Abstract
Regulation of cellular functions can be accomplished by many mechanisms, including transcriptional regulation, alternative splicing, translational regulation, phosphorylation and other posttranslational covalent modifications, degradation, localization, protein–protein interactions, and small-molecule allosteric effectors. Largely because of advances in the techniques of molecular biology in the past few decades, our knowledge of regulation by most of these mechanisms has expanded enormously. Regulation by small-molecule, allosteric interactions is an exception. Many of the best-known allosteric regulators were discovered decades ago, when we knew little about all of these other forms of regulation. Allostery is the most direct, rapid, and efficient regulatory mechanism to sense changes in the concentration of small molecules and alter cellular responses to maintain homeostasis. In this perspective, we present the argument that allosteric regulation is underappreciated in the systems biology world and that many allosteric effectors remain to be discovered.
Two of the earliest examples of allosteric regulation are the control of oxygen binding to hemoglobin by protons (1) and AMP activation of glycogen phosphorylase (2). During the heyday of metabolic research, allosteric regulators were actively sought out. By the time that the “Central Dogma of Molecular Biology” was proposed (3), several examples of “end-product” or “feedback” inhibition were already known (4–7). In each case, the end product of a metabolic pathway was shown to inhibit an enzyme early in the pathway. Surprisingly, in each case the inhibitor was “not a steric analogue of the substrate,” and in 1961 Monod and Jacob (8) coined the term “allosteric inhibition” to indicate inhibitory interactions at a site distinct from the active site. This first discussion of allosteric enzymes was prescient in many respects; they proposed the following: (i) that allostery need not be restricted to end-product inhibition and could be a general form of regulation, (ii) that it might be very difficult to predict the chemical nature of allosteric effectors and therefore to discover them, and (iii) that allosteric sites might be even more useful as drug targets than active sites. By 1965, there were at least 31 allosteric effectors known (9), but only 1 protein kinase/substrate pair (phosphorylase kinase/glycogen phosphorylase) had been identified (10, 11).
How common is allosteric regulation? Is it primarily restricted to the committed step of metabolic pathways, or is it a widespread, perhaps even universal, phenomenon? The answers to these questions are not yet known, but there are many reasons to think that allosteric regulation is not rare. First, allostery is an ideal way for proteins (and RNAs) to sense the changing milieu of a cell and respond to maintain cellular homeostasis. Allostery provides a mechanism to directly sense the levels of small molecules. It can be very rapid, on the order of the rate of diffusion (≈108 M−1·s−1), and readily reversible. And unlike changes in covalent protein modification such as phosphorylation/dephosphorylation cycles, no cellular energy is used up in allosteric signaling. Second, a relatively large percentage of the proteins studied 40 years ago were known to be allosterically regulated, and, although the number of known proteins has skyrocketed, the number of known allosteric regulators has not. Forty years ago, the effort required to discover a new protein was at least that required to discover a small-molecule regulator. Today, gene and protein discovery is systematic, whereas small-molecule effector discovery still occurs one protein at a time and typically only on the rare occasions when the investigator has a reason to look for such regulators.
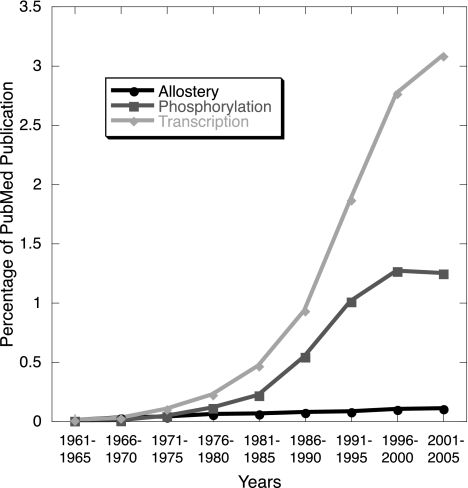
The mechanisms of regulation that have primarily been revealed in the last two decades are those that could be discovered by the tools of molecular biology. Our abilities to synthesize, manipulate, and amplify nucleic acids, as well as to create specific antibodies, have made it possible to discover transcriptional and posttranscriptional modes of regulation on a large scale. Transcriptional regulation can be studied by using promoter fusion constructs, chromatin immunoprecipitation, microarrays, quantitative RT-PCR, Northern blots, etc. Likewise, the analysis of cellular regulation by splicing, mRNA stability, translation, phosphorylation, proteolysis, etc. has become possible because of similar tools. In contrast, these tools have not yet resulted in widely used, routine methods to discover small, allosteric effectors. One consequence is that the number of studies describing modes of regulation amenable to molecular biology techniques, such as transcription or phosphorylation, have increased dramatically over the past 4 decades, whereas those describing allosteric regulation have not (see Fig. 1). Although scientists have described transcriptomes, proteomes, interactomes, and kinomes, the “allosterome” is still a mystery.
