Abstract
Endonuclease G, a protein historically thought to be involved in mitochondrial DNA (mtDNA) replication, repair, recombination and degradation, has recently been reported to be involved in nuclear DNA degradation during the apoptotic process. As a result, its involvement in mtDNA homeostasis has been called into question and has necessitated detailed analyses of its precise location within the mitochondrion. Data is presented localizing rat liver endonuclease G activity exclusively to the mitochondrial intermembrane space with no activity associated with either the interior face of the inner mitochondrial membrane or with the mitochondrial matrix. Additionally, it is shown that endonuclease G can be selectively released from the mitochondrion via induction of a Ca2+-induced mitochondrial permeability transition and that, upon its release, a further nuclease activity loosely associated with the interior face of the inner mitochondrial membrane and distinct in its properties from that of endonuclease G can be detected.
INTRODUCTION
Endonuclease G (endoG) is a mitochondrial enzyme that has been proposed to play a role in the maintenance of mitochondrial DNA (mtDNA). The exact nature of its function, however, has remained somewhat enigmatic with it alternatively being implicated in mtDNA repair (1,2), recombination (3) and replication (4). Recently, a role in apoptosis has been suggested with Li et al. (5) demonstrating that endoG is released from mouse fibroblast mitochondria when they are treated with caspase-8-activated BID (tBID). In cells, release of endoG by the mitochondria is followed by its translocation to the nucleus where, in conjunction with DNase I and exonucleases, it initiates DNA fragmentation (6). This involvement of endoG in apoptosis leaves open the question of whether it also plays a part in mtDNA homeostasis. It would not be the first mitochondrial protein to be involved in both apoptosis and essential mitochondrial processes, e.g. cytochrome c. If endoG has a role in mtDNA maintenance, it would be expected to reside inside the mitochondrial matrix or to be associated with the interior face of the inner mitochondrial membrane. If, however, it is exclusively involved in the apoptotic process, it is likely to be confined to the intermembrane space, similar to other pro-apoptotic proteins such as cytochrome c (7), procaspase 9 (8), apoptosis-inducing factor (9) and others (10,11).
Here we present a detailed characterization of rat liver endoG. Contrary to previously published data where it was reported to be a monomeric protein of 55 kDa (12), we find endoG to be purified as a dimer comprised of 27–29 kDa subunits. Utilizing a 400 bp, double-stranded fragment of linearized DNA with a specific endoG cleavage site, we localize its activity exclusively to the mitochondrial intermembrane space. Additionally, we show that endoG can be released along with cytochrome c during Ca2+-mediated induction of a mitochondrial permeability transition (MPT) and that this release can be prevented by pretreatment of the mitochondria with cyclosporin A. Further, upon removal of endoG activity from the mitochondria, we reveal the presence of a nuclease activity associated with the interior face of the inner mitochondrial membrane that cleaves DNA in a manner distinct from that of endoG. Taken together, our data suggest that endoG activity is involved exclusively in apoptosis and that other nucleases are likely to be responsible for mtDNA maintenance and degradation.
MATERIALS AND METHODS
Mitoplast preparation
Hepatic mitochondria were isolated from male Sprague–Dawley rats in a sucrose–HEPES buffer by differential centrifugation as described by Cohen et al. (13). Mitoplasts were prepared from the mitochondria by the addition of digitonin (0.11 mg digitonin/mg mitochondrial protein), gently stirred on ice for 15 min, diluted with 20 vol buffer A (2 mM HEPES, pH 7.4, 0.25 M sucrose) and centrifuged at 20 000 g for 20 min at 4°C. The pellet was washed in buffer A and resuspended in buffer B (5 mM Tris–HCl, pH 7.4, containing protease inhibitors; Roche Molecular Biochemicals, Indianapolis, IN).
Extraction of endonuclease activity from the inner mitochondrial membrane by salt washing
Mitoplasts were subjected to six freeze–thaw cycles at –80°C followed by centrifugation at 100 000 g for 1 h at 4°C. The supernatant containing released mitochondrial matrix was carefully decanted. The pellet was resuspended using a Potter homogenizer in water containing protease inhibitors and subjected to a second round of six freeze–thaw cycles. Unbroken mitoplasts were removed by centrifugation at 12 000 g for 10 min and inner mitochondrial membrane fragments recovered by centrifugation at 100 000 g for 1 h. The pellet was resuspended in 5 ml of buffer B containing 150 mM KCl and recentrifuged at 100 000 g for 1 h at 4°C. The supernatant was carefully decanted and placed on ice and the pellet was again salt extracted. The salt-extracted proteins were pooled, precipitated by the addition of ammonium sulfate to 80% saturation and resuspended in 10 mM Tris–HCl, pH 7.4, 5% glycerol in the presence of protease inhibitors.
Size exclusion column chromatography
Salt-extracted proteins from the inner mitochondrial membrane were separated by size exclusion chromatography using a 20 × 0.5 cm Sephacryl HR-200 or Sephacryl HR-100 (Sigma, St Louis, MO) column. The column was equilibrated overnight with 40 mM HEPES, pH 7.5, 0.5 mM EDTA, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 15% glycerol and 400 mM NaCl. The elution buffer was passed through the column at <50 µl/min under gravity.
Localization of endoG activity: salt extraction of the exterior and interior face of the inner mitochondrial membrane
Mitoplasts were washed twice in either buffer A or buffer C (2 mM HEPES, pH 7.4, 150 mM KCl) and re-pelleted at 20 000 g for 20 min at 4°C. The supernatants were carefully decanted and stored on ice and the pellets resuspended in 5 ml of buffer B and subjected to six freeze–thaw cycles at –80°C. Unbroken mitoplasts were removed by centrifugation at 20 000 g for 10 min at 4°C. The supernatant, containing fragmented inner mitochondrial membrane and soluble matrix proteins, was carefully decanted and the pellet subjected to a second round of six freeze–thaw cycles. Following removal of unbroken mitoplasts, the supernatants were pooled and centrifuged at 100 000 g for 1 h at 4°C. Membrane fragments obtained from the salt-extracted and unextracted mitoplasts were washed with 5 ml of buffer B and recentrifuged at 100 000 g for 1 h at 4°C.
The membrane pellets were resuspended in 10 ml of buffer A and divided into two aliquots each. An equal volume of buffer or buffer containing 300 mM KCl was added to one aliquot from each preparation. After gentle inversion, the membrane preparations were centrifuged at 100 000 g for 1 h at 4°C. The supernatant was carefully decanted and the membrane pellet resuspended in buffer B.
