Abstract
Polyamides are a class of heterocyclic small molecules with the potential of controlling gene expression by binding to the minor groove of DNA in a sequence-specific manner. To evaluate the feasibility of this class of compounds as antiviral therapeutics, molecules were designed to essential sequence elements occurring numerous times in the HPV genome. This sequence element is bound by a virus-encoded transcription and replication factor E2, which binds to a 12 bp recognition site as a homodimeric protein. Here, we take advantage of polyamide:DNA and E2:DNA co-crystal structural information and advances in polyamide synthetic chemistry to design tandem hairpin polyamides that are capable of displacing the major groove-binding E2 homodimer from its DNA binding site. The binding of tandem hairpin polyamides and the E2 DNA binding protein to the DNA site is mutually exclusive even though the two ligands occupy opposite faces of the DNA double helix. We show with circular permutation studies that the tandem hairpin polyamide prevents the intrinsic bending of the E2 DNA site important for binding of the protein. Taken together, these results illustrate the feasibility of inhibiting the binding of homodimeric, major groove-binding transcription factors by altering the local DNA geometry using minor groove-binding tandem hairpin polyamides.
INTRODUCTION
Human papilloma viruses (HPV) belong to a large family of double-stranded DNA viruses that can induce benign epithelial or fibroepithelial papillomas (warts) of the skin. To date, over 70 subtypes of HPV have been described. Only a small subgroup of these viruses, the ‘high risk genital papillomas’ (namely HPV-16, HPV-18, HPV-31 and HPV-33), have been found to be associated with squamous cell carcinomas of the cervix and anogenital tumors (1–5). HPV can infect the basal layer of the squamous epithelial cells even though viral replication is restricted to the terminally differentiated keratinocytes (for a review see 6). In malignant tumors, these viruses overexpress two virus-encoded proteins, E6 and E7, which bind to and inactivate the host tumor suppressor proteins p53 and the retinoblastoma gene product Rb, respectively (7–14). These observations strongly suggest that the E6/E7 gene products participate directly in the oncogenic progression in these cancers (for reviews see 10,12,14,15).
All HPV subtypes contain a highly conserved region called the long control region (LCR) encoding the DNA elements responsible for both replication of the viral genome and regulated transcription of the viral gene products. In HPV-18 expression of the E6/E7 genes is tightly regulated by the upstream promoter P105 embedded within the LCR. The P105 promoter activity is regulated by the virus-encoded transcription factor E2 and a complex of cellular host proteins in response to tissue-specific and physiological signals. HPV-18 E2 regulates transcription by binding specifically to four conserved sites in the LCR termed BS4, BS3, BS2 and BS1 (16–18). Interestingly, E2 functions both as an activator and repressor of transcription depending on the cellular context and the particular sites occupied within the promoter. Depending on the cellular context, E2 binding to the promoter-distal sites (BS4 and BS3) activates transcription, whereas binding to the promoter-proximal sites (BS2 and BS1) results in repression of transcription (17–19). The BS2 and BS1 cis elements overlap binding sites for the general transcription factors Sp1 and TBP, respectively. The trans-repression effects mediated by the E2 gene product are thought to occur by disrupting preinitiation complex formation via displacement of Sp1 and TBP from their proximal promoter elements within the viral LCR (20). In contrast, E2-dependent trans-activation involves interaction with the co-activator protein CBP and the potential involvement of chromatin remodeling activities (21).
In addition to its role in regulating transcription, E2 is also essential for viral replication. In this role, E2 functions by interacting with and recruiting the virally encoded DNA helicase E1 (22) to an adjacent AT-rich site in the LCR. E2 appears to stimulate replication by recruiting host replication factors to the origin such as the host single-strand DNA binding protein, replication protein A (23–27). Replication factor A can bind to acidic trans-activation domains similar to those found in E2 (28). Recently, E2 has also been shown to mediate the assembly of the pre-initiation replication complex at the origin, but it does not play a direct role in the replication activity per se (23). Thus, any approach that inhibits the binding of HPV-18 E2 to its consensus binding site could potentially affect both transcription and replication of the virus. This approach could provide a novel and unique venue for therapeutic intervention against HPV infection.
The E2 gene product derived from all known HPV strains binds to a consensus site ACCgN4cGGT (lower case letters are preferred but not required) as a homodimer and makes contacts exclusively in the major groove of the DNA helix. The E2-DBD (DNA binding domain) represents a novel structural class of DNA binding proteins. It forms a dimeric β-barrel with each subunit contributing an antiparallel four-stranded β-sheet (29). Upon binding to its site, E2 bends the DNA by ∼43–51° towards the body of the protein leading to the compression and narrowing of the minor groove to 8.5 Å (30–32). The central N4 nucleotides are not conserved in sequence but tend to be A/T-rich in base composition. In particular, E2 binding sites within the genomes of high-risk HPVs contain A/T sequences predisposed to inherent DNA deformation. A large body of crystal structure data of E2-DNA binding domains derived from several HPV strains and its cognate binding site in both the bound and unbound form suggests that the sequence-dependent deformation of the DNA is crucial for E2-DNA binding specificity (29,30,32,33).
In this report, we demonstrate that E2-DNA binding activity can be specifically disrupted by small synthetic pyrrole– imidazole polyamides. Polyamides are synthetic ligands that bind in the minor groove of DNA with affinities and specificities comparable to those of DNA binding proteins (34) and can therefore potentially serve as promoter- and transcription factor-specific inhibitors of gene expression (35). Polyamides can be rationally designed to bind to almost any predetermined DNA sequence using the ‘Dervan pairing rules’. Hairpin-shaped polyamides that contain the aromatic rings N-methylimidazole (Im) and N-methylpyrrole (Py) can bind in the minor groove in a side-by-side, antiparallel fashion to specifically distinguish G·C (Im/Py) from C·G (Py/Im) base pairs. Py/Py pairs are partially degenerate and bind both A·T and T·A pairs (for reviews on the pairing rules, see 36,37). A consecutive run of more than five aromatic rings is overwound relative to the curvature and helical phasing of the DNA helix. To compensate for the overwinding a β-alanine unit is inserted to alleviate the strain in polyamides with longer than five consecutive aromatic residues. Moreover, β-alanine (β) has proven to be a conformationally flexible functional analog of a pyrrole carboxamide unit (38). A β/Py pair can replace a Py/Py pair and allow for recognition of longer DNA sequences while maintaining the degenerate specificity for A·T or T·A base pairs. Using the pairing rules, polyamides designed to interfere with TFIIIA binding to its promoter-response element were shown to be potent and specific inhibitors of 5S RNA gene transcription (35). Such designed polyamides have also been shown to specifically inhibit the transcription of human immunodeficiency virus type I LTR within the genome of human peripheral blood mononuclear cells (39). Recently, polyamides have been used as small molecule transcription activators in vitro by covalently tethering a small peptide activation domain to the C-terminus of a hairpin polyamide (40,41). In addition to targeting specific transcription factor binding sites, polyamides have also been used to indirectly activate and repress particular genes by targeting large blocks of repetitive DNA sequences found in chromosome satellite regions in the Drosophila melanogaster genome. Both cytological (42) and genetic (43) evidence suggests that polyamides mediate opening of heterochromatin resulting in specific phenotypic changes reminiscent of well characterized D.melanogaster homeotic mutants (for a review see 44).
