Abstract
CDK1 is a nonredundant cyclin-dependent kinase (CDK) with an essential role in mitosis, but its multiple functions still are poorly understood at a molecular level. Here we identify a selective small-molecule inhibitor of CDK1 that reversibly arrests human cells at the G2/M border of the cell cycle and allows for effective cell synchronization in early mitosis. Inhibition of CDK1 during cell division revealed that its activity is necessary and sufficient for maintaining the mitotic state of the cells, preventing replication origin licensing and premature cytokinesis. Although CDK1 inhibition for up to 24 h is well tolerated, longer exposure to the inhibitor induces apoptosis in tumor cells, suggesting that selective CDK1 inhibitors may have utility in cancer therapy.
Keywords: apoptosis, cancer therapy, cell cycle, cytokinesis, replication origin
The cell cycle is a precisely controlled set of biochemical and morphological events driven by the sequential activation of cyclin-dependent kinases (CDKs) (1). Three CDKs and their activating cyclins (A, B, D, and E) play key roles in mammalian cell cycle regulation (2). It has been established that CDK4/cyclin D and CDK2/cyclin E/A promote the passage through G1 and S phases, whereas CDK1/cyclin B regulates the transition through late G2 and mitosis (3). However, recent genetic and RNA interference studies in mammalian cells have revealed that CDK2 and CDK4 are not essential for cell cycle progression, thus leaving CDK1 as the only nonredundant cell cycle driver (4–6). Genetic studies in yeast and mammalian cells have established the critical role of CDK1 in mitosis, but the long list of its putative substrate proteins still is growing, and its precise functions in the context of the dividing cell are poorly understood (7, 8). This deficiency is due, in part, to the lack of specific molecular tools for reversible modulation of CDK1 activity in vivo. Several potent small-molecule inhibitors have been reported, but their activity and cell cycle profiles are not consistent with specific CDK1 inhibition (9, 10). Here, we identify a selective and reversible inhibitor of the catalytic activity of human CDK1/cyclin B1 and CDK1/cyclin A complexes that allows synchronization of proliferating cells in the late G2 phase and probing of CDK1 function in the cellular context.
Results and Discussion
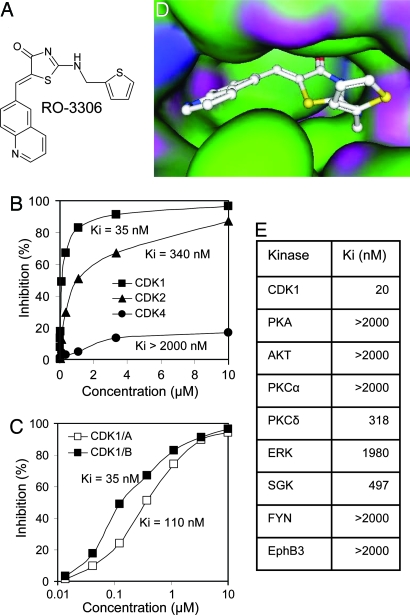
We screened a diverse library of organic compounds for their ability to inhibit the catalytic activity of human CDK1/cyclin B1. The hits then were tested for selectivity against CDK2/cyclin E and CDK4/cyclin D. A class of thiazolinone analogs emerged as a source of ATP-competitive CDK1 inhibitors that were then further optimized for potency, selectivity, and ability to modulate CDK1 in proliferating cells. One quinolinyl thiazolinone derivative, RO-3306, showed good potency, in vitro selectivity, and a cell cycle profile (G2/M arrest) consistent with CDK1 inhibition (Fig. 1A). RO-3306 inhibited CDK1/cyclin B1 activity with Ki of 35 nM, nearly 10-fold selectivity relative to CDK2/cyclin E and >50-fold relative to CDK4/cyclin D (Fig. 1B). It also inhibited CDK1/cyclin A complexes with Ki of 110 nM (Fig. 1C). RO-3306 is an ATP-competitive inhibitor and, therefore, is likely to bind in the ATP pocket of CDK1. The crystal structure of CDK1 has not been resolved yet, but the high degree of primary structure homology (86%) between the ATP-binding domains of CDK1 and CDK2 permits modeling of compound binding by using the CDK2 structure (11). Molecular docking experiments positioned RO-3306 in the ATP-binding pocket of CDK2 (Fig. 1D). The fact that RO-3306 is fairly selective against the most closely related kinase, CDK2, suggested that it may be selective against most cellular kinases. Indeed, RO-3306 showed >15-fold selectivity against a diverse panel of eight human kinases (Fig. 1E).
Fig. 1.
RO-3306 is a selective CDK1 inhibitor. (A) Chemical structure of RO-3306. (B) Activity of RO-3306 against CDK1/cyclin B1, CDK2/cyclin E, and CDK4/cyclin D. (C) Activity of RO-3306 against CDK1 complexes with cyclin A or cyclin B1. (D) A presumed model of RO-3306 binding at the ATP pocket of CDK2 using computation data and structure coordinates. (E) Activity of RO-3306 against eight diverse kinases measured by the IMAP assay.
