, so that distance per se is unlikely to determine the path of a linker polypeptide that could stretch up to 150 Å. The exterior β-strand of the proximal SRF subunit is left unblocked by crystal contacts for both copies of the complex, and a path for the linker free of steric hindrance from symmetry copies in the crystal lattice also exists. The most plausible explanation of the single-sided binding is that the linker path to the distal SRF monomer does not wrap around the complex and can proceed to the direct addition of an anti-parallel strand to the β-sheet. The position of the SRF proximal monomer requires the linker segment to cross over the flexible end of the N-terminal extension and the DNA backbone and then make a U-turn in order to add a strand to the β-sheet. The linker sequences in the three human TCF proteins Elk-1, SAP-1 and NET are not significantly homologous in sequence, although they all have a relatively high content of glycine, proline and serine. They range in linker length from 38 to 53 amino acids, a feature that may affect their ability to bind cooperatively with SRF on different SRE-containing promoters.
Enhancement of promoter specificity via ETS–MADS domain interaction
The three human TCF proteins Elk-1, SAP-1 and NET display different affinities for the SRE in the c-fos promoter. These differences are mediated via direct contacts between the ETS and MADS domains in addition to protein–DNA interaction. Mutagenesis and in vitro binding assays have shown that amino acid V68 of the SAP-1 ETS domain is a critical determinant of specificity on the c-fos promoter (Shore et al., 1996). Changing the analogous amino acid D69 in Elk-1 to valine is sufficient to confer SAP-1 binding specificity on the Elk-1 protein. This site is in contact with SRF L155, and a carboxylic acid side chain extending from the TCF would weaken this interaction on the c-fos SRE (Figure 5A). The X-ray structure of an Elk-1–E74–DNA complex supports a different role for this TCF amino acid in promoter recognition (Mo et al., 2000). In the absence of SRF, D69 makes an intermolecular contact with K70, apparently causing Y66 to have a different side chain conformation than in SAP-1–SRF–DNA. This rearrangement is coupled to a series of major groove interactions altered from those seen in the ternary complex. The SAP-1–SRF and Elk-1–SRF DNA-binding domain interactions are unlikely to be conserved throughout evolution, however, as the spacing between TCF- and SRF-binding sites is divergent in avian and amphibian SRE sequences.
The interplay of SAP-1, Elk-1 and NET at various promoters containing EBS and CArG sequences varies depending on the actual DNA sequences and spacing between sites. For some promoters such as c-fos, SRF is seen to aid the recruitment of the TCF (Shaw et al., 1989), while for others such as pip92 and nur77, the TCF protein is required to engage SRF on non-optimal CArG sites (Williams and Lau, 1993; Latinkic et al., 1996). The length of the flexible linker between the B-box and ETS domain and the path it must take owing to site spacing will in part determine the binding affinity of the whole ternary complex. The structural results also indicate that site spacing partially governs the effectiveness of SAP-1 versus the other TCF proteins in forming a ternary complex, by enabling the direct interaction of the ETS and MADS domains. Cooperativity and binding affinity are not the only determinants of activation, however, as the susceptibility of the different TCF proteins to phosphorylation by signal transduction kinase enzymes also plays an important role (Janknecht and Hunter, 1997).
