Abstract
A major rate-limiting step in transcription initiation by RNA polymerase II is recognition and binding of the TATA element by the transcription factor TFIID. TFIID is composed of TATA binding protein (TBP) and approximately a dozen TBP-associated factors (TAFs). Emerging consensus regarding the role of TAFs is that TFIID assumes a gene specific activity that is regulated by interaction with other factors. In spite of many studies demonstrating the essential nature of TAFs in transcription, very little is known about the subunit contacts within TFIID. To understand fully the functional role of TAFs, it is imperative to define TAF–TAF interactions and their topological arrangement within TFIID. We performed a systematic two-hybrid analysis using the 13 essential TAFs of the Saccharomyces cerevisiae TFIID complex and TBP. Specific interactions were defined for each component, and the biological significance of these interactions is supported by numerous genetic and biochemical studies. By combining the interaction profiles presented here, and the available studies utilizing specific TAFs, we propose a working hypothesis for the arrangement of components in the TFIID complex. Thus, these results serve as a foundation for understanding the overall architecture of yeast TFIID.
INTRODUCTION
Transcription initiation by eukaryotic RNA polymerase II (pol II) requires the concerted action of various general transcription factors such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH in addition to pol II (1–3). The first step in forming a functional pre-initiation complex is the interaction of TFIID with the promoter. TFIID is a multi-protein complex that contains TATA binding protein (TBP) and over a dozen different TBP associated factors (TAFs) (4–8). In yeast, 13 of these TAFs are essential for cell viability and, as such, must play critical and non-redundant roles in the regulation of gene expression.
TAFs are not restricted to the TFIID complex, and can also be found in the yeast SAGA complex (9) and mammalian PCAF complex (10). In vivo depletion of these shared TAFs results in broad defects on transcription by pol II (11–13). In contrast, depletion of TAFs specific to the TFIID complex appears to affect transcription in a promoter-specific manner (11,12,14,15). In keeping with gene-specific roles in transcriptional regulation, certain TAFs are critical for TATA-less promoter transcription due to important TAF interactions with initiator and downstream promoter elements (16,17). Finally, although activated transcription is not generally dependent on TAFs in the yeast system (14,15), activator-specific recruitment of TFIID is important for the regulation of ribosomal protein gene transcription in yeast (18), as well as the derepression of DNA damage-regulated genes (19). Likewise, activated transcription in higher eukaryotic systems can be dependent on TAFs, presumably due to direct interactions between TAFs and activators (2,3,20,21). Thus, TAFs play important roles in many different aspects of the regulation of gene expression.
Although the stoichiometry of the various TFIID components has been characterized (22,23), very little is known about the overall architecture of the TFIID complex. Electron microscopy and digital image analyses of single TFIID particles indicate a C-shaped form with a central cavity (24,25), and some of the individual TAFs have been mapped within the complex using TAF-specific antibodies (26). However, a complete understanding of the TFIID complex necessitates a cataloging of the specific TAF–TAF interactions within the complex. Here we describe an extensive two-hybrid analysis of protein–protein interactions between all of the essential components of yeast TFIID. The majority of these interactions are consistent with previously reported biochemical and/or genetic data dealing with the individual components. The two-hybrid results, coupled with the large volume of biochemical and genetic data regarding the individual subunits of TFIID, were used to construct a model of the topological arrangement of TAFs in the TFIID complex. This TFIID interaction map serves as an important step in a comprehensive analysis of TFIID.
MATERIALS AND METHODS
DNA constructs
Activation domain (AD) hybrids were cloned into the 2µ LEU2 marked vector, pACT2.2 (27), which contains the ADH1 promoter, a nuclear localization sequence, the hemagglutinin (HA) epitope and the Gal4 AD (residues 768–881). All the AD–TAF constructs were created using PCR and homologous recombination cloning in yeast. Full length DNA binding domain (DB)–TAF hybrids were created by subcloning from the respective AD construct into the pPC97-TRP1 vector (CEN, TRP1), which contains the ADH1 promoter, a nuclear localization sequence and the Gal4 DB (residues 1–147) (28). For those DB–TAF fusions that were positive for transcriptional activation when expressed in yeast cells (i.e. that could artificially recruit the transcription machinery), deletions in the open reading frame (ORF) of the particular TAF were cloned into the DB fusion vectors. Three derivatives were designed for TAF1, corresponding to amino acids 1–208, 208–367 and 367–1066. For TAF12, two regions containing amino acid residues 1–280 and 280–539 were tested. Deletions for TAF3 were created by cloning amino acids 1–81 and 81–353 into pPC97-TRP1 vector.
Yeast strains
All Saccharomyces cerevisiae strains used were transformants of either MaV103 (28) or CG1945 (Clontech). MaV103 contains the GAL1 promoter (with four Gal4 binding sites) fused to the HIS3 TATA element and coding sequence; GAL4 and GAL80 are both deleted in the strain. CG1945 contains the GAL1 promoter and the GAL1 TATA element fused to the HIS3 coding sequence; GAL4 and GAL80 are both deleted in the strain. CG1945 cells have much tighter control of background levels of expression from the HIS3 gene.
Protein expression of two-hybrid constructs
Extracts were prepared from strains harboring the indicated TAF fusion constructs essentially as described in Cormack and Struhl (29). Approximately 25–30 µg of protein was loaded onto the SDS–PAGE gels. Immunoblots were performed using monoclonal antibodies directed against HA (BAbCO) at 1:1000 dilution, and the anti-mouse secondary antibody (Promega) was prepared at 1:20 000. Signals were developed using a chemiluminiscence kit (Pierce).