Fig. 1.
Percent of papers listed in PubMed (www.ncbi.nlm.nih.gov/entrez/query.fcgi) describing transcription, phosphorylation, and allostery during the past 4 decades. Titles and abstracts were searched for the following terms: “alloster*,” “protein kinase” OR “tyrosine kinase” OR “ser/thr kinase” OR “phosphorylase kinase” OR “phosphoprotein” OR “protein phosphorylation,” and “transcription*.” The number of papers identified was divided by the total number of papers published during the time interval.
How are allosteric regulators generally found? In the past, they were primarily found either by serendipity or metabolic intuition followed by experimental verification. For example, the discovery of the allosteric activation of the AMP-activated protein kinase (AMPK) was a combination of these methods. Kinetic studies showed that this kinase had maximal activity at 400 μM ATP and barely detectable activity at 4 mM ATP (12). The protein substrate of AMPK that was being studied, acetyl CoA carboxylase, catalyzes the committed step in fatty acid synthesis and is inhibited by phosphorylation. It made sense that fatty acid synthesis should be inhibited when the energy charge of the cell is low, providing the rationale for testing whether AMP activates the acetyl CoA carboxylase-inhibiting kinase. A relatively high AMP/ATP ratio is now known to activate AMPK, and this enzyme is often called the cellular “energy gauge” or “fuel sensor” (13).
More recently, several allosteric effectors have been discovered from high-throughput chemical screens in search of specific enzyme activators or inactivators. For example, metabolic studies provided no reason to suspect that monomeric glucokinase would be regulated by allostery. However, it was known that increased glucokinase activity could decrease blood glucose levels, and hence there was a strong motivation to search for activators of this enzyme as potential diabetes therapeutics. A high-throughput chemical screen revealed a class of small-molecule activators that both increases the kcat and decreases the S0.5 for glucose and lowers the blood glucose concentration in rodent models of diabetes (14). Crystal structures of the enzyme plus and minus the activator and glucose indicate that the activator binds ≈20 Å from the active site and appears to stabilize an active conformation of the enzyme (15). Five known activating mutations of glucokinase, discovered from patients with hypoglycemia, map to the same site (16). Additionally, a glucokinase mutant (V62M) discovered in a patient with maturity-onset diabetes of the young, type 2, also resides at this site. The purified V26M glucokinase has a decreased S0.5 for glucose, the opposite of what one would expect for a patient with hyperglycemia (and from the other known MODY2 mutants) (16). Intriguingly, the V26M mutant does not respond to stimulation by the pharmacological allosteric activator (16). This combination of findings about the V26M mutant (less active in vivo, more active in vitro, and resistant to the pharmacological allosteric activator) strongly suggests the existence of an endogenous allosteric regulator of glucokinase.
A second example of “where there’s a will there’s a way” to discover allosteric regulators is the analysis of caspases by Wells and colleagues (17–19). These researchers have used a high-throughput, disulfide-tethering technique to find small-molecule allosteric inhibitors of caspase-1, -3, and -7. For caspase-1 and -7, the allosteric sites are located at the dimer interfaces, ≈15 Å from the active sites. Binding of the allosteric inhibitors stabilizes conformations of the caspases that are catalytically inactive and are similar to the zymogen forms of the proteases; in other words, the inhibitors functionally reverse zymogen activation (17, 19). The authors postulate the existence of an endogenous allosteric inhibitor that would act as a safeguard against spurious caspase activation.