Induction of the mitochondrial permeability transition
Mitochondria (75 µg/ml) were suspended in 2 ml of buffer D (2 mM HEPES, pH 7.5, 0.25 M sucrose, 10 mM succinate, 1 mM potassium phosphate) and a MPT initiated by the addition of calcium (1 µM final concentration) (14). The progression of the MPT was monitored by the change in absorbance at 540 nm over 15 min at room temperature. The mitochondria were then centrifuged at 10 000 g for 10 min and the supernatant carefully decanted. Mitochondria were resuspended in buffer E (10 mM Tris, pH 7.5, 10% glycerol in the presence of protease inhibitors) while the proteins released into the supernatant were precipitated by the addition of ammonium sulfate (80% saturation) and stored overnight at 4°C. The precipitated proteins were pelleted by centrifugation at 13 000 g for 30 min at 4°C and dissolved in 100 µl of buffer E. This procedure was also repeated in the absence of calcium chloride and with mitochondria that had been preincubated with cyclosporin A (4.5 µg/ml for 5 min).
EndoG activity gel
Protein fractions (25 µg) in Laemmli buffer were loaded, without prior boiling, onto a 12% polyacrylamide gel containing 20 µg/ml BamHI endonuclease-restricted pBR322 plasmid DNA and electrophoresed at 12 mA/gel for 90 min as described by Low and Gerschenson (15). The SDS in the gel was removed by washing with 50 mM Tris–HCl, pH 8.0, 0.1 mM EDTA, 1 mM DTT and 0.5% (w/v) Triton X-100 at room temperature for 1 h. The gel was incubated in 50 mM Tris–HCl, pH 8.0, 0.1 mM EDTA, 5 mM DTT, 0.25 mM Mg(OAc)2 and 0.02% (v/v) Tween-20 at 37°C overnight and stained with ethidium bromide (0.5 µg/ml) for 15 min. The digested DNA was visualized by UV transillumination and the image captured using a Kodak image station 440CF (Eastman Kodak Co., New Haven, CT).
Western blotting of cytochrome c and endoG
For cytochrome c, protein fractions (20 µg) in Laemmli buffer were loaded onto a 12% polyacrylamide gel and electrophoresed at 200 V for 45 min. The proteins were transferred to nitrocellulose (200 mA for 2 h) and the membrane was blocked with dried milk (5%) in Tris-buffered saline (TBS), pH 7.4, containing Tween-20 at 0.5% (TBST) for 1 h at room temperature. Cytochrome c was probed with mouse anti-cytochrome c (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:2000 dilution in TBST for 1 h and then anti-mouse IgG–horseradish peroxidase conjugate (Santa Cruz Biotechnology) at 1:5000 dilution for 1 h in TBST. Between each stage the membrane was washed (three times) in TBST for 5 min. Visualization was by addition of chemiluminescent substrate (SuperSignal; Pierce Biotechnology Inc., Rockville, IL) and quantification was carried out using a Kodak image station 440CF. For detection of endoG by western blotting, activity gels containing regions of digested DNA were incubated in 10% SDS for 1 h and the proteins present transferred to nitrocellulose membranes at 300 mA for 80 min. The membranes were then blocked with phosphate-buffered saline (PBS) containing 0.1% Tween-20, 5% milk and 5% fetal calf serum for 24 h at 4°C with gentle rocking. Membranes were incubated with a primary antibody to endoG (a kind gift from Dr X. Wang, 1:1000 dilution) overnight at 4°C, washed (3 × 30 min) with PBS containing 0.1% Tween-20, then incubated with secondary antibody (1:5000 goat anti-rabbit IgG conjugated to horseradish peroxidase; Sigma) in blocking solution for 45 min at room temperature. The membranes were washed with PBS/0.1% Tween-20 for 3 × 30 min and endoG detected by chemiluminescence.
Preparation of endoG-specific substrate and determination of endoG activity
Rat liver mtDNA was amplified (forward primer, 5′-gacatctcgatggtaacggg-3′; reverse primer, 5′-tcgggaaattttaccaatgc-3′) between nucleotides 15828 and 16231. This region of the mitochondrial D-loop encompasses a poly(dG)n residue tract that has been shown to be susceptible to site-specific cleavage by endoG (3). In some cases mtDNA was amplified using a 5′-fluorescein-labeled forward primer. Endonuclease G activity was determined by incubating 100 ng of amplified DNA with 250 µg/ml of each of the protein fractions in a 20 µl reaction volume (25 mM HEPES, 5 mM MgCl2, pH 7.5) for varying time periods. The reactions were stopped by the addition of 2 µl of agarose loading buffer and the products immediately separated by electrophoresis through a 1.7% agarose gel, containing 50 µg/ml ethidium bromide, in TBE (0.1 M Trizma base, 90 mM boric acid, 1 mM disodium ethyldiaminotetraacetic acid) buffer at an applied voltage of 110 V. In some cases, linearized pBR322 plasmid was used as a substrate for endoG activity. The plasmid possesses a number of poly(dG)n tracts and site-specific cleavage by endoG results in a characteristic banding pattern. The DNA was visualized by UV transillumination and quantification was carried out using a Kodak image station 440CF (Eastman Kodak Co.). In some cases the DNA was transferred to uncharged nylon membranes (Duralon-UV; Stratagene, La Jolla, CA) and detected by Southern hybridization as described previously (16).
Protein and DNA determinations
Protein was determined using the Bio-Rad Detergent Compatible protein assay (Bio-Rad Laboratories, Hercules, CA) or the Micro BCA protein assay (Pierce Biotechnology Inc.). DNA was assayed by fluorescence using Picogreen (Molecular Probes, Eugene, OR).
Enzyme assays
Malate dehydrogenase activity was assayed by following the decrease in NADH absorbance at A340 nm in the presence of rotenone as described by Huang et al. (17).