Artificial recruitment and two-hybrid assays
Each DB–TAF fusion was transformed into yeast strain MaV103 or CG1945 using a standard lithium acetate transformation. The ability to activate transcription by each DB–TAF fusion was tested by streaking the strain onto media containing 3-aminotriazole (AT), at a concentration of 40 mM for MaV103 or 3–5 mM for CG1945. Cells were grown at 30°C for 3–7 days. Growth on this media indicated the ability to artificially activate the transcription of a HIS3 reporter gene. For those DB–TAF hybrids that were positive for artificial recruitment in both MaV103 and CG1945, truncations were designed in the ORFs of the TAF and these constructs were tested in the artificial recruitment assay. Fusions that were negative in the artificial recruitment assay were transformed with the AD–TAF fusions and assayed for two-hybrid interactions by streaking to media containing AT. Cell growth on AT-containing media indicated a positive two-hybrid interaction between the two TAFs that were assayed. Scoring from + to +++ was based on the relative amount of cell growth for each DB–TAF strain containing the different AD–TAF derivatives. ‘+’ indicates growth in the primary streak only, and denotes a weak two-hybrid interaction which was significantly more than the growth level observed with the AD vector alone. ‘++’ indicates intermediate level of growth which is indicative of a stronger interaction, whereas ‘+++’ indicates robust growth and the strongest interaction. TAF2 and the deletion constructs for TAF1 and TAF3 were assayed in strain MaV103 for the two-hybrid studies. The remaining TAFs were assayed in strain CG1945.
RESULTS
Characterization of TFIID components in an artificial recruitment assay
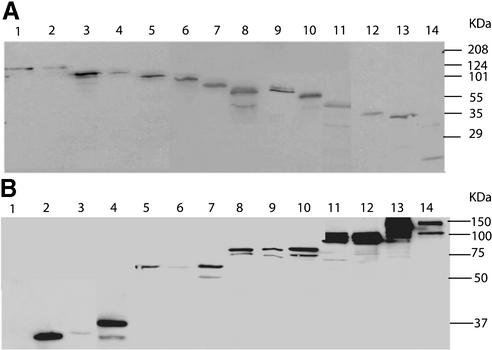
Two-hybrid analysis is a useful tool for mapping protein– protein interactions between subunits of transcription-related complexes (30), and defining and disrupting interactions between transcription factors (28). We set out to map the protein–protein interactions in the yeast TFIID complex using a systematic two-hybrid assay with all of the essential components of this complex. The 13 essential yeast TAFs (Table 1) and TBP were individually fused to the Gal4 DB (residues 1–147 of Gal4) and the Gal4 AD (residues 768–881). The fusions were tagged with the HA epitope and expression of each was confirmed by immunoblot analyses (Fig. 1).
Table 1. Pol II TAF nomenclature used in the present study.
| Name | S.cerevisiae | Homo sapiens |
|---|---|---|
| TAF1 | TAF145/130 | TAF250 |
| TAF2 | TAF150 | TAF150 |
| TAF3 | TAF47 | TAF140 |
| TAF4 | TAF48 | TAF130/135 |
| TAF5 | TAF90 | TAF100 |
| TAF6 | TAF60 | TAF80 |
| TAF7 | TAF67 | TAF55 |
| TAF8 | TAF65 | (BAB71460) |
| TAF9 | TAF17 | TAF32/31 |
| TAF10 | TAF25 | TAF30 |
| TAF11 | TAF40 | TAF28 |
| TAF12 | TAF61/68 | TAF20/15 |
| TAF13 | TAF19 | TAF18 |
Reference (71).
Figure 1.
(A) Expression of AD–TAF fusions in vivo. Protein extracts (25 µg) from strains harboring the indicated derivatives were loaded on 10% SDS–PAGE gels, and AD fusions were detected using antibodies directed against the HA epitope. Lanes (1–14) show the AD–TAF fusion constructs in descending order of molecular weight. Lane 1, AD–TAF2; lane 2, AD–TAF1; lane 3, AD–TAF5; lane 4, AD–TAF7; lane 5, AD–TAF8; lane 6, AD–TAF12; lane 7, AD–TAF6; lane 8, AD–TAF4; lane 9, AD–TAF3; lane 10, AD–TAF11; lane 11, AD–TAF10; lane 12, AD–TAF13; lane 13, AD–TAF9; lane 14, AD. (B) Expression of DB–TAF fusions in vivo. Protein extracts (25 µg) from strains harboring the indicated derivatives were loaded on 12% SDS–PAGE gels, and DB fusions were detected using antibodies directed against the HA epitope. Lanes (1–14) show the DB–TAF fusion constructs in ascending order of molecular weight. Lane 1, DB (no HA tag); lane 2, DB–TAF9; lane 3, DB–TAF13; lane 4, DB–TAF10; lane 5, DB–TAF11; lane 6, DB–TAF3; lane 7, DB–TAF4; lane 8, DB–TAF6; lane 9, DB–TAF12; lane 10, DB–TAF8; lane 11, DB–TAF7; lane 12, DB–TAF5; lane 13, DB–TAF1; lane 14, DB–TAF2. Although each DB–TAF fusion construct was easily detectable, the expression levels varied. Since each DB–TAF strain is transformed with the set of AD–TAFs and these are compared only with each other for relative growth on AT (and not to other DB–TAF strains), these differences in DB–TAF levels should have little effect on the scoring process for TAF–TAF interactions.
DB-fusions were tested for activation by artificial recruitment in strain MaV103 (28), in which binding sites for Gal4 control the transcription of the HIS3 gene. Cells can be assayed for HIS3 expression by growth on 3-AT, a competitive inhibitor of the HIS3 gene product. Like DB–TBP (31–33), 11 of the DB–TAF constructs conferred the ability to grow on AT (Table 2 and Fig. 2). Thus, tethering these particular TAFs to a promoter allows for artificial recruitment of the transcription machinery, and expression of the HIS3 gene. Although very useful for demonstrating functional activity of the DB–TAF fusions, inherent activation precludes the use of this strain background in a two-hybrid assay, since the DB-fusions are positive for growth in the absence of a Gal4 AD fusion.
Table 2. Results of artificial recruitment assays.
| DB constructs | MaV103 | CG1945 |
|---|---|---|
| DB | –a | – |
| DB–TBP | +b | – |
| DB–TAF1 | + | + |
| DB–TAF2 | – | – |
| DB–TAF3 | + | + |
| DB–TAF4 | + | – |
| DB–TAF5 | + | – |
| DB–TAF6 | + | – |
| DB–TAF7 | + | – |
| DB–TAF8 | + | – |
| DB–TAF9 | + | – |
| DB–TAF10 | + | – |
| DB–TAF11 | + | – |
| DB–TAF12 | + | + |
| DB–TAF13 | – | – |
a‘–’ Denotes no growth on AT and thus was negative for activated transcription from the HIS3 reporter gene.
b‘+’ Denotes that DB–TAF fusion construct was positive for growth on AT and thus could activate transcription from the HIS3 reporter gene.
Figure 2.