In all examples reported, hairpin polyamides have been employed to displace transcription factors or DNA binding proteins that make essential minor groove contacts with the DNA such as TFIIIA, TBP, LEF-1, ets-related factor (ESX) and deadpan (35,39,45). In contrast, displacement of GCN4, which makes exclusively major groove contacts, has been accomplished only by the attachment of positively charged residues (Arg-Pro-Arg) at the tail end of polyamides which can compete with the protein for a shared phosphate backbone contact (46). Here, we demonstrate the inhibition of DNA binding by HPV-18 E2 protein, an exclusively major groove-binding protein, by a DNA minor groove-binding tandem hairpin polyamide. Using structural information derived from published X-ray co-crystal structures of the E2–DNA complex (47), we have designed linker moieties that transverse into the minor groove at the dimerization interface of the homodimer. This bound ligand prevents compression of the minor groove necessary for E2 binding and thus destabilizes the E2–DNA complex. The success of this approach opens the possibility of interfering with other major groove interacting proteins by the rational design of polyamides that can alter and/or prevent the alteration of local DNA geometry.
MATERIALS AND METHODS
Synthesis of polyamides
Polyamides were synthesized linearly via semi-automated solid phase synthesis using Argonaut Quest technology and well established synthesis and purification protocols (42,48,49). FMOC protected 8-amino-3,6-dioxaoctanoic acid linkage was purchased from Nova. Deprotection and functionalization from the DABA amino turn was carried out as previously described (42) followed by standard solid phase peptide synthesis couplings.
Oligonucleotides
Footprinting sense, 5′-AGCTTGCAACCGATTTCGGTTGCCGCAACCGATTTCGGTTGCCA-3′; footprinting antisense, 5′-AGCTTGGCAACCGAAATCGGTTGCGGCAAC CGAAATCGGTTGCA-3′; BS4 gel shift sense, 5′-GTG CAACCGATTTCGGTTGCCT-3′; BS4 gel shift antisense, 5′-AGGCAACCGAAAGCGGTTGCAC-3′; BS4-Bend-TS, 5′-CTAGTGCAACCGATTTCGGTTGCC-3′; BS4-Bend-BS, 5′-TCGAGGCAACCGAAATCGGTTGCA-3′; E2dbdNdeI, 5′-GCAGCCATATGCTCTGTAGTGGTAACACTACGCC-3′; E2dbdBamHI, 5′-GATCGGGATCCTTACATTGTCATGTATCCCACC-3′.
Plasmid constructs
Plasmid 6XBS4-pBLCAT2, containing six multimerized HPV-18 E2 binding sites was constructed as follows. Two complementary oligonucleotides (footprinting sense and antisense), encoding two BS4 sites each, were annealed and ligated using T4 ligase. The resulting concatamers of the appropriate size were gel isolated and subcloned into the HindIII site of pBLCAT2. Transformants were screened by sequencing transformants for clones that encoded three inserts in the same orientation. The resulting concatamerized insert has the sequence 5′-(AAGCTTGCAACCGATTTCGGTTGCCGCAACCGATTTCGGTTGCC)3-3′ with the E2 BS4 site in bold type. Plasmid E2dbd-pET15b was constructed as follows. The DNA binding domain of HPV-18 E2 (amino acids 1–137) was amplified with primers E2dbdNdeI and E2dbdBamHI from plasmid pHPV18-E2 (Michael Botchan, University of California–Berkeley) by PCR and subcloned into the NdeI and BamHI sites of plasmid pET15b (Novagen). Clones were verified by DNA sequencing. Plasmid pBS4-Bend5 was constructed by cloning annealed oligonucleotides BS4-Bend-TS and BS4-Bend-BS into the XbaI and SalI sites of plasmid pBend5 (ATCC).
Protein purification
The E2dbd-pET15b transformed BL21(DE3)pLysS cells (Invitrogen) were used for the induction and overexpression of the 6× His-tagged E2dbd protein. Proteins were purified from a 250 ml culture using Ni–NTA resin using protocols and buffers recommended by the manufacturer (Qiagen) for the batch purification of native proteins from Escherichia coli. Following elution from the resin, the protein was dialyzed against buffer D (50 mM Tris–HCl, 1 M NaCl, 0.5 mM EDTA, 1 mM DTT, 10% glycerol, pH 8.0) using 5 kDa molecular weight cut-off dialysis tubing (Spectra). The specific activity of E2dbd was determined by EMSA using a stoichiometric binding analysis with an excess (100 nM) of a double-stranded oligonucleotide containing a single BS4 site (50). The protein preparation was determined to be 60% active. The equilibrium dissociation constant (Kd) for E2dbd was also determined to be 4.5 nM using EMSA with a limiting concentration (50 pM) of the same oligonucleotide duplex.
Quantitative DNase I footprinting titration experiments
DNase I footprinting was performed as described by Trauger and Dervan (51). Reactions were performed with a 32P 5′-end-labeled 250 bp restriction fragment (10 pM) derived from plasmid 6XBS4-pBLCAT2, in a final volume of 400 µl in EMSA buffer (100 mM Tris–HCl, pH 7.0, 50 mM NaCl, 10 mM MgCl2, 10 mM CaCl2, 10% glycerol, 1 mM DTT, 100 µg/ml BSA and 0.01% NP-40). Polyamide stock solution was added to this assay mixture and allowed to equilibrate at 22°C for 16 h. For reactions containing E2dbd, 10 µl of protein sample was added at increasing concentration and allowed to incubate for an additional hour. Footprinting reactions were initiated by the addition of 10 µl of DNase I (0.03 U final concentration) containing 1 mM DTT and allowed to proceed for 7 min at 22°C. The reaction was terminated by the addition of 50 µl of a stop solution (1.25 M NaCl, 100 mM EDTA, 0.2 mg/ml glycogen and 28 µM base pair calf thymus DNA) and ethanol precipitated. Reactions were resuspended in 98% formamide, 10 mM EDTA loading buffer, denatured by heating at 85°C for 10 min, and placed on ice. The reaction products were separated by electrophoresis on an 8% polyacrylamide–7 M urea gel. Fixed and dried gels were exposed to the Molecular Dynamics phosphorimager plate and data analysis was performed using the equipped ImageQuant software.
Gel mobility shift assays
Complementary oligonucleotides containing the BS4 binding site were synthesized (Integrated DNA Technologies) for use as double-stranded probes in electrophoretic mobility shift assays. Equimolar amounts (10 pmol) of two complementary oligonucleotides were annealed and end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. After labeling, unincorporated nucleotides were removed using the QiaQuick nucleotide removal kit (Qiagen) following the manufacturer’s instructions. Mobility shift assays were performed in 20 µl reaction volumes consisting of 50 pM labeled oligonucleotides in EMSA buffer (100 mM Tris–HCl, pH 7.0, 50 mM NaCl, 10 mM MgCl2, 10 mM CaCl2, 10% glycerol, 1 mM DTT, 100 µg/ml BSA and 0.01% NP-40). Polyamide stock solutions were serially diluted in DMSO and added to the reaction mixture and allowed to equilibrate at 37°C for 2 h. E2dbd (10 µg/ml) was diluted 1:4 in EMSA buffer immediately before adding 1.0 µl to the samples (final concentration of E2dbd ∼10 nM). The samples were further incubated at 37°C for 90 min. The order of addition of polyamide and E2dbd was reversed for the experiment shown in Figure 3B. The bound and free DNA forms were resolved on 5% non-denaturing polyacrylamide gels in 1× TBE buffer. Gels were dried and exposed to the Molecular Dynamics phosphorimager plate and data analysis was performed using the equipped ImageQuant software.