Common basis for human SAP-1–SRF and yeast MATα2–MCM1 interaction
Human SRF and yeast MCM1 proteins are 80% identical in their sequence region, which comprises the MADS-box domain and the C-terminal coil and α-helix that immediately follow it. These sequences together are necessary and sufficient for MCM1 function in vivo (Primig et al., 1991). The SAP-1 ETS domain and MATα2 homeodomain proteins belong to structurally distinct families of transcription factor, but nevertheless share the common feature of a long flexible chain that connects to an activation or repression domain, respectively. Comparison of the structures of the SAP-1 B-box with the MATα2 linker in the context of their ternary complexes reveals striking similarity for the amino acids of central importance in the interaction, despite the fact that SAP-1 adds an additional antiparallel strand to the MADS-box β-sheet whereas MATα2 adds a parallel β-strand (Tan and Richmond, 1998). The principal contributing side chains from the B-box of SAP-1 and the MCM1 binding motif (MBM) found in the MATα2 flexible linker correspond well in contribution to interaction along the eight sequential positions as viewed from the outer β-strand of SRF or MCM1 (Figure 6A). Foremost of these are positions 1 and 3 containing SAP-1 L152 and F150 and MATα2 L114 and F116, respectively, which adopt side chain conformations that fill homologous binding pockets in SRF and MCM1. Positions 2–6 in the bound β-strands of SAP-1 and MATα2 have similar conformations for isostructural aliphatic atom groups. Position 8 is divergent as SAP-1 G145 does not contact SRF, while MATα2 Q121 lies along the surface of MCM1. However, the overall interaction surface of SAP-1 with SRF is substantially larger than for MATα2 with MCM1. The SAP-1 side chain of L146 in position 7 lies along the surface of SRF, and G145 leads into the 3103 helix, which makes additional contacts.
The yeast factor MCM1 can bind human TCF proteins (Dalton and Treisman, 1992). Superposition of the SAP-1– SRF–DNA and MATα2–MCM1 structures shows how this can occur. The counterpart of SRF R200 in MCM1 is P75. The proline side chain provides a hydrophobic ‘knob’ that would interact with the Cδ atoms of the TCF leucine (e.g. SAP-1 L146) at the end of the β-strand. The side chain of SRF R200 points away from the leucine side chain making no contact, but this is compensated for by the interactions of 3103 with SRF. Yeast proteins such as STE12 that are interaction partners with MCM1 do not stably bind SRF without alteration of the SRF sequence (Mueller and Nordheim, 1991). A mutant SRF protein containing the changes A198S, R200P, L202F and Q203E mimicking MCM1 at these positions can bind STE12 (Figure 6B). The importance of the precise conformation of this region, the loop connecting the MADS-box βII strand with the coil folded back over it, is emphasized by the fact that yeast Arg RI, which has only two conservative differences with MCM1 (S198T and F202L), could not bind Elk-1 (Mueller and Nordheim, 1991). The molecular surface of SRF that accommodates the TCF 3103 helix (SRF M169, K170, Y173) and the 3104 helix (SRF L219, N220) is conserved between SRF, MCM and ARG RI (Figure 6B).
MBM sequence in Fli-1
Based on our SAP-1–SRF–DNA and MATα2– MCM1–DNA structures, we have used the B-box and MBM sequences to seek homologous regions in other transcription factors known to interact with either SRF or MCM1 core regions. For example, human Fli-1 is an ETS domain transcription factor not included in the TCF family; however, it and its oncogene variant EWS Fli-1 form complexes with SRF (Magnaghi-Jaulin et al., 1996). Fli-1 contains two distinct SRF-binding sequences, called SBM1 and SBM2, contained in regions found by deletion analysis of amino acids 220–251 N-terminal and amino acids 372–451 C-terminal to the ETS domain (Dalgleish and Sharrocks, 2000). Although alignment of the SBM2 sequence with the SAP-1 B-box or MATα2 MBM shows no obvious correspondence, alignment of Fli-1 as a reverse sequence reveals a striking match to the MATα2 MBM (Figure 6A). As a consequence, we propose that amino acids 395–402 of Fli-1 SBM2 add an antiparallel β-strand to the SRF β-sheet, filling all eight positions along the SRF β-sheet edge. The region containing SBM1 has no phenylalanine, so that proposals based on, for example, tyrosines, are significantly more speculative. The occurrence of two SRF interaction sequences within Fli-1, one N-terminal and one C-terminal to the ETS domain, suggests that both monomers of SRF can be bound by a single Fli-1 molecule. This possibility can explain how for the Egr-1 promoter, which has an EBS flanking a CArG-box on both sides, Elk-1 and SAP-1 form quaternary complexes with SRF, but Fli-1 is observed to make only a ternary complex (Watson et al., 1997).