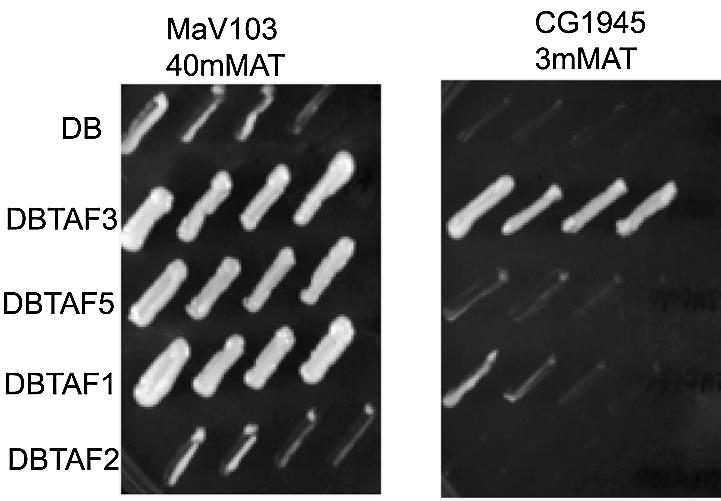
Representative study of the artificial recruitment assay. Two strains of yeast, MaV103 and CG1945, which differ in promoter context for the HIS3 reporter gene, were transformed with each of the indicated DB–TAF fusion constructs and assayed for the ability to grow on AT containing media. Cell growth on AT (40 mM for MaV103 or 3 mM for CG1945) is indicative of activated expression of HIS3 reporter gene. Yeast growth in each panel is a serial dilution of slashes from a single colony. All strains grew robustly on control plates of synthetic complete media lacking leucine and tryptophan (not shown).
We have previously shown that the activity of artificially recruited TBP is dependent on the reporter promoter (34). Others have also demonstrated that activation by artificial recruitment is highly sensitive to promoter sequence and context (35–37). For example, in MaV103 cells, in which the Gal4-driven promoter contains the HIS3 TATA element and the HIS3 structural gene, DB–TBP activates transcription (Table 2) (34). In contrast, DB–TBP fails to activate transcription in the yeast strain CG1945 (Clontech), which contains a Gal4-driven promoter with the GAL1 TATA element fused to the HIS3 structural gene. Due to the differential function of DB–TBP in the two strains, we tested the DB–TAF fusions in CG1945. Like TBP, 10 of the TAFs failed to activate HIS3 gene expression when recruited to the promoter in this strain (Table 2). Thus, these DB–TAF fusion strains were appropriate for use in a systematic two-hybrid analysis.
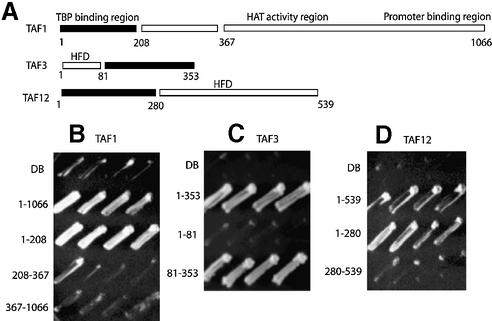
Delineation of regions competent for artificial recruitment in TAF1, TAF3 and TAF12
Three of the DB–TAF derivatives remained competent for activation in yeast strain CG1945. In order to define, and attempt to excise, a region of each of these TAFs that conferred the potent artificial recruitment function, we designed a set of deletion derivatives for each (Fig. 3). Yeast TAF1 corresponds to an ORF of 1066 amino acids, and has several interesting domains including a TBP-interacting domain (38–41), a central conserved region containing the histone acetyltransferase activity (42) and a promoter- interacting region near the C-terminus (43). We found that the artificial recruitment function for TAF1 was restricted to the N-terminal 208 amino acids, whereas the more C-terminal regions (encompassing residues from 208 to 367 or from 367 to 1066) were negative for artificial recruitment. Yeast TAF3 is encoded by an ORF of 353 amino acids, and has a histone fold domain (HFD) located near the N-terminus (44). The artificial recruitment competent region of TAF3 was mapped to the region containing residues 81–353, while the DB fusion to residues 1–81 of TAF3 failed to activate transcription. TAF12, a protein of 539 amino acids, has a non-conserved N-terminus, and a C-terminal region that is essential for cell viability and also contains a HFD (45). DB fusions to an N-terminal region (residues 1–280) and a C-terminal region (residues 280–539) of TAF12 were tested. We found that the N-terminal domain was positive for the artificial recruitment, whereas the C-terminal region containing the histone fold motif was negative in the artificial recruitment assays. Interestingly, the HFDs of TAF3 and TAF12 were negative for artificial recruitment, which could be a common characteristic of HFDs, since none of the other TAFs containing the HFDs (TAF4, TAF6, TAF8, TAF9, TAF10, TAF11 and TAF13) was positive in the artificial recruitment assay in the CG1945 strain background.
Figure 3.
Artificial recruitment assay for deletion derivatives of TAF1, TAF3 and TAF12. (A) Linear representation of each of the TAFs with the regions indicated that were used in the deletion constructs. For TAF1, the N-terminal TBP binding domain, a central conserved domain and a C-terminal promoter binding region are indicated. For TAF3 and TAF12, the location of the HFD is indicated. The black box in each panel denotes the region that was positive in an artificial recruitment assay. (B) Artificial recruitment assay for deletion derivatives of TAF1. Strains containing vector alone with the DNA binding domain of Gal4 activator (DB) (control), full length DB–TAF1 (residues 1–1066) as well as DB fused deletions constructs of TAF1 as indicated were assayed for their ability to grow on AT. Cell growth on AT is indicative of activated expression of HIS3 reporter gene. (C) Artificial recruitment assay for deletion derivatives of TAF3. Strains containing the vector alone (control), full length DB–TAF3 (residues 1–353) as well as DB fused deletion constructs of TAF3 as indicated were assayed for their ability to grow on AT, indicative of activated expression of the HIS3 reporter gene. (D) Artificial recruitment assay for deletion derivatives of TAF12. Strains containing the vector alone (control), full length DB–TAF12 (residues 1–539) as well as DB fused deletions constructs of TAF12 as indicated were assayed for their ability to grow on AT, indicative of activated expression of HIS3 reporter gene.