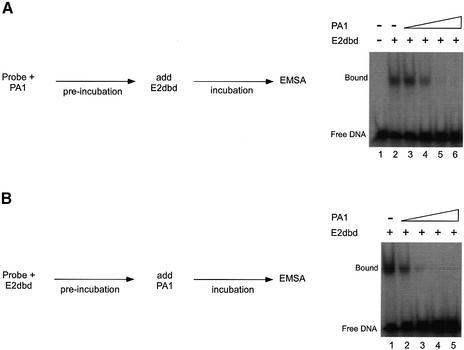
Figure 3.
Order of addition experiments with E2dbd and polyamides. (A) An EMSA was performed in which the radiolabeled HPV E2 BS4 probe was preincubated with polyamide (or the vehicle DMSO) under equilibrium conditions prior to the addition of E2dbd as described in Materials and Methods. DNA was incubated in the absence of either polyamide (lane 1) or E2dbd (lanes 2–6) or preincubated with DMSO (lane 2) or 0.13 (lane 3), 0. 64 (lane 4), 3.2 (lane 5) or 16 nM PA1 (lane 6) prior to the addition of a fixed concentration of the DNA binding domain of E2 (10 nM). (B) An EMSA was performed in parallel in which the radiolabeled HPV E2 BS4 probe was preincubated with the DNA binding domain of E2 under equilibrium conditions prior to the addition of 10 nM polyamide PA1 (or the vehicle DMSO) as described in Materials and Methods. DNA was preincubated with E2dbd (lanes 1–5) prior to the addition of a variable concentration of DMSO (lane 1) or 0.13 (lane 2), 0. 64 (lane 3), 3.2 (lane 4) or 16 nM PA1 (lane 5).
Circular permutation assay
Double-stranded oligonucleotides containing the BS4 site were synthesized (IDT), annealed and ligated into the XbaI– SalI site of pBend5 (ATCC) to generate plasmid pBS4-Bend5. Orientation of the insert was verified by DNA sequencing. Isometric DNA fragments containing the BS4 site at various positions in the fragment were generated by digesting pBS4-Bend5 with MluI, NheI, SpeI, PvuII, XhoI, EcoRV, NruI, RsaI and BamHI restriction endonucleases. DNA fragments were purified by agarose gel electrophoresis and recovered with the QiaQuick gel extraction kit (Qiagen) following the manufacturer’s instructions. DNA fragments were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. After labeling, unincorporated nucleotides were removed using the QiaQuick nucleotide removal kit (Qiagen) following the manufacturer’s instructions. The eluted samples were applied to 10% polyacrylamide gels and electrophoresed at 22°C and 17 V/cm for 6 h. The bend angle α was calculated using the equation, µM/µE = cos(α/2), where µM and µE represent the mobility of the protein–DNA complex with the bend at the middle and at the end of the DNA fragment, respectively (52).
RESULTS
Tandem hairpin polyamides efficiently displace E2 compared to the single hairpin polyamides
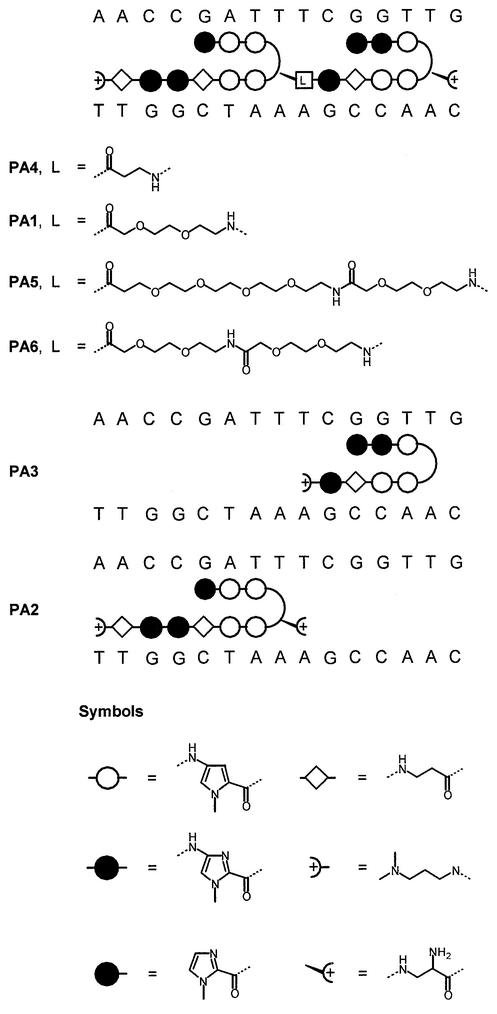
In order to examine the ability of tandem linked or single hairpin polyamides to displace E2, we designed molecules targeted to the E2 binding site BS4 (Fig. 1). The pseudo-symmetry of this site presents a unique advantage in the ability to design polyamides that could bind in either forward or reverse orientation. We designed two single hairpin polyamides: PA2 to the 5′ half-site and PA3 to the 3′ half-site of BS4. A tandem hairpin polyamide, PA1, was synthesized by linking the two hairpins with a diethylene linker (42,53). This strategy of linking two hairpin designs by a flexible linker was previously shown to provide enhanced binding affinity and binding specificity to DNA (49,53). The DNA binding affinities of these tandem hairpin molecules designed for the DNA E2 binding site were measured by quantitative DNase I footprinting assay (Fig. 2). In addition, they were assayed for an ability to displace E2 from its DNA binding site as determined by both electrophoretic mobility shift assays (EMSAs) and quantitative DNase I footprinting (Fig. 4).
Figure 1.
Schematic representation of single and tandem hairpin polyamides designed to target the HPV E2-DNA binding site. The N-methylpyrrole and N-methylimidazole rings are represented by open circles and filled circles, respectively. The flexible β-alanine monomer is represented by a diamond symbol. The γ-turn monomer, R-2,4-diaminobutyric acid, is indicated by a curved line. The dimethylaminopropylamide (Dp) at the C-terminus is represented by a plus sign enclosed within a half-circle. The position of the various flexible linkers (mono-, di-, tetra- and hexaethylene) used in this study is indicated by an open rectangle. The relationship between the polyamide nomenclature and type of linker is indicated in Table 1.
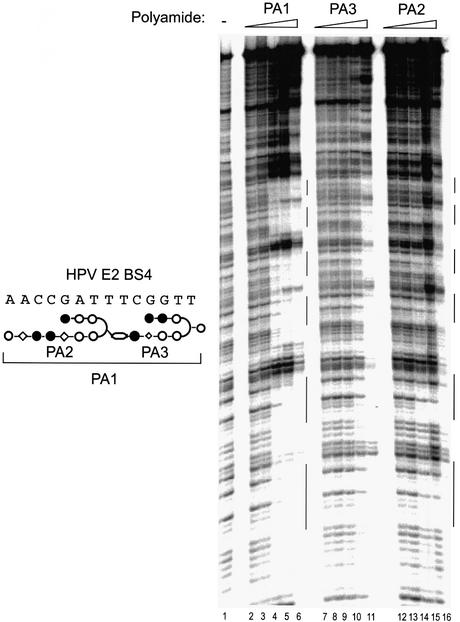
Figure 2.