RESULTS
Isolation and characterization of endonuclease activity
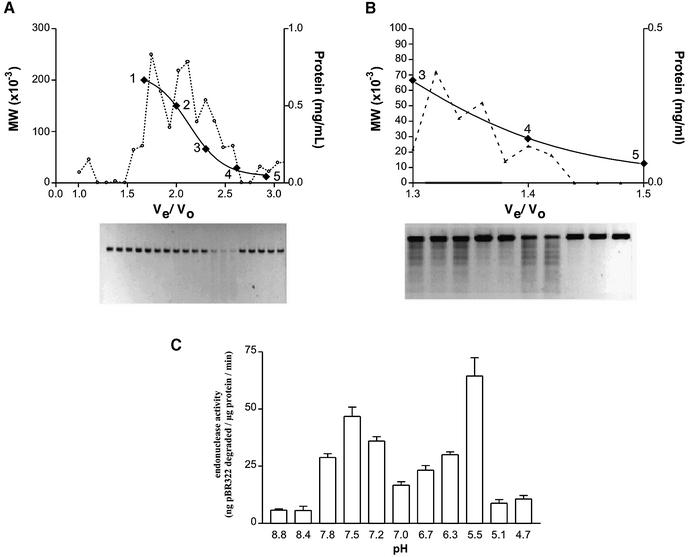
An endonuclease activity was extracted from the inner mitochondrial membrane of rat hepatocytes and extensively characterized. In order to minimize the possibility of nuclear contamination, mitochondria were first treated with digitonin to remove the outer membrane. The resulting mitoplasts were then disrupted by repeated freeze–thawing in a hypotonic buffer and the matrix removed. The remaining membranes were extensively washed before being salt washed. This procedure results in the selective extraction of proteins (yield 2.8 mg/liver) with endonuclease activity (0.52 ng linearized pBR322 degraded/min/µg protein) associated with, but not integral to, the inner mitochondrial membrane. Figure 1A shows the Sephacryl HR-200 elution profile of the salt-extracted proteins along with the associated endonuclease activity as determined by the degradation of linearized pBR322 plasmid DNA. Activity was found to be associated with proteins of an apparent molecular mass range of 27–54 kDa (yield 0.8 mg/liver; endonuclease activity 1.47 ng/min/µg). The active fractions were pooled, concentrated by ammonium sulfate precipitation and passed over a Sephacryl HR-100 column (endonuclease activity 10 ng/min/µg). Figure 1B shows two peaks of activity at apparent molecular mass ranges of 48–64 and 24–29 kDa, respectively.
Figure 1.
Size exclusion chromatography and endonuclease activity of salt-extracted mitochondrial inner membrane proteins. Separation of proteins extracted from the inner mitochondrial membrane was achieved by means of size exclusion chromatography. The elution of known standards was used to identify the approximate size of the proteins in the active fractions: 1, β-amylase (200 kDa); 2, alcohol dehydrogenase (150 kDa); 3, bovine serum albumin (66 kDa); 4, carbonic anyhdrase (29 kDa); 5, cytochrome c (12.4 kDa). The relative activity of each fraction was determined by the loss of 20 ng of linearized pBR322 incubated in 20 µl of buffer containing proteins extracted from the inner mitochondrial membrane at a concentration of 0.02 mg/ml. (A) Sephacryl HR-200 separation; proteins were incubated with DNA for 1 h at 37°C. (B) Sephacryl HR-100 separation; proteins and DNA were incubated for 10 min at 37°C. (C) pH dependence of endonuclease activity.
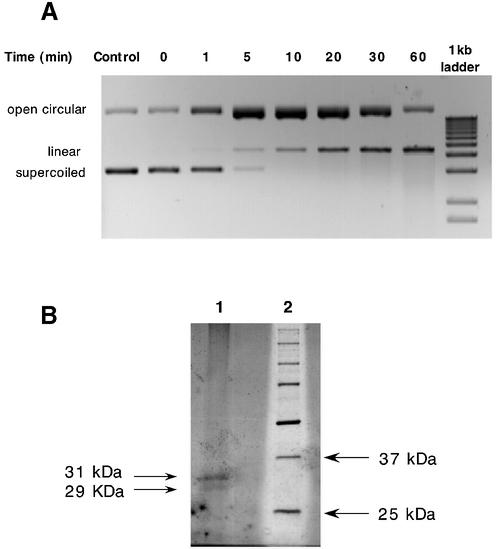
The endonuclease activity was found to be dependent on divalent cations with the optimum Mg2+ and Mn2+ concentrations being 5 and 0.7 mM, respectively (data not shown). Activity was not detected in the presence of Na+ or Ca2+ but was present when either of these ions was used in conjunction with 5 mM Mg2+ (data not shown), indicating that neither of these ions was inhibitory. Examination of pH dependence in the presence of 5 mM MgCl2 reveals two pH optima for activity at pH 5.5 and pH 7.5 (Fig. 1C). These pH optima were found to be identical for both the 48–64 and 24–29 kDa fractions. Further, the activity of both fractions was found to be site-specific with cleavage of linearized pBR322 plasmid DNA producing a characteristic banding pattern, pictured in Figure 1B. Incubation of supercoiled pBR322 with the active endonuclease extracts resulted in a progressive linearization of the plasmid via an open circular intermediate (Fig. 2A). As the relaxed circular DNA is an intermediate substrate in the conversion of supercoiled to linear DNA, it was predicted that the protein(s) associated with the endonuclease activity would still remain attached to this conformation in order to convert it to the linearized product. The open circular plasmid DNA shown in Figure 2A was therefore excised from the agarose gel, electroeluted and ethanol precipitated. The DNA, along with any associated proteins, was resuspended in Laemmli buffer, boiled and run on a 12% SDS–polyacrylamide gel. The large plasmid DNA remains in the wells while any associated proteins are able to enter the gel. Figure 2B shows one such gel that, upon silver staining, reveals two proteins of 29 and 31 kDa, respectively. Further incubation of this open circular DNA–protein complex at 37°C in the presence of 50 mM HEPES, pH 7.4, and 5 mM MgCl2 resulted in degradation of the DNA (data not shown). No degradation was seen when supercoiled plasmid DNA, not previously incubated with protein, was excised from an agarose gel and incubated in the same buffer (data not shown), suggesting that one or both of the associated proteins (29 or 31 kDa) was responsible for the degradation. All the properties outlined above are consistent with the endonuclease activity isolated from the inner mitochondrial membrane being that of endoG.
Figure 2.
Endonuclease digestion of supercoiled pBR322 plasmid DNA and analysis of proteins associated with open circular intermediate. (A) Supercoiled pBR322 plasmid DNA (1 µg) was incubated at 37°C in the presence of the active 48–64 kDa endonuclease preparation (0.01 mg/ml) from Figure 1B. Aliquots (100 ng) were removed at the time points indicated and analyzed by agarose gel electrophoresis. Control lane indicates plasmid DNA incubated in the absence of protein for 60 min. (B) SDS–PAGE analysis of proteins associated with open circular plasmid DNA separated in (A).