Mapping yeast TFIID interactions using a comprehensive two-hybrid approach
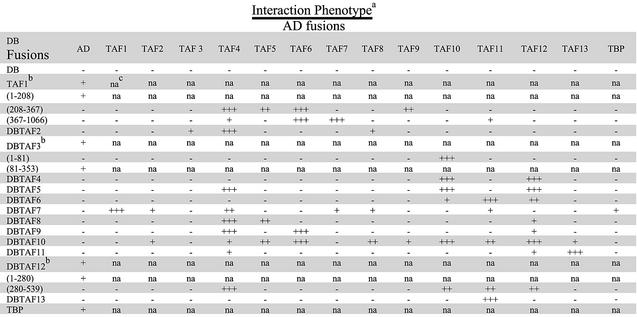
Strains expressing the DB–TAF derivatives (either full length or deletions) that were negative for activated transcription alone were independently transformed with each of the 13 AD–TAF fusion constructs, AD–TBP and the AD vector. These strains were assayed for expression of HIS3 by growth on AT (Fig. 4) and the results were scored for each individual strain. A total of 196 combinations were assayed (Table 3). We observed a total of 33 strong and intermediate interactions, which represents 17% of the interactions tested. If weak interactions are also included, 49 positive interactions were detected, which represents 25% of the assays. It is quite clear from the results that each TAF exhibited a unique interaction profile.
Figure 4.
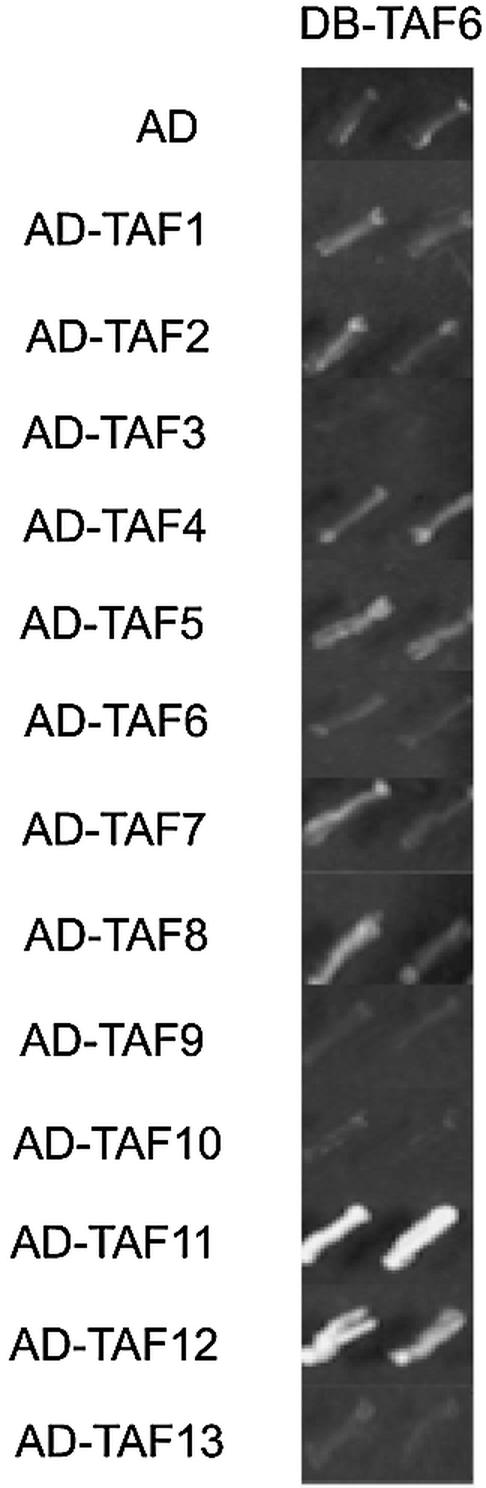
Representative panel for the systematic two-hybrid assay. Yeast strains expressing DB–TAF6 were independently transformed with different AD–TAF constructs and their ability to interact and activate the reporter HIS3 gene was assayed by growth on media containing AT (5 mM). Yeast growth in each panel is a serial dilution of slashes from a single colony. TAF6–TAF11 was scored as strongly positive (+++) and TAF6–TAF12 interaction was scored as (++). An interaction between TAF6 and TAF10 was detectable (+) when strains were streaked for single colony growth (data not shown).
Table 3. Summary of two-hybrid interactions.
aRelative growth rate on AT of strains harboring the indicated AD and DB fusions. Robust growth on AT is scored as ‘+++’ and is result of activation of HIS3 reporter gene. Intermediate and weak growths on AT are represented as ‘++’ and ‘+’, respectively.
bDeletion constructs were used instead of the full length TAFs. Numbers in parentheses indicate the amino acid residues that were used in the construct for the two-hybrid analysis.
cThe DB–TAF fusion constructs that were not appropriate for use in the two-hybrid assay since these constructs themselves activated transcription in the absence of the AD–TAF fusions.
TAF1 showed strong interactions with a number of TAFs (TAF4, TAF5, TAF6, TAF7 and TAF9), which is concordant with its portrayal as a critical scaffolding TAF for TFIID architecture in higher eukaryotes (46). In addition, we found that the 208–367 region of TAF1 exhibited both distinct and overlapping interaction profiles when compared with the 367–1066 region. Residues 208–367 specifically interacted strongly with TAF4, TAF5 and TAF9, whereas TAF6 interacted with both this region and the more C-terminal region of TAF1 (residues 367–1066). Interestingly, a TAF1 derivative with most of this region deleted (residues 208–303) is not competent for interactions in vivo with TBP or other TAFs (43), suggesting that the integrity of the TFIID complex is dependent on interactions specific to this region. TAF7 interacted exclusively with the more C-terminal region of TAF1 (residues 367–1066). An intact C-terminal region of TAF1 is essential for interaction with promoter DNA (43), and it is interesting to speculate that interaction with this TAF may be important for the functional role of TFIID in promoter interactions. It should be noted that TAF1 interactions with TAF4, TAF5 and TAF7 have also been reported for their higher eukaryotic homologs (47–49).