Quantitative DNase I footprinting titrations with the tandem hairpin PA1 and its component single hairpin polyamides. The composite schematic relationship between these molecules is shown to the left of the autoradiogram. The footprinting analysis was performed on a DNA restriction fragment encoding six identical copies of the HPV-18 E2 binding site 4 (BS4) (18). Lane 1 contained no polyamide. The DNA was incubated with increasing concentrations of tandem hairpin PA1: 0.01 (lane 2), 0.1 (lane 3), 1 (lane 4), 10 (lane 5) and 100 nM (lane 6). Concentrations of polyamides PA3 and PA2 surveyed were 0.1 (lanes 7 and 12), 1 (lanes 8 and 13), 10 (lanes 9 and 14), 100 (lanes 10 and 15) and 1000 nM (lanes 11 and 16). Note that the concentration ranges tested for the tandem hairpin and single hairpins differ by an order of magnitude. Vertical bars on the right of the figures represent the six tandem BS4 sites.
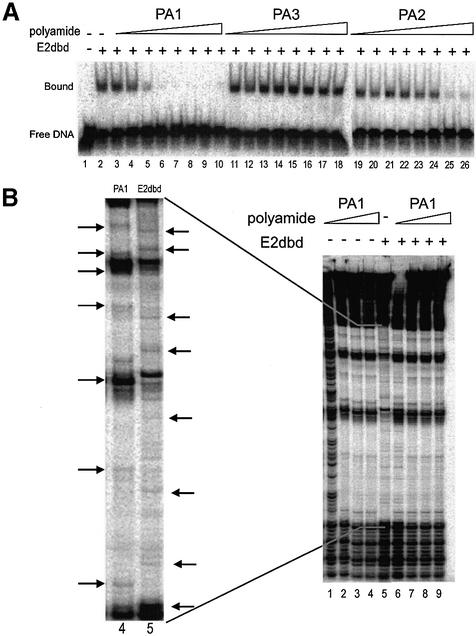
Figure 4.
Competition experiments with E2dbd and polyamides. (A) EMSA was performed with a radiolabeled HPV E2 BS4 probe in reactions lacking E2dbd (lane 1) or containing a constant concentration of E2dbd (10 nM; lanes 2–25). Reactions were supplemented with the vehicle DMSO (lane 2) or polyamide PA1 (lanes 3–10), PA3 (lanes 11–18) or PA2 (lanes 19–26) diluted in DMSO. Polyamide concentrations tested were 0.13 (lanes 3, 11 and 19), 0.64 (lanes 4, 12 and 20), 3.2 (lanes 5, 13 and 21), 16 (lanes 6, 14 and 22), 80 (lanes 7, 15 and 23) and 400 nM (lanes 8, 16 and 24) and 2 (lanes 9, 17 and 25) and 10 µM (lanes 10, 18 and 26). (B) Quantitative DNase I footprinting assays were performed with the radiolabeled restriction fragment described in the legend to Figure 2. Reactions were performed in the absence of E2dbd (lanes 1–4) or in the presence of a constant concentration of E2dbd (lanes 5–9). Polyamide PA1 was titrated at the following concentrations: 0 (lane 5), 0.1 (lanes 1 and 6), 1 (lanes 2 and 7), 10 (lanes 3 and 8) and 100 nM (lanes 4 and 9). A magnification of lanes 4 and 5 in the main autoradiogram is shown as an inset to the left. Horizontal arrows indicate unique hypersensitivity bands characteristic of polyamide PA1 binding (lane 4) or E2dbd binding (lane 5) to a BS4 DNA site.
Quantitative DNase I footprinting experiments were performed by incubating individual polyamides with a radiolabeled DNA probe encoding six identical high affinity E2 binding sites (BS4) under equilibrium conditions (see Materials and Methods). The tandem hairpin polyamide PA1 has a 100-fold order higher DNA binding affinity than the single hairpin, PA3 (Fig. 2, compare lanes 2–6 to lanes 7–11), and 1000-fold higher affinity than PA2 (Fig. 2, compare lanes 2–6 to lanes 12–16). Consistent with earlier reports by Herman et al. (49) and Maeshima et al. (53), these observations suggest that linking hairpin polyamides can lead to a dramatic enhancement in DNA binding affinity.
EMSA experiments were conducted to test the ability of these three polyamides to displace E2 (Figs 3 and 4A). For these experiments, we generated a derivative of the E2 protein, E2dbd, which encodes both its DNA binding and dimerization domains (see Materials and Methods). The measured dissociation constant (Kd = 4.5 nM) of E2dbd is similar to the previously reported Kd for this protein. These experiments were performed under equilibrium conditions as evidenced by the observation that the ability of PA1 to displace E2 from the BS4 site is independent of whether PA1 is added to the DNA prior to or after the addition of protein (Fig. 3). The tandem hairpin PA1 displaced E2 at a concentration of ∼2 nM, the same concentration range as PA1’s measured DNA binding affinity (Kd = 1 nM). In sharp contrast, the single hairpin PA3 is unable to displace E2 even at high concentrations (10 µM) (Fig. 4A, lanes 11–18) despite an apparent binding affinity of 100 nM. The single hairpin polyamide PA2 displaces E2 at ∼2 µM, similar to its binding affinity (Kd = 1 µM) (Fig. 4A, lanes 19–26). The equilibrium dissociation constant (Kd) and equilibrium inhibition constant (Ki) of each polyamide are summarized in Table 1. These data suggest that the tandem hairpin polyamide motif is more efficient at displacing E2 than either of the single hairpin polyamides.
Table 1. Equilibrium binding constants of polyamides to the E2-DNA binding site.
| Molecule | Kd (nM) | Kia (nM) | Linkerb | Comment |
|---|---|---|---|---|
| PA4 | 1000 | 1000 | One | Tandem hairpin |
| PA1 | 1 | 2 | Two | Tandem hairpin |
| PA3 | 100 | >10 000 | NA | Single hairpin |
| PA2 | 1000 | 2000 | NA | Single hairpin |
| PA5 | 1 | 16 | Six | Tandem hairpin |
| PA6 | 3 | 3.2 | Four | Tandem hairpin |
| Netropsin | 100 | >10 000 | NA | Natural product |
NA, not applicable.
aKi is the concentration of polyamide required to reduce the amount of shifted (E2–DNA) complex by half in the presence of a fixed amount of E2dbd protein under equilibrium conditions.
bNumber of ethylene repeats in the flexible linker of a tandem hairpin polyamide.
To directly demonstrate that PA1 displaces E2 by mutual competition for the same site, we performed a DNase I footprinting assay using a DNA probe containing six identical copies of BS4. Although the footprinting patterns of both PA1 and E2dbd are very similar, they each produce a unique identifiable DNase I hypersensitivity pattern as indicated by arrows in Figure 4B. Strikingly, in the presence of increasing concentrations of PA1, the DNase I hypersensitivity pattern characteristic of E2dbd is replaced by a DNase I hypersensitivity pattern attributable to PA1 (see inset in Fig. 4B comparing lanes 4 and 5). The inhibition of E2 binding by PA1 was confirmed in parallel samples by EMSA demonstrating the same polyamide concentration dependence (data not shown). These data indicate that the binding of tandem hairpin PA1 and E2dbd protein to BS4 is mutually exclusive.