Implications for other MADS-box-binding proteins
In addition to Fli-1, the transcription factors PEA-3, C/EBPβ, HTLV Tax, MATα1, STE12 and SFF(Fkh2) have been reported to interact with SRF or MCM1 (Primig et al., 1991; Sengupta and Cochran, 1991; Liu et al., 1995; Sealy et al., 1997; Pic et al., 2000; Shuh and Derse, 2000). However, in these cases, the SBM and MBM regions have not been mapped. Only weak sequence homologies with the SRF B-box and MCM1 MBM occur for these proteins in either orientation. Assuming the β-strand interaction again occurs for these factors, the influence of domain structure, linker flexibility and length, and the contribution of immediately adjacent sequences, such as the 310 structures in SAP-1, weaken the effectiveness of searching for sequence homology. Furthermore, we can not exclude the possibility that other secondary structure motifs interact with SRF and MCM1, and that the hydrophobic groove formed by the β-loop and βII extends the interaction surface.
MADS-box proteins of Type I (e.g. SRF and MCM1) apparently diverged evolutionarily from those of Type II (MEF2) prior to the divergence of plants and animals (Alvarez-Buylla et al., 2000). Most notably, the sequence and structure of the domain connected immediately C-terminal to the MADS-box are different and designated the SAM and MEF2 domains for Type I and II, respectively (Shore and Sharrocks, 1995). A structural comparison revealed that these domains, which both contain two roughly parallel α-helices lying on the MADS-box β-sheet, are oriented at ∼90° to each other (Huang et al., 2000; Santelli and Richmond, 2000). The hydrophobic groove formed in SRF by βII and the SAM domain with which SAP-1 F150 interacts are not conserved in MEF2A. Furthermore, the interaction of the SAP-1 B-box with the phosphate backbone of both DNA strands is only possible because of the local bending of the DNA by 35° induced by SRF. The overall DNA bending of only 17° by MEF2A does not enable these interactions. These results are consistent with the finding that MEF2 transcription factors are not able to interact with either TCF or Fli-1 (Shore and Sharrocks, 1995). In contrast, the heterodimeric bHLH protein E12/MyoD does bind both SRF and MEF2, suggesting that its interactions are made with a conserved region of the MADS-box and possibly independently of DNA bending (Molkentin et al., 1995; Groisman et al., 1996).
SRF provides a versatile binding partner in the determination of promoter specificity for a variety of genes. The B-box, and as suggested here, MBM-like sequences, can play a role in the cooperative binding of factors interacting with SRF, possibly even in the absence of a factor-specific DNA-binding site. The SAP-1–SRF–DNA and MATα2– MCM1–DNA structures indicate that significant interaction can also occur between the MADS-box and heterologous, DNA-binding domains. This mode of binding is possibly the principal interaction in cases of the homeodomain protein Nkx2.5 and the zinc finger protein GATA-4 (Chen and Schwartz, 1996; Belaguli et al., 2000). Molecular structures of transcription factor complexes containing MADS-box proteins have illustrated the importance of specific interaction not only between folded domains, but also between these domains and flexible segments of the polypeptide chain. High resolution structures of MADS-box proteins in combination with members of further transcription factor families should be equally revealing.