The distinctive nature of the interaction profile for each TAF was especially striking for a few of the TAFs with a HFD. For example, HFD-containing TAF13 interacted strongly with HFD-containing TAF11. The TAF11–TAF13 interaction is supported by genetic studies in yeast (50) and structural studies of the human homologs of these two TAFs (51). In contrast to the specificity of TAF13 for TAF11, TAF11 exhibited strong interactions with several other HFD- containing TAFs: TAF6, TAF10 and TAF12 and, at least in the case of TAF12, this interaction maps to the region (residues 280–539) that contains the HFD (Table 3).
HFD-containing TAF4 (H2A-like) and TAF12 (H2B-like), and TAF9 (H3-like) and TAF6 (H4-like), exhibited strong reciprocating interactions. These interactions are entirely consistent with the predictive specificity from their histone counterparts (6), biochemical data describing an octamer complex containing these particular TAFs (52) and studies on their human counterparts (53,54). Our results yield further information in that we detected cross interactions between TAFs that resemble core histones other than their predicted heterodimeric partners. We show that TAF4 interacts with TAF9, and TAF6 interacts with TAF12. In fact, the region of TAF12 harboring the HFD not only interacts with its histone-like partner TAF4 (54,55), but also with HFD-containing TAF10 and TAF11. Interestingly, the TAF12–TAF6 interaction appears to map to the N-terminal region of TAF12 lacking the HFD since elimination of this region results in elimination of the interaction observed when the two full-length TAFs are utilized. This result underscores the important consideration that just because two HFD-containing TAFs interact, this does not indicate that the interaction is mediated via the HFD.
TBP makes very few TAF interactions
Although DB–TBP does not artificially recruit the transcription machinery in the CG1945 strain, co-expressions of the Gal4 AD vector resulted in robust growth on AT (Table 3). We presume this effect is the result of an interaction between TBP and the AD. Since ADs can interact with multiple targets (reviewed in 56), it is likely that a second surface of the AD is then recruiting the remainder of the transcription machinery. Because of this background activation, DB–TBP could not be assayed with the AD–TAF fusion constructs. When TBP was fused to the AD of Gal4 (AD–TBP) and assayed with the DB–TAF derivatives, an interaction was observed with TAF7 and no other DB–TAF derivatives. This suggests that TBP may not make extensive contacts within TFIID and/or that the truncation derivatives of certain TAFs have removed TBP-interacting regions (discussed below).
DISCUSSION
We have used a systematic two-hybrid approach to map the interactions within the yeast TFIID complex. Each TFIID component has a unique interaction profile, and a majority of these interactions have supporting evidence from other studies. For example, we observed interactions between yeast TAF5 and three other TAFs (TAF1, TAF4 and TAF12). In keeping with our observations, a number of studies with human TAF5 show that it interacts with human TAF1, TAF4, TAF6, TAF9, TAF11 and TAF12 (47,57). Thus, the human homolog appears to share some of the interaction properties of the yeast counterpart. A functional interaction between yeast TAF5 and TAF1 is also supported by biochemical studies in which temperature sensitive mutations in TAF5 exhibit a concomitant depletion of TAF1 (58). As another example, we found that yeast TAF6 interacts with yeast TAF1, TAF9 and TAF12 in the two-hybrid assay. Biochemical studies on the metazoan counterpart of yeast TAF6 show that it interacts with human TAF9 and TAF12 as well (59). Further support for the veracity of the interaction profiles we observed is also garnered from biochemical and genetic studies on the HFD-containing TAFs (6,50–53). The corroboration of existing genetic and biochemical data with our interaction profiles strongly supports the robustness of utilizing the two-hybrid assay to define the protein–protein interactions within the TFIID complex.
However, there are possible limitations of the two-hybrid assay for exploring the protein–protein interactions within the TFIID complex. As is common with this assay, there is a potential for false positives and/or false negatives. Since this is an in vivo assay and all the endogenous TAFs (and other transcription factors) are present, false positives might arise from the presence of a bridging molecule acting between the two TAFs being tested. In this case, the interaction between the two fusions would not be direct. For a number of reasons, we think this unlikely. First, if bridging molecules play a major role then we would expect to see a plethora of TAF–TAF interactions. In fact, out of 196 combinations, we only observed 33 strong and intermediate interactions, which represents only 17% of those tested. Secondly, if bridging is a major confounding factor, we would predict to observe it when analyzing two TAFs for which there is a known intermediate. For example, we show that TAF11 interacts with TAF6, TAF12 and TAF13. An interaction between TAF11 and TAF13 has also been reported in other genetic and biochemical studies (50,51). If bridging were a common occurrence, we would expect that endogenous TAF11 would act as an intermediate to facilitate TAF13 and TAF6, or TAF13 and TAF12 interactions. Yet, we detect no interactions between TAF13 and TAF6, or TAF13 and TAF12, in the assay. Thus, we see no evidence supporting the contention that bridging is a major factor in the interpretation of our results. With regard to false negatives, there are certain to be interactions that are not detectable in the two-hybrid assay. These interactions could be too weak for detection or they may be transient. In addition, our fusion design, which places the DB or the AD at the N-terminus of the TAF, could interfere with certain TAF–TAF interactions. This could explain the observation that some of the reciprocal interactions are not detected in the two-hybrid assays. In spite of these limitations, our interaction profiles are a useful step in understanding the overall organization of the TFIID complex.
It is interesting to note that TBP interacts only with TAF7 in this assay. Interestingly, TAF7 is specific to the TFIID complex and not found in other TAF-containing complexes (23,60). As such, this interaction would contribute to the specificity of the association of TBP with TFIID. One possibility for the paucity of interactions for TBP is that our truncation derivatives have eliminated not only the artificial recruitment competent regions of three of the TAFs, but these also may be direct interaction sites with TBP. For example, it is well established that there is an association between the N-terminus of TAF1 and TBP (38,43,61). We observed that this N-terminal region of TAF1 harbored the artificial recruitment functions of the full length TAF, thus we had to eliminate this region from the DB–TAF1 derivative used in the two-hybrid assay. Likewise, it is possible that the regions of TAF3 and TAF12 that were positive for activated transcription, may interact directly with TBP. Finally, since TBP-free TAF-containing complexes (62), and TAF-independent TBP-containing complexes have been described (63–65), it is also possible that TBP does not make extensive and strong contacts with TAFs in the yeast TFIID complex. This hypothesis is consistent with recent observations indicating that TBP dynamically associates with the stable TFIID–TAF complex and is freely exchanged with excess competitor TBP (22).