Effect of linker length on inhibition of DNA binding
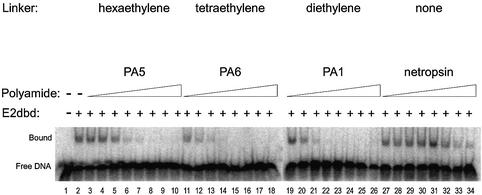
In addition to increasing the DNA binding affinity of the PA1 molecule, the linker moiety in the tandem hairpin appears to displace E2dbd protein more efficiently. To further address the role of linker moieties, we systemically tested the effectiveness of different linker lengths. We generated analogs of PA1 with different length linkers in the context of single hairpin modules (PA3 and PA2) and tested their ability to displace E2 protein. Tandem hairpin polyamides linked by a tetraethylene linker (PA6) (Fig. 5), hexaethylene linker (PA5) (Fig. 5) and a single ethylene linker (PA4) (data not shown) were synthesized and their ability to displace E2 was characterized using the EMSA. The tetraethylene-linked PA6 was equally as effective as the diethylene-linked PA1 (Ki ≈ 2 nM). In contrast, the hexaethylene-linked PA5 was 8-fold less effective (Ki ≈ 16 nM). Importantly, the DNA binding affinities (Kd values) for all three of these polyamides is between 1 and 3 nM (Table 1). In contrast, a tandem hairpin with a shorter linker (PA4) has dramatically poorer DNA binding affinity (Table 1). It is likely that the shorter linker puts the two DNA binding modules ‘out of register’ with the bases at their respective cognate half-sites. Although a shorter linker than a diethylene linker is not well tolerated, there does not appear to be a strict requirement for a particular length linker above the apparent minimal linker length (i.e., the diethylene linker in PA1).
Figure 5.
The effect of linker length on tandem hairpin functionality was assayed by EMSA. The assay was performed as described in the legend to Figure 3A. Control reactions were performed in the absence or the presence of E2dbd (lanes 1 and 2, respectively). Competition experiments with tandem polyamides PA5 (lanes 3–10), PA6 (lanes 11–18) and PA1 (lanes 19–26) are shown. Netropsin, a natural product containing two pyrroles, is used as a reference standard in this experiment (lanes 27–34). Polyamide concentrations surveyed were 0.13 (lanes 3, 11, 19 and 27), 0.64 (lanes 4, 12, 20 and 28), 3.2 (lanes 5, 13, 21 and 29), 16 (lanes 6, 14, 22 and 30), 80 (lanes 7, 15, 23 and 31) and 400 nM (lanes 8, 16, 24 and 32) and 2 (lanes 9, 17, 25 and 33) and 10 µM (lanes 10, 18, 26 and 34).
Tandem hairpin PA1 unbends the E2 binding site
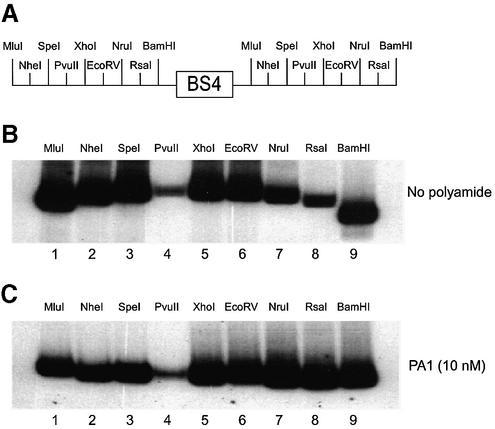
The EMSA and DNase I footprinting displacement experiments (Figs 2 and 4) indicate that binding of PA1 and E2 protein to the BS4 site is mutually exclusive. This was an unexpected observation given that PA1 is a minor groove DNA binding ligand whereas E2 is a major groove DNA binding protein. This observation raises the question as to how PA1 inhibits E2-DNA binding activity. Recent biochemical and structural data have indicated that the E2 binding site is intrinsically bent presumably due to the central A/T-tract in the binding site (31,33,47). Computer-assisted algorithms have also been used to model the curvature and step-wise helicoidal parameters of predetermined nucleic acid sequences. Interestingly, using the software Curves developed by Lavery and Sklenar, the minor groove width of BS4 at the central A/T-tract is predicted to be narrowed to 4.5 Å (54–56). This bend in the DNA is further accentuated by E2 protein binding and bending the DNA towards the body of the protein. The elimination of the DNA bend by site-directed mutagenesis of the A/T-tract results in lower DNA binding affinity for E2 protein even though E2 protein does not make any base-specific contacts in this region. It is postulated that the pre-bent structure provides a favorable energetic interface for stabilizing the E2–DNA complex. Thus, we reasoned that PA1 might inhibit the binding of E2 protein by unbending the BS4 site and destabilizing the E2–DNA complex. To test this possibility, we performed circular permutation assays in the absence and presence of PA1 bound to the BS4 site. In this assay (57), the BS4 site is flanked on both sides by tandemly duplicated restriction enzyme sites, and restriction fragments of identical lengths can be generated with the BS4 site positioned at different locations relative to the ends of the DNA fragment (Fig. 6A). DNA molecules with a bend in the center of the molecule have slower electrophoretic mobilities through native polyacrylamide gels than DNA molecules with a bend located towards the ends. Using this assay, we confirm the intrinsic bend of the BS4 site as indicated by the anomalous migration of DNA fragments with the site in the center of the DNA fragment (Fig. 6B, compare XhoI and EcoRV to MluI and BamHI). However, in the presence of PA1 the anomalous migration of the DNA fragments is eliminated, strongly suggesting that the DNA fragment is unbent (Fig. 6C, compare XhoI and EcoRV to MluI and BamHI). The α values (bend angles), calculated by the equation described by Thompson and Landy (52), for the BS4 site were 18° in the absence of PA1 and only 3° in the presence of PA1. Thus, PA1 is capable of unbending the BS4 site. A model for the mechanism of tandem hairpin polyamide-mediated inhibition of binding of the homodimeric transcription factor E2 by unbending the DNA binding site is shown in Figure 7. In addition to the DNA unbending mechanism shown in Figure 7, another possible mechanism of inhibition of E2 binding might be steric inhibition by the linker of the tandem hairpin polyamide at the dimerization interface of the E2 homodimer in the minor groove of the DNA. Distinguishing between these non-mutually exclusive possibilities will likely require structural studies (NMR or X-ray) of the tandem hairpin polyamide–DNA complex.
Figure 6.
A circular permutation assay was utilized to determine the effect of tandem hairpin PA1 on the intrinsic bend of a probe encoding the E2-DNA binding site. In this variation on an EMSA, a DNA fragment with a centralized bend will result in a lower mobility fragment whereas a DNA with a terminally positioned bend will result in a higher mobility fragment (57). (A) A schematic showing the relative positions of the tandemly duplicated restriction sites found on the radiolabeled probe encoding the BS4 E2-DNA binding site. (B) A circular permutation assay showing the radiolabeled probe restricted with the following enzymes: MluI (lane 1), NheI (lane 2), SpeI (lane 3), PvuII (lane 4), XhoI (lane 5), EcoRV (lane 6), NruI (lane 7), RsaI (lane 8) and BamHI (lane 9). (C) The circular permutation assay described in (B) is supplemented with 10 nM tandem hairpin polyamide PA1.