Site-specific cleavage of DNA by endoG
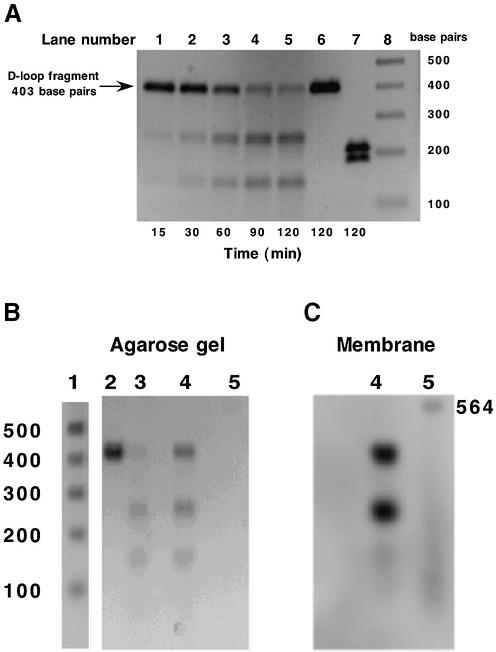
Previous studies have demonstrated the ability of endoG to selectively cleave mtDNA at a poly(dG)n site within the mitochondrial D-loop (18–20), the region of the genome responsible for control of replication. This preferred site of cleavage lies within a region of the mitochondrial genome that is highly conserved across multiple organisms, i.e. conserved sequence block 2 (CSB 2). In the rat, this block contains two tracts of successive dG bases (five and seven, respectively) separated by a thymidine residue. The ability of the isolated endonuclease to specifically cleave this sequence was utilized in the development of an endoG-specific cleavage assay. Figure 3A shows a time course of endonuclease cleavage of the 403 bp amplified fragment by endoG. Only one site of cleavage can be detected resulting in two smaller fragments of ∼150 and 250 bp. As with linearized pBR322 plasmid DNA (Fig. 1C), the site-specific cleavage of the amplified fragment was maximal at pH values 5.5 and 7.5 (data not shown), the pH optima characteristic of endoG. In order to determine the exact location of the cleavage site, the forward primer was labeled with fluorescein at the 5′-end. Amplification using this primer along with the unlabeled reverse primer results in a 403 bp fragment that is selectively labeled at one end. Incubation of this fragment with endoG resulted in cleavage at a site 250 bp away from the site of labeling (Fig. 3B and C). This places the cleavage site in the middle of the poly(dG)n tract, within the CSB2 region, and provides further evidence to support the fact that the predominant nucleolytic activity in our preparations is that of endoG.
Figure 3.
Analysis of endonuclease-mediated cleavage of PCR-amplified mtDNA D-loop fragment. (A) Time course of D-loop fragment cleavage. PCR amplified mtDNA D-loop fragment was incubated in the presence of a 48–64 kDa (see Fig. 1B) active endonuclease fraction (0.05 mg/ml) at 37°C for the times indicated. Aliquots (100 ng) were removed and analyzed by agarose gel electrophoresis. Lanes 1–5, time course; lane 6, mtDNA D-loop incubated for 120 min in the absence of protein; lane 7, SspI cleavage of mtDNA D-loop fragment; lane 8, 1 kb DNA ladder (Life Technologies, Rockville, MA). (B and C) D-loop fragment was amplified using primer 1 5′-end-labeled with fluorescein. Lane 1, 1 kb DNA ladder; lane 2, unlabeled D-loop fragment; lane 3, unlabeled D-loop fragment incubated with active endonuclease; lane 4, labeled D-loop fragment incubated with active endonuclease; lane 5, fluorescein-labeled λ/HindIII fragments. (B) Agarose gel, ethidium bromide stained. (C) DNA transferred to membrane and developed using anti-fluorescein antibody.
Location of endoG activity within the mitochondrion
In order to determine the exact location of the endoG activity within the mitochondrion, mitoplasts were incubated in the presence of the amplified endoG-specific substrate. Figure 4 (lanes 2–4) shows that mitoplasts washed with sucrose buffer possess significant endoG activity. This activity can be removed upon washing with 150 mM KCl (Fig. 4, lanes 5–7), indicating that endoG is not an integral inner mitochondrial membrane protein but is loosely associated with the exterior face of the membrane, presumably via ionic interactions. No leakage of mtDNA or malate dehydrogenase (solubilized mitoplasts, 4.3 µmol NADH/min/mg protein; digitonin extracted outer membrane and intermembrane space, 0.07 µmol NADH/min/mg protein; KCl wash of mitoplasts, not detected) was detected during these washing procedures, an indication that the structural integrity of the mitoplasts was retained. The possibility that a percentage of the endoG activity is associated with the internal face of the inner mitochondrial membrane was investigated by taking salt-washed mitoplasts and disrupting them with numerous freeze–thaw cycles. The membranes were then collected by centrifugation and washed extensively in either buffer A or 150 mM KCl. No increase in endoG activity was seen upon exposure of the internal face of the mitoplast membrane and no activity was detected in the matrix fraction, isolated upon disruption of the mitoplasts (data not shown). To further confirm that endoG activity is absent from the interior face of the inner membrane, salt-washed mitoplasts were sonicated and submitochondrial particles (SMP, inverted inner membrane vesicles where the cristae lie on the external face) prepared. The SMP were either assayed directly for endoG activity or solubilized first and then assayed. While there was some loss of DNA when compared to control incubations, there was no endoG-specific cleavage of the DNA when either the mitochondrial D-loop fragment or linearized pBR322 was used as substrate, demonstrating an absence of enzyme activity associated with the inner face of the mitochondrial inner membrane. In addition, mitochondrial matrix, derived from mitoplast sonication, was incubated with the DNA. As with the SMP, some loss of DNA occurred but no endoG-specific activity was detected. Taken as a whole, these experiments suggest that rat liver endoG activity is located exclusively on the outer face of the inner mitochondrial membrane.
Figure 4.
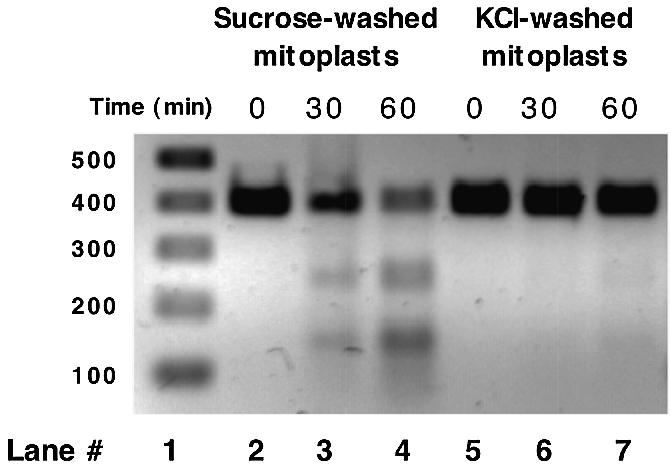
EndoG activity associated with inner mitochondrial membrane detected by degradation of 403 bp fragment of mtDNA. Mitoplasts (0.25 mg/ml) were incubated with amplified D-loop fragment (100 ng) for the times indicated at 25°C. Lane 1, molecular weight markers; lanes 2–4, time course using sucrose-washed mitoplasts; lanes 5–7, time course using salt-washed mitoplasts.