In a comparison of the interaction profiles, it is evident that several TAFs appear to occupy key positions in the TFIID complex. For example, HFD containing TAFs, TAF4, TAF6, TAF9, TAF10 and TAF12, make multiple and strong interactions with a significant number of other TAFs. Others have also shown multiple two-hybrid interactions for a subset of these TAFs (66), as well as TAF10 interactions with TAF3 and TAF8 (44). Consistent with the pivotal role of these HFD-containing TAFs in the TFIID complex, previous studies have shown that mutations in TAF6, TAF9 or TAF12 (13), or TAF10 (67), result in defects in the overall integrity of TFIID.
Based on the results presented here and evidence from other biochemical and genetic studies, we present an interaction map for TFIID of S.cerevisiae (Fig. 5). Since in vitro studies have indicated the presence of a core octamer-like substructure formed by TAFs that resemble histones (52,53), we nucleated our model using these TAF–TAF interactions, and formed the rest of the map using the strong and intermediate interactions observed.
Figure 5.
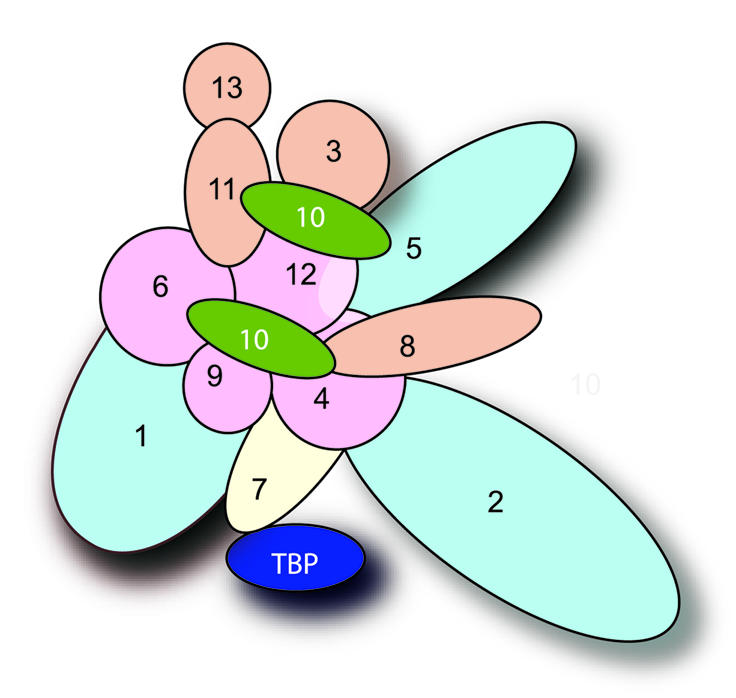
TFIID interaction map. Each oval represents a different TAF as indicated. The histone-like TAFs implicated in an octamer-like complex (52,53) are represented in pink. Other histone fold domain containing TAFs are shown in tan, except for TAF10 (shown in green). TAFs in the background are represented in light blue, TBP in dark blue. To keep the model simple, homodimeric interactions for TAF10 and TAF12 were excluded. The strongest interactions (denoted by +++ or ++ in Table 3) were most heavily weighted for inclusion in the interaction map. Weak interactions (denoted by +) were not generally utilized in the interaction map. Exceptions to this were in the cases where reciprocal interactions were strong, or for interactions with TBP. For TBP there were few total interactions, thus all interactions were accommodated in the model.
There are several striking aspects of the proposed TFIID model. First, though we found no interactions between TBP and TAF1 in the two-hybrid assay, these two components fall in close proximity to each other in the interaction map. Thus, the known interaction between TBP and the N-terminal domain of TAF1 (38–41), can be easily accommodated in the model. In fact, since the more C-terminal regions of TAF1 interact with TAF4, TAF6 and TAF9, this further orients the N-terminal domain of TAF1 toward the proposed location of TBP. It is important to note that we did not consider the interaction between the N-terminal domain of TAF1 and TBP when building the map since this domain of TAF1 activated transcription on its own, which precluded its use in the two-hybrid assay. A second interesting feature of the interaction map is that TBP interacts with TAF7, and TAF7 interacts strongly with TAF1. Thus, there is the potential for three-way interactions between TAF1, TAF7 and TBP. Since TAF1 and TAF7 are specific to the TFIID complex (23,60), this would ensure that TBP is specific for TFIID and not other TAF-containing complexes. In keeping with this idea, TAF7 interacts with HFD-containing TAF4 and, although it is partnered with TAFs shared between TFIID and SAGA, TAF4 itself is TFIID specific (5). Third, TAF2 is also situated nearby TBP, and it is tempting to speculate that this would be the region of TFIID that contacts the promoter. Studies have shown that in addition to TBP, both TAF1 and TAF2 make intimate contacts with the promoter DNA, play roles in core promoter discrimination and aid in recognizing the initiator sequences (17,68–70).
There were challenges in deriving the TFIID interaction map. For example, TAF10 was a complicated TAF to incorporate in the model. It interacts strongly with five other TAFs, and it also self-associates. In addition, it is known to be present in more than one copy in the TFIID complex (22), and appears to be located in two distinct lobes of TFIID (26). Thus, two copies of TAF10 were placed in the map to accommodate all of the strong interactions for TAF10. The fact that TAF10 has unique interaction properties is also evidenced by the observation that different mutations in TAF10 result in loss of distinct TAFs from the TFIID complex (67). As such, further studies regarding this particular TAF and its interaction partners will be required to understand fully the functional role of TAF10 in the TFIID complex.
In spite of the recent advances in biological techniques used to decipher microdetails of a protein complex, the organization of TFIID remains an enigma to date. There is a growing understanding of the components within the TFIID complex (22,23), and low-resolution images (35 Å) have been determined for TFIID (24,25). Furthermore, the nine HFD-containing TAFs have been mapped on this image (26). However, there are some apparent differences between the stoichiometry of the individual subunits (22,23,26), as some TAFs appear to be present in multiple copies via one method but not the other. These minor differences simply serve to illustrate the point that multiple approaches using distinct techniques will be critical for a complete understanding of the TFIID complex. The study presented here provides a level of detail regarding the subunit interactions in TFIID that is novel and lacking from previous studies, and offers a unique insight into the overall architecture of the yeast TFIID complex.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Karen Lehman for technical assistance with several of the two-hybrid assays. This work was supported by a grant to L.A.S. from the National Institutes of Health (GM56884).