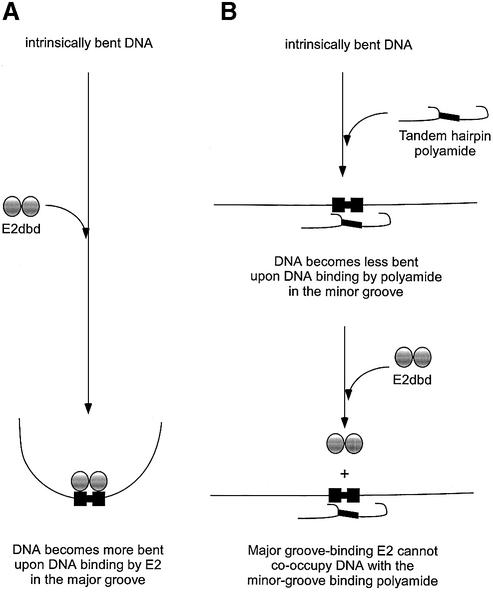
Figure 7.
Model for the inhibition of DNA binding by the major groove-binding HPV-18 E2 homodimer by a reversal of an intrinsic bend in the DNA by a minor groove-binding tandem hairpin polyamide. (A) A hypothetical model is shown for the HPV-18 E2 accentuation of an intrinsic bend in the DNA binding site is based on the enhanced affinity of HPV-16 E2 protein for pre-bent DNA sequences containing adenosine tracts (31) and the increased DNA bending observed in X-ray crystallography studies on BPV-1 E2:DNA co-crystals (33,47). (B) A model for the reversal of the intrinsic bend of the adenosine tract-containing the E2-DNA binding site by the minor groove-binding tandem hairpin polyamide is based on the inhibition of E2 binding experiments shown in Figure 3 and the circular permutation experiment shown in Figure 6.
DISCUSSION
Over the last decade, the pioneering work by Peter Dervan and his colleagues has described the design of synthetic polyamides with DNA binding affinities and specificities comparable to that of natural DNA binding transcriptional regulatory proteins. The initial work was borne out of synthesizing analogs of the A/T-selective minor groove-binding ligand distamycin, which consists of three N-methylpyrrole rings. Subsequent efforts have led to the incorporation of novel heterocycles and chemical moieties paired with alternate structural motifs such as hairpins, slipped dimers, cycles, H-pins and tandem linked hairpins (58). With these developments, polyamides have been synthesized to recognize DNA sequences of increasing length and sequence composition.
Single eight-ring hairpin polyamides have been used successfully to inhibit several DNA binding proteins, such as TBP, LEF-1, ets-1, deadpan, TFIIIA, Herpes simplex viral repressor IE-80 and HTLV-1 Tax. However, in all of these cases the proteins make either exclusively minor groove contacts or combined major and minor groove contacts in the DNA helix. Thus, in many of these examples, polyamides that overlap completely (or partially) with the protein binding sites are effective at displacing the DNA binding protein by disrupting essential amino acid residue–base pair contacts.
In this study, we have extended the utility of the tandem linked hairpin polyamide motif by demonstrating the ability to displace HPV-18 E2, an exclusively major groove-interacting protein. Comparison of a linked tandem hairpin polyamide (PA1) to single hairpin polyamides (PA3 and PA2) designed to the BS4 site showed a dramatic increase in DNA binding affinity of the linked hairpin polyamide as shown in Figure 2. This observation is entirely consistent with earlier reports of Herman et al. (49) that showed that linking two identical six-ring polyamides increased DNA binding affinity by more than 200-fold. In this case, the two six-ring hairpin polyamides are tethered by a five-carbon linker, 5-aminovaleric acid. This polyamide also demonstrated more than 4500-fold specificity for the match site versus a double mismatch site. More recently, Janssen et al. (42) have shown that linking two oligopyrrole type polyamides by an eight-atom linker (8-amino-3,6-dioxaoctanoic acid) also substantially increases the DNA affinity and specificity of the linked molecules compared to the individual modules. Using the same linker, we show that the binding of PA1 and E2 protein to the BS4 site is mutually exclusive as evidenced by our DNase I footprinting experiments. Importantly, the equilibrium dissociation constant (Kd) of PA1 is similar to its equilibrium inhibition constant (Ki). The inability of the single hairpin polyamide (PA3) to displace E2 suggests the possibility of co-occupancy of the site by both polyamide and E2 protein as the two molecules bind on opposite faces of the DNA double helix. This was also evidenced in an earlier study from the Dervan group that showed that an eight-ring polyamide could co-occupy a DNA fragment along with the major groove-binding transcription factor GCN4 (46).
We find that the ability of PA1 to displace E2 from the BS4 site is independent of whether PA1 is added to the DNA prior to or after the addition of protein. Even though the affinities of PA1 and E2 for BS4 are similar (2 nM for PA1 and 4.5 nM for E2), the order of addition of the compound does not affect the Ki. This finding further underscores the suggestion that binding of PA1 and E2 protein is mutually exclusive. Also, this result is consistent with the distinct binding sites of protein and polyamide: PA1 can access its minor groove site even when E2 is bound in the major groove; once bound the polyamide may straighten the DNA and destabilize the E2–DNA complex.
The ability of PA1 to displace E2 prompted us to systematically examine the role of linker lengths in displacement activity. We found that a shorter linker than a diethylene linker greatly affected the DNA binding affinity of the polyamide whereas increasing the length of the linker only moderately affected DNA binding and displacement activity. Unlike the longer linkers, the shorter linker most likely does not provide the conformational flexibility to align the individual polyamide modules to bind to their cognate sites on the duplex.
How does PA1 displace E2? E2 protein, like many other DNA binding proteins that bind either as homodimers or as heterodimers in conjunction with other family members, imparts a bend upon binding. Unlike TBP, which bends the DNA away from the body of the protein, most proteins bend the DNA towards the body of the protein. In many instances, DNA bending appears to play a pivotal role in stabilizing the protein–DNA complex. E2 binds to its core consensus, ACCgN4cGGT, as a homodimer and makes contacts exclusively in the major groove of the DNA helix sequence bending the DNA towards the body of the protein thereby compressing the minor groove at the dimerization interface. The most likely consequence of the binding of polyamide within the dimerization interface might be to prevent the compression of the minor groove and destabilize the E2–DNA complex. The ability of minor groove DNA binding ligands to alter local DNA topology has been extensively documented (59–63). In addition, consistent with the biochemical data presented here, the crystal structure of polyamide complexed with DNA shows a widening of the minor groove by 1–2 Å (64,65). However, the possibility that the linker moieties physically interfere with protein binding at the interface site cannot be ruled out at this time.
Many minor groove DNA binding ligands (such as distamycin, netropsin, echinomycin, etc.) have been endowed with powerful antimicrobial and anticancer properties. Since these molecules lack DNA sequence binding specificity, it is conceivable that at high concentrations these types of molecules might interfere with numerous DNA-directed processes such as transcription, replication, recombination and repair. The ability of these molecules to alter global DNA topology such as negative DNA supercoiling and perturbation of compacted nucleosomal DNA has also been documented (66–70). Thus these molecules might elicit effects on gene expression by indirectly affecting global DNA properties. Not surprisingly, these molecules have prominent cytotoxic effects. The ability of polyamides to bind to DNA in a sequence-dependent context represents a major improvement in utilizing minor groove DNA binding ligands as specific modulators of gene expression. Significantly, the demonstration that these tandem hairpins can be utilized to displace major groove DNA binding proteins considerably increases the repertoire of DNA binding proteins that can be targeted by polyamides.