Materials and methods
Preparation and crystallization of the SAP-1–SRF–DNA ternary complex
The SRF core polypeptide (amino acids 132–223) was expressed in bacteria culture and purified as described (Pellegrini et al., 1995). A segment of pSAP1A provided by R.Treisman encoding SAP-1 (amino acids 1–156) was PCR amplified and subcloned into a pET3a vector already containing the coding sequence for an N-terminal His6 tag followed by thioredoxin (TRX), a poly-glycine linker and a cleavage site for TEV NIa protease (Gibco Corp.). The resulting His6TRXSAP1a fusion polypeptide was expressed in Escherichia coli BL21 (DE3) pLysS at 37°C and affinity purified from whole-cell extract made 7 M in urea by TALON (Clontech) chromatography. The protein was refolded by dialysis against a solution [250 mM NaCl, 20 mM Tris pH 7.5, 0.5 mM EDTA, 1 mM dithiothreitol (DTT)] containing 75 mM arginine in the first step and without in the second. The fusion polypeptide was cleaved by incubating the protein for 24 h at 4°C with TEV NIa protease. SAP-1 was purified to homogeneity by cation exchange and hydrophobic chromatography at 4°C (SP-5PW and Phenyl-5PW TSK) and concentrated to 2.5 mg/ml yielding 1.1 mg per liter of culture. N-terminal sequencing and MALDI-TOF confirmed the integrity of the cleavage product. Oligonucleotides were synthesized with an Applied Biosystems 380B synthesizer and purified by reverse phase HPLC. After mixing of SRF core, SAP-1 and DNA in a 1:1:1 ratio at 1.5 mg/ml, the ternary complex was purified by gel electrophoresis (Bio-Rad) at 4°C using a 10% (w/v) acrylamide (acrylamide/bis-acrylamide, 40:1, w/w) gel containing 44.5 mM Tris base/boric acid, 1.25 mM EDTA, eluted into 5 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and concentrated to 5 mg/ml. Diffraction quality crystals were obtained using 26 bp double-stranded DNA oligonucleotides having G to C complementary ends. Crystals appeared at 4°C within 1–2 weeks using vapor diffusion with 10 µl of hanging drops containing 2.5 mg/ml ternary complex, 8–10% PEG1500, 25 mM bis-Tris pH 6.3–6.8, 50 mM NH4NO3, 0.5 mM DTT and 500 µl of reservoir solutions lacking complex but containing twice the drop concentration of salt, buffer and precipitant.
Crystallographic structure determination
Crystals (0.3 × 0.3 × 0.7 mm) were harvested into their reservoir solution containing 20% PEG1500, and through addition of glycerol in 2.5% steps to 30% followed by flash-cooling into liquid nitrogen- cooled, liquid propane, the diffraction pattern was improved from 6 Å at 4°C to 3.5 Å at –180°C, as measured using a Rigaku RU200 X-ray generator equipped with a MAR180 image plate detector. A complete dataset to 3.0 Å was collected at the ESRF, Grenoble, France, beam-line ID-14-2 using a Quantum 4 CCD detector and processed to structure factors with DENZO and SCALEPACK (Otwinowski, 1993). The structure was solved by molecular replacement using a combined model of SRF core and SAP-1 DNA-binding domains [Protein Data Bank (PDB) codes 1srs and 1bc7] in conjunction with the programs AMoRe (Navaza, 1994) and CNS (Brunger et al., 1998). An initial 2Fo – Fc map calculated using only the SRF–DNA structure showed electron density for the additional DNA in the ternary complex, the α-helices of the SAP-1 ETS domain and B-box. The molecular replacement solution was further confirmed with SIR phases obtained from a methylmercury derivative (24 h soak in 1 mM CH3HgNO3) collected at ID-14-2. The molecular replacement solution phases were used to calculate a difference Fourier map incorporating the derivative data that clearly identified the positions of four heavy atom sites which were refined and included in the overall phase calculation with MLPHARE (Otwinowski, 1991). A single round of solvent flattening and NCS averaging using the program DM (CCP4, 1994; Cowtan, 1994) were used for SIR phase improvement and model building proceeded using sigmaA-weighted 2Fo – Fc and Fo – Fc maps displayed with the program O (Jones et al., 1991). Structure refinement was carried out using data from 51.0 to 3.15 Å resolution with NCS restraints between two independent ternary complexes in the asymmetric unit applied during the initial positional refinement. The model was improved further by simulated annealing refinement using CNS and combined energy minimization and B-factor refinement using REFMAC (Murshudov et al., 1997). NCS restraints were released during this process, which converged to 28.5% R-free and 25.5% R-working (Table I). The final model was checked for errors with simulated annealing omit maps (Brunger et al., 1998). The N-terminal four amino acids in all four copies of the SRF core monomer and amino acids 0–2 and 94–136 of both SAP-1 copies in the asymmetric unit were not included in the model due to disorder. The electron density map is weakest in the region of the SAP-1 ETS domain that contributes all the outliers in the Ramachandran plot (Table I). The B-factors for SAP-1 are 1.5 to two times higher than for the DNA and SRF core less the N-extension. Protein and DNA parameters were calculated using CNS (Brunger et al., 1998), PROCHECK (Laskowski et al., 1993) and CURVES (Lavery and Sklenar, 1988). Molecular images were generated using programs MidasPlus (Ferrin et al., 1988), O (Jones et al., 1991) and WebLabViewerPro 3.5 (Molecular Simulations Inc.). Both the atomic coordinates and the structure-factor amplitudes have been deposited in the PDB database (code 1hbx).