REFERENCES
- 1.Reinberg D., Orphanides,G., Ebright,R., Akoulitchev,S., Carcamo,J., Cho,H., Cortes,P., Drapkin,R., Flores,O., Ha,I., Inostroza,J.A., Kim,S., Kim,T.K., Kumar,P., Lagrange,T., LeRoy,G., Lu,H., Ma,D.M., Maldonado,E., Merino,A., Mermelstein,F., Olave,I., Sheldon,M., Shiekhattar,R. and Zawel,L. (1998) The RNA polymerase II general transcription factors: past, present and future. Cold Spring Harb. Symp. Quant. Biol., 63, 83–103. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 3.Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 4.Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- 5.Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- 6.Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- 7.Goodrich J.A. and Tijan,R. (1994) TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr. Opin. Cell Biol., 6, 403–409. [DOI] [PubMed] [Google Scholar]
- 8.Verrijzer C.P. and Tijan,R. (1996) TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci., 21, 338–342. [PubMed] [Google Scholar]
- 9.Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D., Reese,J.C., Yates,J.R.,III and Workman,J.L. (1998) A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko V.V., Kotani,T., Zhang,X., Schiltz,R.L., Howard,T., Yang,X.-J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- 11.Moqtaderi Z., Keaveney,M. and Struhl,K. (1998) The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell, 2, 675–682. [DOI] [PubMed] [Google Scholar]
- 12.Apone L.M., Virbasius,C.-A., Holstege,F.C., Wang,J., Young,R.A. and Green,M.R. (1998) Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell, 2, 653–661. [DOI] [PubMed] [Google Scholar]
- 13.Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–672. [DOI] [PubMed] [Google Scholar]
- 14.Apone L.M., Virbasius,C.A., Reese,J.C. and Green,M.R. (1996) Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev., 10, 2368–2380. [DOI] [PubMed] [Google Scholar]
- 15.Moqtaderi Z., Bai,Y., Poon,D., Weil,P.A. and Struhl,K. (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature, 383, 188–191. [DOI] [PubMed] [Google Scholar]
- 16.Burke T. and Kadonaga,J. (1997) The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev., 11, 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chalkley G.E. and Verrijzer,C.P. (1999) DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J., 18, 4835–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mencia M., Moqtaderi,Z., Geisberg,J.V., Kuras,L. and Struhl,K. (2002) Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell, 9, 823–833. [DOI] [PubMed] [Google Scholar]
- 19.Li B. and Reese,J.C. (2000) Derepression of DNA damage-regulated genes requires yeast TAF(II)s. EMBO J., 19, 4091–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Q., Lieberman,P.M., Boyer,T.G. and Berk,A.J. (1992) Holo TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev., 6, 1964–1974. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann A., Oelgeschlager,T. and Roeder,R.G. (1997) Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc. Natl Acad. Sci. USA, 94, 8928–8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders S.L., Garbett,K.A. and Weil,P.A. (2002) Molecular Characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol., 22, 6000–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders S.L., Jennings,J., Canutescu,A., Link,A.J. and Weil,P.A. (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol., 22, 4723–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andell F. III, Ladurner,A.G., Inouye,C., Tjian,R. and Nogales,E. (1999) Three-dimensional structures of the human TFIID-IIA-IIB complex. Science, 286, 2153–2156. [DOI] [PubMed] [Google Scholar]
- 25.Brand M., Leurent,C., Mallouh,V., Tora,L. and Schultz,P. (1999) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science, 286, 2151–2153. [DOI] [PubMed] [Google Scholar]
- 26.Leurent C., Sanders,S., Ruhlmann,C., Mallouh,V., Weil,P.A., Kirschner,D.B., Tora,L. and Schultz,P. (2002) Mapping histone fold TAFs within yeast TFIID. EMBO J., 21, 3424–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durfee T., Becherer,K., Chen,P.-L., Yeh,S.-H., Yang,Y., Kilburn,A.E., Lee,W.-H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- 28.Vidal M., Brachmann,R.K., Fattaey,A., Harlow,E. and Boeke,J.D. (1996) Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein interactions. Proc. Natl Acad. Sci. USA, 93, 10315–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cormack B.P. and Struhl,K. (1992) The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell, 69, 685–696. [DOI] [PubMed] [Google Scholar]
- 30.Flores A., Briand,J., Gadal,O., Andrau,J., Rubbi,L., Van Mullem,V., Boschiero,C., Goussot,M., Marck,C., Carles,C., Thuriaux,P., Sentenac,A. and Werner,M. (1999) A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA, 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee S. and Struhl,K. (1995) Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature, 374, 820–822. [DOI] [PubMed] [Google Scholar]
- 32.Klages N. and Strubin,M. (1995) Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature, 374, 822–823. [DOI] [PubMed] [Google Scholar]
- 33.Xiao H., Friesen,J.D. and Lis,J.T. (1995) Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol., 15, 5757–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stargell L.A., Ogg,R.C., Adkins,J.N., Robinson,M.M. and Lumb,K.J. (2001) Transcriptional activity of the TFIIA four-helix bundle in vivo. Proteins, 43, 227–232. [DOI] [PubMed] [Google Scholar]
- 35.Gaudreau L., Keaveney,M., Nevado,J., Zaman,Z., Bryant,G., Struhl,K. and Ptashne,M. (1999) Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc. Natl Acad. Sci. USA, 96, 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevado J., Gaudreau,L., Adam,M. and Ptashne,M. (1999) Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl Acad. Sci. USA, 96, 2674–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan M.P., Stafford,G.A., Yu,L. and Morse,R.H. (2000) Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol., 20, 5847–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai Y., Perez,G.M., Beechem,J.M. and Weil,P.A. (1997) Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-Binding Protein interaction domain in the N terminus of yeast TAFII130. Mol. Cell. Biol., 17, 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kokubo T., Swanson,M.J., Nishikawa,J.-I., Hinnebusch,A.G. and Nakatani,Y. (1998) The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol., 18, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotani T., Miyake,T., Tsukihashi,Y., Hinnebusch,A., Nakatani,Y., Kawaichi,M. and Kokubo,T. (1998) Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem., 273, 32254–32264. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi A., Miyake,T., Ohyama,Y., Kawaichi,M. and Kokubo,T. (2001) Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J. Biol. Chem., 276, 395–405. [DOI] [PubMed] [Google Scholar]