The feasibility of using these tandem hairpin polyamides as future therapeutics will greatly depend on the determination of the intracellular location of action of these molecules. This is especially important given the recently published observation that in most cell types (with the exception of certain T-cell types), single hairpin polyamides conjugated with fluorescent dyes are localized to the cytoplasm and not the nucleus (71). Thus a major effort in the development of polyamides is focused on screening cell lines that are more amenable to polyamide uptake and understanding the mechanisms for nuclear exclusion of these molecules. The development of a polyamide-based therapeutic will clearly require a multi-disciplinary approach involving medicinal chemists, biologists and pharmacologists.
HPV infection represents a serious unmet medical need. Latest estimates of HPV prevalence among sexually active women without symptoms range from 20 to nearly 50%. It is believed more than 50 million Americans are currently infected with the virus, with the number growing by almost 1 million every year. Although a great majority of HPV infections produce no overt symptoms, a significant portion do develop genital warts (condylomas). Additionally, 90% of all anogential cancers are associated with HPV DNA. Currently, there are no cures for HPV infection. Treatment generally involves surgical removal of warts or application of caustic compounds such as trichloroacetic acid or podophyllin. The ability to interfere with E2-DNA binding activity by small synthetic molecules reported here represents a step in the design of rational molecules targeting HPV.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Michael Botchan (University of California–Berkeley) for helpful discussions and providing essential reagents. We thank the members of the GeneSoft scientific advisory board, especially Ulrich Laemmli (University of Geneva) and Tom Maniatis (Harvard University), for important input into this work. We also thank our valued colleagues at GeneSoft Inc. for many helpful discussions during the course of this project. We are grateful to the referees of the manuscript for suggesting several important improvements to the final version.
REFERENCES
- 1.Howley P.M. (1991) Role of the human papillomaviruses in human cancer. Cancer Res., 51, 5019–5022. [PubMed] [Google Scholar]
- 2.de Villiers E.M. (1994) Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol., 186, 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Zur Hausen H. (1996) Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta Rev. Cancer, 1288, 55–78. [DOI] [PubMed] [Google Scholar]
- 4.Zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nature Rev. Cancer, 2, 342–350. [DOI] [PubMed] [Google Scholar]
- 5.Munger K. (2002) The role of human papillomaviruses in human cancers. Front. Biosci., 7, 641–649. [DOI] [PubMed] [Google Scholar]
- 6.Chow L. and Broker T. (1994) Papillomavirus DNA replication. Intervirology, 37, 150–158. [DOI] [PubMed] [Google Scholar]
- 7.Werness B.A., Levine,A.J. and Howley,P.M. (1990) Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science, 248, 76–79. [DOI] [PubMed] [Google Scholar]
- 8.Dyson N., Howley,P.M., Munger,K. and Harlow,E. (1989) The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science, 243, 934–937. [DOI] [PubMed] [Google Scholar]
- 9.Hawley-Nelson P., Vousden,K.H., Hubbert,N.L., Lowy,D.R. and Schiller,J.T. (1989) HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J., 8, 3905–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howley P.M., Munger,K., Romanczuk,H., Scheffner,M. and Huibregtse,J.M. (1991) Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses. Princess Takamatsu Symp., 22, 239–248. [PubMed] [Google Scholar]
- 11.Munger K., Phelps,W.C., Bubb,V., Howley,P.M. and Schlegel,R. (1989) The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol., 63, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munger K., Scheffner,M., Huibregtse,J.M. and Howley,P.M. (1992) Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products. Cancer Surv., 12, 197–217. [PubMed] [Google Scholar]
- 13.Munger K., Werness,B.A., Dyson,N., Phelps,W.C., Harlow,E. and Howley,P.M. (1989) Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J., 8, 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howley P.M., Scheffner,M., Huibregtse,J. and Munger,K. (1991) Oncoproteins encoded by the cancer-associated human papillomaviruses target the products of the retinoblastoma and p53 tumor suppressor genes. Cold Spring Harb. Symp. Quant. Biol., 56, 149–155. [DOI] [PubMed] [Google Scholar]
- 15.Vousden K. (1993) Interactions of human papillomavirus transforming proteins with the products of tumor suppressor genes. FASEB J., 7, 872–879. [DOI] [PubMed] [Google Scholar]
- 16.Demeret C., Desaintes,C., Yaniv,M. and Thierry,F. (1997) Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol., 71, 9343–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thierry F. and Howley,P.M. (1991) Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol., 3, 90–100. [PubMed] [Google Scholar]
- 18.Steger G. and Corbach,S. (1997) Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol., 71, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanczuk H., Thierry,F. and Howley,P.M. (1990) Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol., 64, 2849–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan S.-H., Louis,E.-C., Walker,P.A. and Bernard,H.-U. (1994) The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E2 promoter through displacement of Sp1 and TFIID. J. Virol., 68, 6411–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee D., Lee,B., Kim,J., Kim,D.W. and Choe,J. (2000) cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem., 275, 7045–7051. [DOI] [PubMed] [Google Scholar]
- 22.Yasugi T., Benson,J.D., Sakai,H., Vidal,M. and Howley,P.M. (1997) Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J. Virol., 71, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow L.T. and Broker,T.R. (1994) Papillomavirus DNA replication. Intervirology, 37, 150–158. [DOI] [PubMed] [Google Scholar]
- 24.Ham J., Dostatni,N., Gauthier,J.M. and Yaniv,M. (1991) The papillomavirus E2 protein: a factor with many talents. Trends Biochem. Sci., 16, 440–444. [DOI] [PubMed] [Google Scholar]
- 25.Kuo S.R., Liu,J.S., Broker,T.R. and Chow,L.T. (1994) Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem., 269, 24058–24065. [PubMed] [Google Scholar]
- 26.Melendy T., Sedman,J. and Stenlund,A. (1995) Cellular factors required for papillomavirus DNA replication. J. Virol., 69, 7857–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demeret C., Le Moal,M., Yaniv,M. and Thierry,F. (1995) Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res., 23, 4777–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R. and Botchan,M.R. (1993) The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell, 73, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 29.Hegde R.S. (2002) The papillomavirus E2 proteins: structure, function and biology. Annu. Rev. Biophys. Biomol. Struct., 31, 343–360. [DOI] [PubMed] [Google Scholar]
- 30.Kim S.S., Tam,J.K., Wang,A.F. and Hegde,R.S. (2000) The structural basis of DNA target discrimination by papillomavirus E2 proteins. J. Biol. Chem., 275, 31245–31254. [DOI] [PubMed] [Google Scholar]