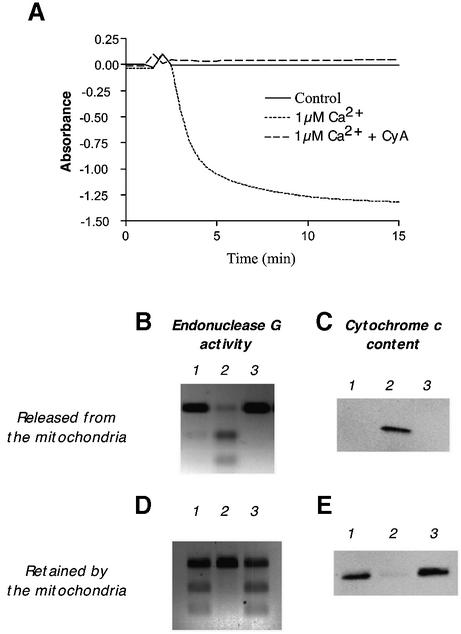
Release of endoG activity upon initiation of a MPT
Since our studies with mitoplast membranes suggest that endoG activity is loosely associated with the outer face of the inner mitochondrial membrane, it may be that, like cytochrome c, it is also present in the intermembrane space from where it would be released upon damage to the outer membrane. In order to investigate this, mitochondria were incubated in the presence of Ca2+ (1 µM) and the release of endoG activity and cytochrome c protein was monitored. Calcium treatment results in the induction of a MPT (Fig. 5A) that can be prevented by preincubation of the mitochondria with cyclosporin A. Figure 5 shows the activity of endoG and amount of cytochrome c released from the mitochondria during induction of a MPT (Fig. 5B and C) and the quantities remaining behind (Fig. 5D and E). Induction of a MPT results in the total loss of endoG activity from the mitochondria along with virtually all of the cytochrome c protein. Pretreatment of the mitochondria with cyclosporin A completely prevents this loss. We conclude that rat hepatic endoG activity, like cytochrome c, is exclusively located in the mitochondrial intermembrane space loosely associated with the outer surface of the inner membrane and is readily released upon rupture of the outer mitochondrial membrane.
Figure 5.
Effect of a MPT upon the retention of endoG activity and cytochrome c protein by rat liver mitochondria. (A) Mitochondria (75 µg/ml) were suspended in 2 mM HEPES, pH 7.5, 0.25 M sucrose, 10 mM succinate and 1 mM potassium phosphate at 25°C, in the presence or absence of cyclosporin A (CyA, 4.5 µg/ml), and a MPT induced by the addition of Ca2+ (1 µM final concentration). Absorbance was monitored at 540 nm. (B and D) EndoG activity released and retained by the mitochondria upon induction of a MPT. (C and E) Cytochrome c protein released and retained by the mitochondria upon induction of a MPT. Lane 1, control mitochondria; lane 2, Ca2+-induced MPT; lane 3, Ca2+-induced MPT following pre-treatment (5 min) with CyA.
Identification of DNase activity
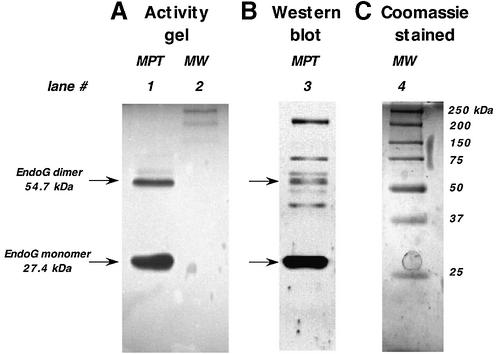
In order to further confirm that the activity released upon induction of a MPT belonged to endoG, an activity gel was prepared. Proteins released following induction of a MPT were separated on a 12% polyacrylamide gel containing linearized pBR322 plasmid DNA. This substrate had previously been shown to be attacked by endoG in an initially site-specific manner with prolonged incubation resulting in widespread degradation (Fig. 1A and B). Figure 6 shows the results of the activity gel. At pH 7.5, one of the enzyme’s two pH optima (Fig. 1C), two clear bands of DNA degradation can be visualized (lane 1) with apparent molecular weights of 27 and 54 kDa. These molecular weights correspond exactly to the expected sizes of the monomeric and dimeric forms of endoG and are in the same size ranges as those isolated by size exclusion chromatography (Fig. 1B). In addition, a very faint activity band can be seen above the 54 kDa band at 63 kDa. Both the 27 and the 54 kDa bands exhibited the same pH activity profile (Fig. 1C) as seen with the 24–29 and 48– 64 kDa fractions isolated by size exclusion chromatography (Fig. 1B). The proteins in the activity gel were denatured, by incubating the gel in SDS, transferred onto nitrocellulose and probed with an antibody to endoG. Lane 3 of Figure 6 shows the resulting western blot. Major bands can be seen at 27 and 54 kDa, the two regions of the gel that also displayed DNase activity (lane 1). A number of other bands can also be detected, including the 63 kDa band. As this band also appeared in the activity gel, it is likely that it is either an alternative conformation of endoG or a complex between the monomeric/dimeric forms and another protein that is unable to be disassociated under the denaturing conditions used. Other proteins detected by the antibody were found to be without activity, suggesting that they are not endoG or that they are inactive forms of the enzyme. Taken together, these data strongly support the contention that the enzyme activity being studied is that of endoG.
Figure 6.
Western blotting of endoG activity gel. An endoG activity gel was run, transferred to nitrocellulose and probed with an antibody to endoG as described in Materials and Methods. (A) Activity gel. Lane 1, proteins released during induction of a MPT; lane 2; molecular weight markers (MW). (B) Western blot of lane 1. (C) Coomassie blue stained MW.