Acknowledgments
Acknowledgements
The authors are grateful to R.Treisman (Imperial Cancer Research Fund, London, UK) for the pSAP1a plasmid. We thank D.Sargent for assistance in X-ray data collection and the staff at ESRF beam line ID14-2 (Grenoble, France) for excellent technical support. This work was supported in part by the European TMR Network ‘Signaling Networks in Development and Disease’ and the Swiss National Science Foundation.
References
- Alvarez-Buylla E.R., Pelaz,S., Liljegren,S.J., Gold,S.E., Burgeff,C., Ditta,G.S., Ribas de Pouplana,L., Martinez-Castilla,L. and Yanofsky,M.F. (2000) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl Acad. Sci. USA, 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaguli N.S., Sepulveda,J.L., Nigam,V., Charron,F., Nemer,M. and Schwartz,R.J. (2000) Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol., 20, 7550–7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cahill M.A., Nordheim,A. and Janknecht,R. (1994) Co-occurrence of CArG boxes and TCF sites within viral genomes. Biochem. Biophys. Res. Commun., 205, 545–551. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. and Schwartz,R.J. (1996) Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol. Cell. Biol., 16, 6372–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) ‘dm’: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, 31, 34–38. [Google Scholar]
- Cruzalegui F.H., Cano,E. and Treisman,R. (1999) ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene, 18, 7948–7957. [DOI] [PubMed] [Google Scholar]
- Dalgleish P. and Sharrocks,A.D. (2000) The mechanism of complex formation between Fli-1 and SRF transcription factors. Nucleic Acids Res., 28, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S. and Treisman,R. (1992) Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell, 68, 597–612. [DOI] [PubMed] [Google Scholar]
- Donaldson L.W., Petersen,J.M., Graves,B.J. and McIntosh,L.P. (1996) Solution structure of the ETS domain from murine Ets-1: a winged helix–turn–helix DNA binding motif. EMBO J., 15, 125–134. [PMC free article] [PubMed] [Google Scholar]
- El Hassan M.A. and Calladine,C.R. (1997) Conformational character istics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos. Trans. R. Soc., 355, 43–100. [Google Scholar]
- Ferrin T., Huang,C., Jarvis,L. and Langridge,R. (1988) The MIDAS display system. J. Mol. Graph., 6, 13–27. [Google Scholar]
- Fujii M., Tsuchiya,H., Chuhjo,T., Akizawa,T. and Seiki,M. (1992) Interaction of HTLV-1 Tax1 with p67SRF causes the aberrant induction of cellular immediate early genes through CArG boxes. Genes Dev., 6, 2066–2076. [DOI] [PubMed] [Google Scholar]
- Gille H., Kortenjann,M., Strahl,T. and Shaw,P.E. (1996) Phosphoryl ation-dependent formation of a quaternary complex at the c-fos SRE. Mol. Cell. Biol., 16, 1094–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginty D.D., Bonni,A. and Greenberg,M.E. (1994) Nerve growth factor activates a ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell, 77, 713–725. [DOI] [PubMed] [Google Scholar]
- Giovane A., Pintzas,A., Maira,S.M., Sobieszczuk,P. and Wasylyk,B. (1994) Net, a new ets transcription factor that is activated by Ras. Genes Dev., 8, 1502–1513. [DOI] [PubMed] [Google Scholar]
- Groisman R., Masutani,H., Leibovitch,M.P., Robin,P., Soudant,I., Trouche,D. and Harel-Bellan,A. (1996) Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix–loop–helix proteins. J. Biol. Chem., 271, 5258–5264. [DOI] [PubMed] [Google Scholar]
- Grueneberg D.A., Natesan,S., Alexandre,C. and Gilman,M.Z. (1992) Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science, 257, 1089–1095. [DOI] [PubMed] [Google Scholar]
- Heidenreich O. et al. (1999) MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J. Biol. Chem., 274, 14434–14443. [DOI] [PubMed] [Google Scholar]