This high-throughput, disulfide-tethering technique addresses two of the major difficulties in discovering allosteric effectors: unknown chemical composition and low affinity. Although proteins are made from amino acids and nucleic acids are made from nucleosides, small-molecule effectors come from a wide spectrum of chemical classes. Although it is often possible to predict what compounds might bind at the active site based on the enzymatic reaction that occurs there, no such rules apply to allosteric sites. Therefore, an unbiased screen of a diverse, small-molecule library is crucial. The glucokinase activator described above was initially found in a screen of 120,000 structurally diverse compounds, followed by chemical optimization of the initial compound (14). The unique capability of the disulfide-tethering technique is the ability to find low-affinity effectors. Many of the small molecules in a cell that may be sensed by allosteric proteins are present at relatively high concentrations (micromolar to millimolar). For a protein to sense the change in such a metabolite, the affinity would need to be low, and low-affinity interactions are difficult to discover. This novel covalent tethering method side-steps the need for high affinity.
As illustrated by the two examples of newly discovered allosteric regulators described above, pharmacologists have realized the utility of searching for allosteric effectors in addition to, or instead of, active-site ligands. Active-site ligands are constrained by the active-site chemistry, whereas allosteric sites have no such constraints. Active-site structures are often quite similar between distinct proteins, presenting a challenge for drug selectivity. Allosteric sites are not generally conserved between different proteins (19, 20). Although the idea that drugs targeting allosteric sites might be even more effective than those targeting active sites has only recently become a popular notion among pharmacologists, it was proposed in the first paper defining allostery (8).
In addition to the problem that allosteric effectors are not generally easy to identify, the relative lack of interest in discovering new allosteric regulators by biologists may stem from a misconception about what types of proteins are susceptible to allosteric regulation. Allosteric regulation simply means that a protein’s activity is regulated by binding of a ligand at a site other than the active site. By this definition, all proteins could be allosterically regulated, an idea that has been suggested by others (21). However, there is a common perception that only oligomeric proteins, particularly those with multiple identical subunits, can be allosterically regulated. This perception may stem from the history of the discovery of allosteric regulators. The Monod–Wyman–Changeux (MWC) and Koshland–Némethy–Filmer (KNF) models were proposed in the 1960s to explain the allosteric regulation seen in hemoglobin and several metabolic regulatory enzymes (9, 22). Several of the best studied of these systems not only exhibited heterotrophic regulation (meaning regulation by small molecules that were distinct from the substrates or primary ligand) but also exhibited positive cooperativity or positive homotropic regulation. For example, oxygen is both the primary ligand for hemoglobin and, because it binds with positive cooperativity, is a positive homotropic regulator of hemoglobin. It is this positive cooperativity or homotropic regulation that causes the sigmoidal binding and initial velocity curves so commonly associated with allosteric proteins. It is important to realize that the MWC and KNF models were developed as inclusive models that could explain both the positive cooperativity and the heterotropic allostery seen in many of the proteins that have been studied to date. Although these landmark papers have brilliantly guided the subsequent studies on the allosteric regulation of oligomeric proteins, and particularly positive cooperativity, they also have been misinterpreted by some to indicate that only oligomeric proteins can be allosterically regulated. In fact, even a trusted textbook of biochemistry (23), when discussing allosteric enzymes, states that “[t]hese enzymes consist of multiple subunits and multiple active sites.“ Additionally, a recent high-profile review of allosteric mechanisms in signaling pathways focused on the WMC theory and hence only discussed allostery as it applies to oligomers, particularly homooligomers (24).
Although many of the originally discovered allosterically regulated proteins happened to be oligomers, there is no a priori reason why heterotropically regulated proteins need to be multimers, particularly homooligomers. In fact, there are many allosterically regulated monomeric and heterooligomeric proteins that do not show cooperativity in substrate binding or catalysis. Two examples include Ca2+ binding by recoverin (25, 26) and PAS-domain-containing cytoplasmic histidine kinases (27). The cellular fuel sensor AMPK, although not a monomer, has a monomeric AMP/ATP sensing domain (13). Therefore, although one’s favorite protein may not have an oligomeric structure similar to hemoglobin, phosphofructokinase, or aspartate transcarbamoylase, it may still be regulated by allostery.