Characterization of a mitochondrial nuclease distinct from endoG
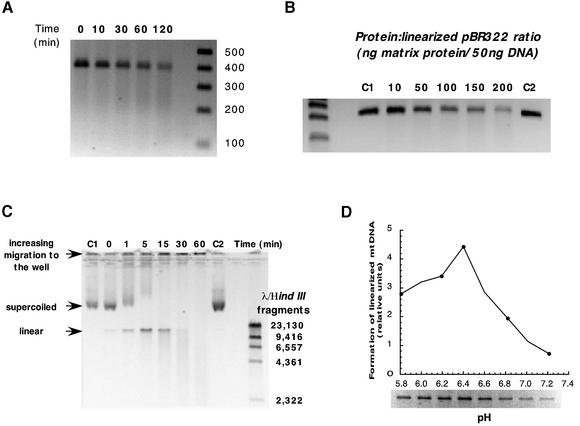
Having determined that endoG activity is localized exclusively to the mitochondrial intermembrane space, the question remains what other nuclease activities exist internal to the mitochondrial inner membrane. It is extremely likely that a nuclease, responsible for the general degradation of irreparably damaged mtDNA, resides within the mitochondrial matrix or in loose association with the inner face of the inner mitochondrial membrane. Up until now it has been difficult to study this because of endoG contamination. Localization of endoG activity to the intermembrane space now allows its selective removal from the mitochondria. Following induction of a MPT and subsequent salt washing of the mitochondria, SMP were prepared and mitochondrial matrix isolated. The matrix was incubated with a number of DNA substrates that had previously been shown to be susceptible to cleavage by endoG and the results are displayed in Figure 7. Both the amplified region of the mitochondrial D-loop and linearized pBR322, two substrates that are cleaved by endoG in a site-specific manner (Figs 1 and 3), were not similarly cleaved by the matrix nuclease activity. Instead, a gradual degradation of both substrates in a manner more consistent with an exonuclease activity was observed (Fig. 7A and B). When mtDNA isolated from rat liver was used as a substrate, both endonuclease and exonuclease activities were detected (Fig. 7C). Supercoiled DNA was cleaved to a linear conformation that was subsequently degraded. Additionally, a time-dependent accumulation of mtDNA at the top of the gel was observed (Fig. 7C). This activity can be further distinguished from that of endoG by its pH profile. Mitochondrial DNA was incubated with matrix protein at varying pH values for 3 min. This time period was found to be sufficient to form linear mtDNA without any further degradation. Endo nucleolytic cleavage of DNA, as measured by formation of linearized mtDNA, was found to have a single pH optimum at 6.4 (Fig. 7D), in marked contrast to the dual pH optima of 5.5 and 7.5 seen with endoG (Fig. 1C).
Figure 7.
Characterization of mitochondrial endo-exonuclease activity. (A) Time course of amplified mitochondrial D-loop degradation by an endo- exonuclease extract isolated from SMP that had been freeze–thawed and the membranes removed by centrifugation. DNA (100 ng) was incubated in the presence of endo-exonuclease extract (0.25 µg/µl) at 37°C for the times indicated. (B) Dose–response degradation of linearized pBR322. All incubations proceeded for 15 min. C1, control DNA; C2, control DNA incubated for 15 min at 37°C. (C) Time course of supercoiled mtDNA cleavage and subsequent degradation. Mitochondrial protein (0.15 µg/µl) was incubated with DNA (100 ng) at 37°C for the times indicated. C1, control mtDNA; C2, control mtDNA incubated for 60 min at 37°C. (D) pH dependence of endonuclease activity. Supercoiled mtDNA (100 ng) was incubated in the presence of endo-exonuclease extract (0.15 µg/µl) for 3 min.
DISCUSSION
For many years endoG has been linked with mtDNA maintenance. Suggested functions have ranged from repair of oxidative damage (1,2), degradation of defective mitochondrial genomes (12), mtDNA recombination (3) and the initiation of mtDNA replication by the generation of RNA primers (4). Recently, however, data by Li et al. (5) have cast some doubt as to its involvement with the mitochondrial genome by demonstrating tBID-induced release of endoG from mitochondria and its subsequent translocation to the nucleus where it can initiate chromatin DNA degradation in a caspase-independent apoptotic process. The question of whether endoG activity resides solely within the mitochondrial intermembrane space is an important one when considering mtDNA homeostasis. We therefore conducted a detailed characterization of rat liver endoG with the aim of determining both its properties and location within the mitochondrion. Previously, it had been reported that rat liver endoG was a monomeric protein of 55 kDa (21). This is in marked contrast to the bovine isoform that has been shown to be a homodimer consisting of subunits of 27–29 kDa (20,22). Our data, however, suggest that the liver isoform is similar to that found in the bovine system, with activity being associated with apparent protein molecular weights of 24–29 and 48– 54 kDa, as determined by size exclusion chromatography (Fig. 1B), or 27 and 54 kDa, as determined by western blotting (Fig. 6). These activities most likely represent the monomeric and homodimeric forms of the enzyme. Further support for this comes from experiments where open circular pBR322 plasmid DNA, an intermediate in the endoG-elicited conversion of supercoiled DNA to linear DNA (Fig. 2A), was electroeluted from an agarose gel and associated proteins separated by SDS–PAGE and detected via silver staining. Only two proteins, with apparent molecular weights of 29 and 31 kDa, were found to be associated with the isolated DNA (Fig. 2B). As the open circular conformation is eventually cleaved to the linear form, it is likely that one of the associated proteins was endoG. Other properties of the rat isoform, i.e. requirement for divalent cations for activity, dual pH optima (Fig. 1C) and preferential cleavage of poly(dG:dC)n tracts (Figs 1B and 3A), are similar to those reported for the bovine enzyme.
In order to assay endoG activity and to distinguish it from any other potential nucleases within the mitochondrion, a region of the mitochondrial D-loop incorporating the CSB 2 tract was amplified and utilized as a specific substrate (Fig. 3A). In the rat, the CSB 2 comprises successive 5 and 7 bp poly(dG)n tracts separated by a single dT residue and has been shown to be selectively susceptible to endoG-mediated cleavage (18–20). This finding, along with the highly conserved nature of the CSB 2 region across multiple species, represents the major contributing data behind the long-held concept that endoG is involved in mtDNA homeostasis. The amplified 403 bp substrate is selectively cleaved by endoG within the poly(dG:dC)n tract into two distinguishable bands of 250 and 150 bp, respectively (Fig. 3). The highly specific nature of the DNA substrate makes it an ideal tool for determining the precise localization of endoG activity within the mitochondrion. Data suggest that endoG activity is localized exclusively to the mitochondrial intermembrane space where it is readily released in its entirety, along with cytochrome c, during onset of a cyclosporin A-sensitive permeability transition (Fig. 5). Studies involving fractionation of mitochondria into mitoplasts and submitochondrial particles support this contention with only intact mitoplasts possessing any endoG activity (Fig. 4). Upon washing with 150 mM KCl all the endoG activity is removed (Fig. 4). Further, both matrix proteins and solubilized SMP, derived from salt-washed mitoplasts, fail to reveal any endoG activity (data not shown), an indication that enzyme activity does not reside on the interior face of the mitochondrial inner membrane or within the mitochondrial matrix. That the enzyme activity being detected is that of endoG is supported by the data in Figure 6. An activity gel performed with proteins released upon induction of a MPT reveals pronounced DNase activity associated with bands of 27, 54 and, to a lesser extent, 63 kDa, all of which are recognized by an antibody specific to endoG. Proteins of similar molecular weight ranges (separated by size exclusion chromatography) were also associated with the site-specific degradation of linearized pBR322 (Fig. 1C), suggesting that both methodologies were detecting the same proteins.