- Hipskind R.A., Baccarini,M. and Nordheim,A. (1994) Transient activation of RAF-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol. Cell. Biol., 14, 6219–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Louis,J.M., Donaldson,L., Lim,F.L., Sharrocks,A.D. and Clore,G.M. (2000) Solution structure of the MEF2A–DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J., 19, 2615–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R. and Hunter,T. (1997) Convergence of MAP kinase pathways on the ternary complex factor Sap-1a. EMBO J., 16, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R. and Nordheim,A. (1996a) MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun., 228, 831–837. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Nordheim,A. (1996b) Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene, 12, 1961–1969. [PubMed] [Google Scholar]
- Janknecht R., Ernst,W.H., Pingoud,V. and Nordheim,A. (1993) Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J., 12, 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Laskowski R., MacArthur,M., Moss,D. and Thornton,J. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Latinkic B.V., Zeremski,M. and Lau,L.F. (1996) Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic Acids Res., 24, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Ling Y., Lakey,J.H., Roberts,C.E. and Sharrocks,A.D. (1997) Molecular characterization of the B-box protein–protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J., 16, 2431–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Peng,B., Ma,J., Liu,Y. and Ng,S. (1995) Serum response element associated transcription factors in mouse embryos: serum response factor, YY1, and PEA3 factor. Dev. Genet., 16, 229–240. [DOI] [PubMed] [Google Scholar]
- Lopez M., Oettgen,P., Akbarali,Y., Dendorfer,U. and Libermann,T.A. (1994) ERP, a new member of the ets transcription factor/oncoprotein family: cloning, characterization, and differential expression during B-lymphocyte development. Mol. Cell. Biol., 14, 3292–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Masutani,H., Robin,P., Lipinski,M. and Harel-Bellan,A. (1996) SRE elements are binding sites for the fusion protein EWS–FLI-1. Nucleic Acids Res., 24, 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R., Wynne,J. and Treisman,R. (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell, 73, 381–393. [DOI] [PubMed] [Google Scholar]
- Masutani H., Magnaghi-Jaulin,L., Ait-Si-Ali,S., Groisman,R., Robin,P., Harel-Bellan,A. and Ait-Si-Ali,S. (1997) Activation of the c-fos SRE through SAP-1a. Oncogene, 15, 1661–1669. [DOI] [PubMed] [Google Scholar]
- Mo Y., Vaessen,B., Johnston,K. and Marmorstein,R. (1998) Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol. Cell, 2, 201–212. [DOI] [PubMed] [Google Scholar]
- Mo Y., Vaessen,B., Johnston,K. and Marmorstein,R. (2000) Structure of the elk-1–DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA. Nature Struct. Biol., 7, 292–297. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Black,B.L., Martin,J.F. and Olson,E.N. (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell, 83, 1125–1136. [DOI] [PubMed] [Google Scholar]
- Montaner S., Perona,R., Saniger,L. and Lacal,J.C. (1999) Activation of serum response factor by RhoA is mediated by the nuclear factor-κB and C/EBP transcription factors. J. Biol. Chem., 274, 8506–8515. [DOI] [PubMed] [Google Scholar]
- Mueller C.G. and Nordheim,A. (1991) A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J., 10, 4219–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Natesan S. and Gilman,M. (1995) YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol. Cell. Biol., 15, 5975–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Norman C., Runswick,M., Pollock,R. and Treisman,R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell, 55, 989–1003. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1991) Isomorphous replacement and anomalous scattering. In Wolf,W., Evans,P.R. and Leslie,A.G.W. (eds), Proceedings of the CCP4 Study Weekend: Data Collection and Processing. SERC Daresbury Laboratory, Warrington, UK, pp. 55–62.