In summary, even though recent technological advances have not facilitated systematic discovery of small-molecule, allosteric regulators, a great many may well exist. It is difficult to imagine that most of the biologically important allosteric regulators were already discovered before the days of PCR and commercially available restriction enzymes; there must be more just waiting to be revealed.
Acknowledgments
We thank Drs. Timothy Formosa, Martin C. Rechsteiner, and Steven L. McKnight for thoughtful critiques of this manuscript.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bohr C. Zentr. Physiol. 1903;17:682. [Google Scholar]
- 2.Cori G., Colowick S., Cori C. J. Biol. Chem. 1938;123:381–389. [Google Scholar]
- 3.Crick F. Symp. Soc. Exp. Biol. 1958;XII:138. [PubMed] [Google Scholar]
- 4.Novick A., Szilard L. In: Dynamics of Growth Processes. Boell E. J., editor. Princeton: Princeton Univ. Press; 1954. pp. 21–32. [Google Scholar]
- 5.Pardee A. B., Yates R. A. J. Biol. Chem. 1956;221:757–770. [PubMed] [Google Scholar]
- 6.Roberts R., Abelson P., Cowie D., Bolton E., Britten R. Studies of Biosynthesis in Escherichia coli. Washington, DC: Carnegie Institution of Washington Publ.; 1955. p. 607. [Google Scholar]
- 7.Umbarger H. E. Science. 1956;123:848. doi: 10.1126/science.123.3202.848. [DOI] [PubMed] [Google Scholar]
- 8.Monod J., Jacob F. Cold Spring Harbor Symp. Quant. Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 9.Monod J., Wyman J., Changeux J. P. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 10.Krebs E. G., Fischer E. H. Biochim. Biophys. Acta. 1956;20:150–157. doi: 10.1016/0006-3002(56)90273-6. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland E. W., Jr., Wosilait W. D. Nature. 1955;175:169–170. doi: 10.1038/175169a0. [DOI] [PubMed] [Google Scholar]
- 12.Yeh L. A., Lee K. H., Kim K. H. J. Biol. Chem. 1980;255:2308–2314. [PubMed] [Google Scholar]
- 13.Kahn B. B., Alquier T., Carling D., Hardie D. G. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Grimsby J., Sarabu R., Corbett W. L., Haynes N. E., Bizzarro F. T., Coffey J. W., Guertin K. R., Hilliard D. W., Kester R. F., Mahaney P. E., et al. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 15.Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. Structure (London) 2004;12:429–438. doi: 10.1016/j.str.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Gloyn A. L., Odili S., Zelent D., Buettger C., Castleden H. A., Steele A. M., Stride A., Shiota C., Magnuson M. A., Lorini R., et al. J. Biol. Chem. 2005;280:14105–14113. doi: 10.1074/jbc.M413146200. [DOI] [PubMed] [Google Scholar]
- 17.Hardy J. A., Lam J., Nguyen J. T., O’Brien T., Wells J. A. Proc. Natl. Acad. Sci. USA. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy J. A., Wells J. A. Curr. Opin. Struct. Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Scheer J. M., Romanowski M. J., Wells J. A. Proc. Natl. Acad. Sci. USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christopoulos A. Nat. Rev. Drug Discov. 2002;1:198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 21.Gunasekaran K., Ma B., Nussinov R. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 22.Koshland D. E., Jr., Nemethy G., Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 23.Berg J. M., Tymoczko J. L., Stryer L. Biochemistry. New York: Freeman; 2002. p. 208. [Google Scholar]
- 24.Changeux J. P., Edelstein S. J. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 25.Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., Ikura M., Tsien R. Y. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T., Ames J. B., Harvey T. S., Stryer L., Ikura M. Nature. 1995;376:444–447. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 27.Ponting C. P., Aravind L. Curr. Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]