The presence of endoG activity within the intermembrane space raises the question of whether its role is solely to assist in apoptosis or if it has some other physiological function. Endonuclease G is a member of an ever-growing family of sugar-non-specific nucleases that are ubiquitous in their occurrence. Included in this group are a number of prokaryotic nucleases including the Anabaena nuclease, the Serratia nuclease and EndA from Streptococcus pneumoniae that are secreted by the bacterium in order to digest exogenous nucleic acid. The nucleotides produced are then taken up by the bacterium and used as precursors for nucleic acid synthesis (23). It may be that endoG performs a role in protecting mitochondria from invasion by extra-mitochondrial nucleic acid and in doing so also provides a source of nucleotides that are taken up into the matrix and used in the synthesis of mtDNA.
An interesting observation is that upon removal of endoG activity from the mitochondria, another nuclease activity can be detected internal to the inner mitochondrial membrane. This nuclease causes a gradual degradation of amplified DNA and linearized pBR322 plasmid DNA (Fig. 7A and B) without the site-specific cleavage seen with endoG (Fig. 1B). In this case it appears to be acting as an exonuclease. When supercoiled mtDNA is used as a substrate, however, both endo- and exonuclease activities can be detected (Fig. 7C). That the endonuclease activity is not due to the presence of endoG activity is shown by the fact that the matrix enzyme has a single activity optimum at pH 6.4 (Fig. 7D) whereas endoG has two optima for activity at pH values 5.5 and 7.5, respectively (Fig. 1C). Whether the endo- and exonucleolytic activities arise from the same enzyme or from separate enzymes has yet to be determined. The bulk of the enzyme appears to reside in the mitochondrial matrix with a much smaller amount detected in solubilized SMP. This may be artifactual, however, since the matrix was isolated during preparation of SMP, which involves sonication of the inner mitochondrial membrane. This can result in proteins becoming disassociated from the membrane and appearing in the matrix fraction. If it is associated with the membrane, it is likely to be either an integral membrane protein or loosely associated with the interior face of the membrane, as it was not removed during salt washing of the mitoplasts, in contrast to endoG activity (Fig. 3). The physiological role of this enzyme has yet to be determined but it may be involved in the general degradation of mtDNA. To date, an enzyme devoted solely to the turnover of the mitochondrial genome has yet to be identified.
The localization of endoG activity to the intermembrane space raises questions about the other endo-exonucleases characterized in mitochondria from other species. Major mitochondrial endo-exonucleases have been identified in species as diverse as Podospora anserina (24), Neurospora crassa (25), Crithidia fasciculata (26) and Saccharomyces cerevisiae (27). In all of these organisms, the enzymes show activity towards both DNA and RNA and have a requirement for divalent cations, most notably Mg2+. Many appear to be homodimeric with the monomers ranging in size from 32 kDa in Crithidia (26) to 49 kDa in Podospora (24) and all have optimum activities around physiological pH with the exception of that found in the kinetoplasts of Crithidia, which exhibits endonuclease activity at pH 6.4, interestingly the same pH value as the optimum for the unknown matrix enzyme presented in this study. The role of these enzymes remains somewhat elusive and, in view of the data presented in this manuscript, it is clear that accurate localization of these enzymes within the mitochondrion is imperative in order to ascribe a function. To date it appears that the DNase content of mammalian mitochondria consist of a number of base excision repair endonucleases, e.g. OGG1 (28), located internal to the inner mitochondrial membrane, in close proximity to the mitochondrial genome, and a potent endo-exonuclease (endoG) residing within the intermembrane space. Whether the only DNase activities residing within the mitochondrial matrix are involved in mtDNA repair or whether there are others involved solely in mtDNA degradation, such as the matrix activity described in this manuscript, remains to be seen.
In conclusion, our data demonstrate that rat hepatic endoG is a homodimeric enzyme with subunits of 27 kDa, according to western blot analysis, or 24–29 kDa, according to size exclusion chromatography. In this respect and in its properties, i.e. pH optima, requirement for divalent cations and preference for homopolymeric (dG:dC)n tracts, it is identical to the bovine isoform already characterized in detail (20,22). Additionally, its activity is localized exclusively to the mitochondrial intermembrane space where it exists in loose association with the mitochondrial inner membrane. No endoG activity could be detected in association with the internal face of the mitochondrial inner membrane or within the mitochondrial matrix. Further, upon release of endoG activity, via induction of a MPT, an alternative nuclease(s) activity can be demonstrated internal to the inner mitochondrial membrane. The precise location and physiological role of this enzyme(s) with regard to mtDNA homeostasis are currently under study. Taken together these data strongly suggest that endoG activity is not involved in mtDNA homeostasis, a finding that may be utilized in the development of new gene therapies for various mitochondrial disease states. Endonuclease G is a potent endonuclease capable of efficient digestion of circular DNA. Its presence on the exterior face of the inner mitochondrial membrane probably contributes significantly to the difficulty of transfection of mitochondria with plasmid DNA. Hence, a selective knockout of endoG may have no deleterious consequences to mtDNA maintenance, allowing for the successful transfection of respirationally impaired mitochondria with plasmids harboring deficient genes.
Acknowledgments
ACKNOWLEDGEMENT
This research was funded by NIAAA grant AA12225.