- 31.Hines C.S., Meghoo,C., Shetty,S., Biburger,M., Brenowitz,M. and Hegde,R.S. (1998) DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins. J. Mol. Biol., 276, 809–818. [DOI] [PubMed] [Google Scholar]
- 32.Rozenberg H., Rabinovich,D., Frolow,F., Hegde,R.S. and Shakked,Z. (1998) Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc. Natl Acad. Sci. USA, 95, 15194–15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde R.S., Grossman,S.R., Laimins,L.A. and Sigler,P.B. (1992) Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature, 359, 505–512. [DOI] [PubMed] [Google Scholar]
- 34.Trauger J.W., Baird,E.E. and Dervan,P.B. (1996) Recognition of DNA by designed ligands at subnanomolar concentrations. Nature, 382, 559–561. [DOI] [PubMed] [Google Scholar]
- 35.Gottesfeld J.M., Neely,L., Trauger,J.W., Baird,E.E. and Dervan,P.B. (1997) Regulation of gene expression by small molecules. Nature, 387, 202–205. [DOI] [PubMed] [Google Scholar]
- 36.White S., Baird,E.E. and Dervan,P.B. (1997) On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem. Biol., 4, 569–578. [DOI] [PubMed] [Google Scholar]
- 37.Dervan P.B. and Burli,R.W. (1999) Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol., 3, 688–693. [DOI] [PubMed] [Google Scholar]
- 38.Kelly J.J., Baird,E.E. and Dervan,P.B. (1996) Binding site size limit of the 2:1 pyrrole-imidazole polyamide-DNA motif. Proc. Natl Acad. Sci. USA, 93, 6981–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickinson L.A., Gulizia,R.J., Trauger,J.W., Baird,E.E., Mosier,D.E., Gottesfeld,J.M. and Dervan,P.B. (1998) Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl Acad. Sci. USA, 95, 12890–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ansari A.Z., Mapp,A.K., Nguyen,D.H., Dervan,P.B. and Ptashne,M. (2001) Towards a minimal motif for artificial transcriptional activators. Chem. Biol., 8, 583–592. [DOI] [PubMed] [Google Scholar]
- 41.Mapp A.K., Ansari,A.Z., Ptashne,M. and Dervan,P.B. (2000) Activation of gene expression by small molecule transcription factors. Proc. Natl Acad. Sci. USA, 97, 3930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssen S., Durussel,T. and Laemmli,U.K. (2000) Chromatin opening of DNA satellites by targeted sequence-specific drugs. Mol. Cell, 6, 999–1011. [DOI] [PubMed] [Google Scholar]
- 43.Janssen S., Cuvier,O., Muller,M. and Laemmli,U.K. (2000) Specific gain- and loss-of-function phenotypes induced by satellite-specific DNA-binding drugs fed to Drosophila melanogaster. Mol. Cell, 6, 1013–1024. [DOI] [PubMed] [Google Scholar]
- 44.Henikoff S. and Vermaak,D. (2000) Bugs on drugs go GAGAA. Cell, 103, 695–698. [DOI] [PubMed] [Google Scholar]
- 45.Winston R.L., Ehley,J.A., Baird,E.E., Dervan,P.B. and Gottesfeld,J.M. (2000) Asymmetric DNA binding by a homodimeric bHLH protein. Biochemistry, 39, 9092–9098. [DOI] [PubMed] [Google Scholar]
- 46.Bremer R.E., Baird,E.E. and Dervan,P.B. (1998) Inhibition of major-groove-binding proteins by pyrrole-imidazole polyamides with an Arg-Pro-Arg positive patch. Chem. Biol., 5, 119–133. [DOI] [PubMed] [Google Scholar]
- 47.Hegde R.S. and Androphy,E.J. (1998) Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. J. Mol. Biol., 284, 1479–1489. [DOI] [PubMed] [Google Scholar]
- 48.Baird E.E. and Dervan,P.B. (1996) Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc., 118, 6141–6146. [DOI] [PubMed] [Google Scholar]
- 49.Herman D.M., Baird,E.E. and Dervan,P.B. (1999) Tandem hairpin motif for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem. Eur. J., 5, 975. [DOI] [PubMed] [Google Scholar]
- 50.Carey M. and Smale,S.T. (2000) Transcription Regulation in Eukaryotes: Concepts, Strategies and Techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Trauger J.W. and Dervan,P.B. (2001) Footprinting methods for analysis of pyrrole-imidazole polyamide/DNA complexes. Methods Enzymol., 340, 450–466. [DOI] [PubMed] [Google Scholar]
- 52.Thompson J.F. and Landy,A. (1988) Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res., 16, 9687–9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeshima K., Janssen,S. and Laemmli,U.K. (2001) Specific targeting of insect and vertebrate telomeres with pyrrole and imidazole polyamides. EMBO J., 20, 3218–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravishanker G., Swaminathan,S., Beveridge,D.L., Lavery,R. and Sklenar,H. (1989) Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves”, dials and windows. J. Biomol. Struct. Dyn., 6, 669–699. [DOI] [PubMed] [Google Scholar]
- 55.Lavery R. and Sklenar,H. (1989) Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 56.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 57.Wu H.M. and Crothers,D.M. (1984) The locus of sequence-directed and protein-induced DNA bending. Nature, 308, 509–513. [DOI] [PubMed] [Google Scholar]
- 58.Dervan P.B. (2001) Molecular recognition of DNA by small molecules. Bioorg. Med. Chem., 9, 2215–2235. [DOI] [PubMed] [Google Scholar]
- 59.Albert F.G., Eckdahl,T.T., Fitzgerald,D.J. and Anderson,J.N. (1999) Heterogeneity in the actions of drugs that bind in the DNA minor groove. Biochemistry, 38, 10135–10146. [DOI] [PubMed] [Google Scholar]
- 60.Hansma H.G., Browne,K.A., Bezanilla,M. and Bruice,T.C. (1994) Bending and straightening of DNA induced by the same ligand: characterization with the atomic force microscope. Biochemistry, 33, 8436–8441. [DOI] [PubMed] [Google Scholar]
- 61.Barcelo F., Muzard,G., Mendoza,R., Revet,B., Roques,B.P. and Le Pecq,J.B. (1991) Removal of DNA curving by DNA ligands: gel electrophoresis study. Biochemistry, 30, 4863–4873. [DOI] [PubMed] [Google Scholar]
- 62.Waring M. (1991) Binding of antibiotics to DNA. Ciba Found. Symp., 158, 128–142. [PubMed] [Google Scholar]
- 63.Cons B.M. and Fox,K.R. (1990) Effects of sequence selective drugs on the gel mobility of a bent DNA fragment. Biochem. Biophys. Res. Commun., 171, 1064–1070. [DOI] [PubMed] [Google Scholar]
- 64.Kielkopf C.L., White,S., Szewczyk,J.W., Turner,J.M., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science, 282, 111–115. [DOI] [PubMed] [Google Scholar]
- 65.Kielkopf C.L., Baird,E.E., Dervan,P.B. and Rees,D.C. (1998) Structural basis for G.C recognition in the DNA minor groove. Nature Struct. Biol., 5, 104–109. [DOI] [PubMed] [Google Scholar]
- 66.Storl K., Burckhardt,G., Lown,J.W. and Zimmer,C. (1993) Studies on the ability of minor groove binders to induce supercoiling in DNA. FEBS Lett., 334, 49–54. [DOI] [PubMed] [Google Scholar]
- 67.Triebel H., Bar,H., Walter,A., Burckhardt,G. and Zimmer,C. (1994) Modulation of DNA supercoiling by interaction with netropsin and other minor groove binders. J. Biomol. Struct. Dyn., 11, 1085–1105. [DOI] [PubMed] [Google Scholar]
- 68.Portugal J. and Waring,M.J. (1986) Antibiotics which can alter the rotational orientation of nucleosome core DNA. Nucleic Acids Res., 14, 8735–8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Portugal J. and Waring,M.J. (1987) Interaction of nucleosome core particles with distamycin and echinomycin: analysis of the effect of DNA sequences. Nucleic Acids Res., 15, 885–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Low C.M., Drew,H.R. and Waring,M.J. (1986) Echinomycin and distamycin induce rotation of nucleosome core DNA. Nucleic Acids Res., 14, 6785–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belitsky J.M., Leslie,S.J., Arora,P.S., Beerman,T.A. and Dervan,P.B. (2002) Cellular uptake of N-methylpyrrole/N-methylimidazole polyamide-dye conjugates. Bioorg. Med. Chem., 10, 3313–3318. [DOI] [PubMed] [Google Scholar]