- Otwinowski Z. (1993) Oscillation data reduction program. In Wolf,W., Evans,P.R. and Leslie,A.G.W. (eds), Proceedings of the CCP4 Study Weekend: Data Collection and Processing. SERC Daresbury Laboratory, Warrington, UK, pp. 55–62.
- Paradis P., MacLellan,W.R., Belaguli,N.S., Schwartz,R.J. and Schneider,M.D. (1996) Serum response factor mediates AP-1-dependent induction of the skeletal α-actin promoter in ventricular myocytes. J. Biol. Chem., 271, 10827–10833. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Tan,S. and Richmond,T.J. (1995) Structure of serum response factor core bound to DNA. Nature, 376, 490–498. [DOI] [PubMed] [Google Scholar]
- Pic A. et al. (2000) The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J., 19, 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.A., Rogers,A.E. and Treisman,R. (1995) Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J., 14, 2589–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Winkler,H. and Ammerer,G. (1991) The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J., 10, 4209–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V.N., Huebner,K., Isobe,M., ar-Rushdi,A., Croce,C.M. and Reddy,E.S. (1989) elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science, 244, 66–70. [DOI] [PubMed] [Google Scholar]
- Santelli E. and Richmond,T.J. (2000) Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J. Mol. Biol., 297, 437–449. [DOI] [PubMed] [Google Scholar]
- Schroeter H., Mueller,C.G.F., Meese,K. and Nordheim,A. (1990) Synergism in ternary complex formation between the dimeric glycoprotein p67-SRF, polypeptide p62-TCF and the c-fos serum response element. EMBO J., 9, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy L., Malone,D. and Pawlak,M. (1997) Regulation of the c-fos serum response element by C/EBPβ. Mol. Cell. Biol., 17, 1744–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P. and Cochran,B.H. (1991) MATα1 can mediate gene activation by a-mating factor. Genes Dev., 5, 1924–1934. [DOI] [PubMed] [Google Scholar]
- Shaw P.E., Schroeter,H. and Nordheim,A. (1989) The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell, 56, 563–572. [DOI] [PubMed] [Google Scholar]
- Shore P. and Sharrocks,A.D. (1994) The transcription factors Elk-1 and serum response factor interact by direct protein–protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol., 14, 3283–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P. and Sharrocks,A.D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Shore P., Whitmarsh,A.J., Bhaskaran,R., Davis,R.J., Waltho,J.P. and Sharrocks,A.D. (1996) Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol., 16, 3338–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuh M. and Derse,D. (2000) Ternary complex factors and cofactors are essential for human T-cell leukemia virus type 1 Tax transactivation of the serum response element. J. Virol., 74, 11394–11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. and Richmond,T.J. (1998) Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature, 391, 660–666. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1990) The SRE: a growth factor responsive transcriptional regulator. Semin. Cancer Biol., 1, 47–58. [PubMed] [Google Scholar]
- Treisman R. (1995) Journey to the surface of the cell: Fos regulation and the SRE. EMBO J., 14, 4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Marais,R. and Wynne,J. (1992) Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J., 11, 4631–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D.K., Robinson,L., Hodge,D.R., Kola,I., Papas,T.S. and Seth,A. (1997) FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene, 14, 213–221. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J., Shore,P., Sharrocks,A.D. and Davis,R.J. (1995) Integration of MAP kinase signal transduction pathways at the serum response element. Science, 269, 403–407. [DOI] [PubMed] [Google Scholar]
- Williams G.T. and Lau,L.F. (1993) Activation of the inducible orphan receptor gene nur77 by serum growth factors: dissociation of immediate-early and delayed-early responses. Mol. Cell. Biol., 13, 6124–6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Shore,P., Willingham,N., Lakey,J.H. and Sharrocks,A.D. (1999) The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J., 18, 5666–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]