REFERENCES
- 1.Gerschenson M., Low,R.L. and Loehr,J. (1994) Levels of the mitochondrial endonuclease during rat cardiac development implicate a role for the enzyme in repair of oxidative damage in mitochondrial DNA. J. Mol. Cell. Cardiol., 26, 31–40. [DOI] [PubMed] [Google Scholar]
- 2.Houmiel K.L., Gerschenson,M. and Low,R.L. (1991) Mitochondrial endonuclease activity in the rat varies markedly among tissues in relation to the rate of tissue metabolism. Biochim. Biophys. Acta, 1079, 197–202. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz-Carrillo A. and Renaud,J. (1987) Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J., 6, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cote J. and Ruiz-Carrillo,A. (1993) Primers for mitochondrial DNA replication generated by endonuclease G. Science, 261, 765–769. [DOI] [PubMed] [Google Scholar]
- 5.Li L.Y., Luo,X. and Wang,X. (2001) Endonuclease G is an apoptotic DNase when released from mitochondria. Nature, 412, 95–99. [DOI] [PubMed] [Google Scholar]
- 6.Widlak P., Li,L.Y., Wang,X. and Garrard,W.T. (2001) Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem., 276, 48404–48409. [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Kim,C.N., Yang,J., Jemmerson,R. and Wang,X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell, 86, 147–157. [DOI] [PubMed] [Google Scholar]
- 8.Krajewski S., Krajewska,M., Ellerby,L.M., Welsh,K., Xie,Z., Deveraux,Q.L., Salvesen,G.S., Bredesen,D.E., Rosenthal,R.E., Fiskum,G. et al. (1999) Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc. Natl Acad. Sci. USA, 96, 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Susin S.A., Lorenzo,H.K., Zamzami,N., Marzo,I., Snow,B.E., Brothers,G.M., Mangion,J., Jacotot,E., Costantini,P., Loeffler,M. et al. (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature, 397, 441–446. [DOI] [PubMed] [Google Scholar]
- 10.Verhagen A.M., Ekert,P.G., Pakusch,M., Silke,J., Connolly,L.M., Reid,G.E., Moritz,R.L., Simpson,R.J. and Vaux,D.L. (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell, 102, 43–53. [DOI] [PubMed] [Google Scholar]
- 11.Du C., Fang,M., Li,Y., Li,L. and Wang,X. (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell, 102, 33–42. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda S. and Ozaki,K. (1997) Action of mitochondrial endonuclease G on DNA damaged by L-ascorbic acid, peplomycin and cis-diamminedichloroplatinum (II). Biochem. Biophys. Res. Commun., 235, 291–294. [DOI] [PubMed] [Google Scholar]
- 13.Cohen N.S., Kyan,F.S., Kyan,S.S., Cheung,C.-W. and Raijman,L. (1985) The apparent Km of ammonia for carbamoyl phosphate synthase (ammonia) in situ. Biochem. J., 229, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastorino J.G., Marcineviciute,A., Cahill,A. and Hoek,J.B. (1999) Potentiation by chronic ethanol treatment of the mitochondrial permeability transition. Biochem. Biophys. Res. Commun., 265, 405–409. [DOI] [PubMed] [Google Scholar]
- 15.Low R.L. and Gerschenson,M. (2002) In Copeland,W.C. (ed.), Mitochondrial DNA: Methods and Protocols. Humana Press, Totowa, NJ, Vol. 197, pp. 331–349.
- 16.Cahill A., Stabley,G.J., Wang,X. and Hoek,J.B. (1999) Chronic ethanol consumption causes alterations in the structural integrity of mitochondrial DNA in aged rats. Hepatology, 30, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C.H., Keyhani,E. and Lee,C.P. (1973) Fractionation by sucrose density gradient centrifugation of membrane fragments derived by sonic disruption of beef heart mitochondria. Biochim. Biophys. Acta, 305, 455–473. [DOI] [PubMed] [Google Scholar]
- 18.Low R.L., Buzan,J.M. and Couper,C.L. (1988) The preference of the mitochondrial endonuclease for a conserved sequence block in mitochondrial DNA is highly conserved during mammalian evolution. Nucleic Acids Res., 16, 6427–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low R.L., Cummings,O.W. and King,T.C. (1987) The bovine mitochondrial endonuclease prefers a conserved sequence in the displacement loop region of mitochondrial DNA. J. Biol. Chem., 262, 16164–16170. [PubMed] [Google Scholar]
- 20.Cote J., Renaud,J. and Ruiz-Carrillo,A. (1989) Recognition of (dG)n.(dC)n sequences by endonuclease G. Characterization of the calf thymus nuclease. J. Biol. Chem., 264, 3301–3310. [PubMed] [Google Scholar]
- 21.Ikeda S., Tanaka,T., Hasegawa,H. and Ozaki,K. (1996) Identification of a 55-KDA endonuclease in rat liver mitochondria with nucleolytic properties similar to endonuclease G. Biochem. Mol. Biol. Int., 38, 1049–1057. [PubMed] [Google Scholar]
- 22.Cummings O.W., King,T.C., Holden,J.A. and Low,R.L. (1987) Purification and characterization of the potent endonuclease in extracts of bovine heart mitochondria. J. Biol. Chem., 262, 2005–2015. [PubMed] [Google Scholar]
- 23.Franke I., Meiss,G. and Pingoud,A. (1999) On the advantage of being a dimer, a case study using the dimeric Serratia nuclease and the monomeric nuclease from Anabaena sp. strain PCC 7120. J. Biol. Chem., 274, 825–832. [DOI] [PubMed] [Google Scholar]
- 24.Bouex P., Sabourin,M., Chaignepain,S., Castroviejo,M. and Laquel-Robert,P. (2002) Purification and characterization of an endo-exonuclease from Podospora anserina mitochondria. Biochim. Biophys. Acta, 1574, 72–84. [DOI] [PubMed] [Google Scholar]
- 25.Chow T.Y. and Fraser,M.J. (1983) Purification and properties of single strand DNA-binding endo- exonuclease of Neurospora crassa. J. Biol. Chem., 258, 12010–12018. [PubMed] [Google Scholar]
- 26.Engel M.L. and Ray,D.S. (1998) A structure-specific DNA endonuclease is enriched in kinetoplasts purified from Crithidia fasciculata. Nucleic Acids Res., 26, 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dake E., Hofmann,T.J., McIntire,S., Hudson,A. and Zassenhaus,H.P. (1988) Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem., 263, 7691–7702. [PubMed] [Google Scholar]
- 28.Croteau D.L., Rhys,C.M.J., Hudson,E.K., Dianov,G.L., Hansford,R.G. and Bohr,V.A. (1997) An oxidative damage-specific endonuclease from rat liver mitochondria. J. Biol. Chem., 272, 27338–27344. [DOI] [PubMed] [Google Scholar]