- 42.Mizzen C.A., Yang,X., Kokubo,T., Brownell,J.E., Bannister,A.J., Owen-Hughes,T., Workman,J., Wang,L., Berger,S.L., Kouzarides,T., Nakatani,Y. and Allis,C.D. (1996) The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- 43.Mencia M. and Struhl,K. (2001) Region of yeast TAF 130 required for TFIID to associate with promoters. Mol. Cell. Biol., 21, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gangloff Y.G., Sanders,S.L., Romier,C., Kirschner,D., Weil,P.A., Tora,L. and Davidson,I. (2001) Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol., 21, 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moqtaderi Z., Yale,J.D., Struhl,K. and Buratowski,S. (1996) Yeast homologues of higher eukaryotic TFIID subunits. Proc. Natl Acad. Sci. USA, 93, 14654–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassarman D.A. and Sauer,F. (2001) TAFII250: a transcription toolbox. J. Cell Sci., 114, 2895–2902. [DOI] [PubMed] [Google Scholar]
- 47.Tao Y., Guermah,M., Martinez,E., Oelgeschlager,T., Hasegawa,S., Takada,R., Yamamoto,T., Horikoshi,M. and Roeder,R.G. (1997) Specific interactions and potential functions of human TAFII100. J. Biol. Chem., 272, 6714–6721. [DOI] [PubMed] [Google Scholar]
- 48.Weinzerl R.O., Dynlacht,B.D. and Tjian,R. (1993) Largest subunit of Drosophila transcription factor IID directs assembly of a complex containing TBP and a coactivator. Nature, 362, 511–517. [DOI] [PubMed] [Google Scholar]
- 49.Lavigne A., Mengus,G., May,M., Dubrovskaya,V., Tora,L., Chambon,P. and Davidson,I. (1996) Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J. Biol. Chem., 271, 19774–19780. [DOI] [PubMed] [Google Scholar]
- 50.Komarnitsky P.B., Michel,B. and Buratowski,S. (1999) TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev., 13, 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birck C., Poch,O., Romier,C., Ruff,M., Mengus,G., Lavigne,A., Davidson,I. and Moras,D. (1998) Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell, 94, 239–249. [DOI] [PubMed] [Google Scholar]
- 52.Selleck W., Howley,R., Fang,Q., Podolny,V., Fried,M.,G., Buratowski,S. and Tan,S. (2001) A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nature Struct. Biol., 8, 695–700. [DOI] [PubMed] [Google Scholar]
- 53.Hoffmann A., Chiang,C., Oelgeschlager,T., Xie,X., Burley,S.K., Nakatani,Y. and Roeder,R.G. (1996) A histone octamer-like structure within TFIID. Nature, 380, 356–359. [DOI] [PubMed] [Google Scholar]
- 54.Werten S., Mitschler,A., Romier,C., Gangloff,Y.,G., Thuault,S., Davidson,I. and Moras,D. (2002) Crystal structure of a subcomplex of human TFIID formed by TBP-associated factors hTAF4 (hTAFII135) and hTAF12 (hTAFII20). J. Biol. Chem., 277, 45502–45509. [DOI] [PubMed] [Google Scholar]
- 55.Gangloff Y.G., Werten,S., Romier,C., Carre,L., Poch,O., Moras,D. and Davidson,I. (2000) The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol., 20, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gill G. (2001) Regulation of the initiation of eukaryotic transcription. Essays Biochem., 37, 33–43. [DOI] [PubMed] [Google Scholar]
- 57.Furukawa T. and Tanese,N. (2000) Assembly of partial TFIID complexes in mammalian cells reveals distinct activitites assoicated with individual TATA box-binding protein-associated factors. J. Biol. Chem., 275, 29847–29856. [DOI] [PubMed] [Google Scholar]
- 58.Durso R.J., Fisher,A.K., Albright-Frey,T.J. and Reese,J.C. (2001) Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol., 21, 7331–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hisatake K., Ohta,T., Takada,R., Guermah,M., Horikoshi,M., Nakatani,Y. and Roeder,R.G. (1995) Evolutionary conservation of human TATA-binding-polypeptide-associated factors TAFII31 and TAFII80 and interactions of TAFII80 with other TAFs and with general transcription factors. Proc. Natl Acad. Sci. USA, 92, 8195–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang C.M. and Roeder,R.G. (1995) Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science, 267, [DOI] [PubMed] [Google Scholar]
- 61.Banik U., Beechem,J.M., Klebanow,E., Schroeder,S. and Weil,P.,A. (2001) Fluorescence-based analyses of the effects of full-length recombinant TAF130p on the interaction of TATA box-binding protein with TATA box DNA. J. Biol. Chem., 276, 49100–49109. [DOI] [PubMed] [Google Scholar]
- 62.Wieczorek E., Brand,M., Jacq,X. and Tora,L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- 63.Kuras L., Kosa,P., Mencia,M. and Struhl,K. (2000) TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science, 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- 64.Li X.Y., Bhaumik,S.R. and Green,M.R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science, 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- 65.Mitsiou D.J. and Stunnenberg,H.G. (2000) TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIA alphabeta precursor and the TFIIA gamma subunit. Mol. Cell, 6, 527–537. [DOI] [PubMed] [Google Scholar]
- 66.Uetz P., Giot,L., Cagney,G., Mansfield,T., Judson,R., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P., Qureshi-Emili,A., Li,Y., Godwin,B., Conover,D., Kalbfleisch,T., Vijayadamodar,G., Yang,M., Johnston,M., Fields,S. and Rothberg,J.M. (2000) A comprehenisive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 67.Kirschner D.B., vom Baur,E., Thibault,C., Sanders,S.L., Gangloff,Y.G., Davidson,I., Weil,P.A. and Tora,L. (2002) Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol., 22, 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sypes M.A. and Gilmour,D.S. (1994) Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res., 22, 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Verrijzer C.P., Yokomori,K., Chen,J.-L. and Tjian,R. (1994) Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science, 264, 933–941. [DOI] [PubMed] [Google Scholar]
- 70.Verrijzer C.P., Chen,J.-L., Yokomori,K. and Tjian,R. (1995) Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell, 81, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 71.Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]