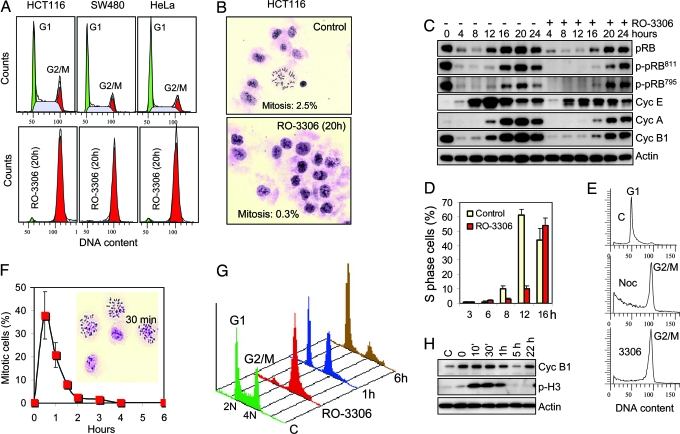
It has been well established that CDK1/cyclin B1 activity is required for cell cycle transition through mitosis (12). Cyclin B1 accumulates in the G2 phase, ensuring peak levels of CDK1 activity at the G2/M border. However, CDK1 activity is controlled by inhibitory phosphorylation until activating phosphatases prime the cells for entry into mitosis (13). Treatment of proliferating cells with a CDK1-specific inhibitor should not affect the transition through G1 and S phases but should confer arrest at the G2/M border. Removal of the chemical inhibitor then should allow rapid and synchronous entry into mitosis. In full agreement with this model, treatment of proliferating human cancer cells (HCT116, SW480, and HeLa) with RO-3306 for 20 h led to a complete block of the cell cycle in the G2/M phase (Fig. 2A). The arrested cells were large in size with a flat morphology typical for G2 cells. Their low mitotic index (<1%) confirmed that the cells were in the G2 phase (Fig. 2B).
Fig. 2.
RO-3306 reversibly arrests cells at the G2/M phase border. (A) Cell cycle profile of proliferating human cells (HCT116, SW480, and HeLa) treated with solvent (Upper) or RO-3306 (9 μM) (Lower) for 20 h. (B) Mitotic spreads of RO-3306-arrested HCT116 cells treated as above. (C) Cell transition through the G1 and S phase in the presence of RO-3306. HeLa cells were synchronized in mitosis by a shake-off after 14 h of incubation in 100 nM nocodazole, washed, and allowed to transition through the G1 and S phase in the presence or absence of RO-3306 (9 μM). Cells were collected at the indicated times after release from nocodazole block, and total pRB, phospho-pRB, and cyclins E, A, and B1 were analyzed by Western blotting. (D) Kinetics of S phase entry in the presence of RO-3306. HeLa cells synchronized in mitosis by nocodazole as above were allowed to transition through the G1 phase in the presence or absence of RO-3306 (9 μM), and the number of S phase cells was determined by flow cytometry of BrdU-labeled cells. (E) Cell cycle distribution of HeLa cells treated as above. Cells were analyzed 3 h after release from nocodazole block (c), 21 h after readdition of 100 nM nocodazole at 3 h after release from nocodazole block (noc), or 24 h after release from nocodazole block in the presence of 9 μM RO-3306 (3306). (F) Cells synchronously enter mitosis after release from RO-3306 block. HCT116 cells were arrested by RO-3306 (9 μM for 20 h), washed, and incubated in fresh prewarmed medium. Mitotic spreads were prepared at the indicated times and used to calculate mitotic index. Inset shows mitotic spreads from a 30-min sample. (G) RO-3306-synchronized cells transition normally through mitosis. Cell cycle profile of HCT116 cells 1 and 6 h after release from a 20-h RO-3306 block compared with untreated (C) and RO-3306-arrested cells. (H) Western blot analysis of cyclin B1 and phospho-histone H3 in HCT116 cells after release from RO-3306 block.
To assess the CDK1 selectivity of RO-3306 in vivo, we examined the phosphorylation status of retinoblastoma protein (pRB) residues Ser-795 and Ser-811, known as preferred cellular substrates of CDK4 and CDK2, respectively (14). Because CDK2 and CDK4 phosphorylate pRB during the late G1 phase, we synchronized HeLa cells in prometaphase by a shake-off in the presence of nocodazole and allowed them to transition through the G1 and S phase in the presence or absence of RO-3306 after the levels of pRB, cyclin E, cyclin A, and cyclin B1 (Fig. 2C). RO-3306 did not prevent the phosphorylation of pRB on either serine residue 20–24 h after release from the nocodazole block. At this time, the majority of the cells in both populations passed through the G1 and S phase. This result suggests that RO-3306 does not significantly affect cellular CDK2 and CDK4 function at concentrations that completely block CDK1 function. However, RO-3306 delayed entry into the S phase by ≈4 h compared with the controls (Fig. 2D). This delay could be due to a slightly reduced cellular CDK2 activity, a possible role of CDK1 in the G1 phase, or an unknown off-target effect. Despite the delayed entry, HeLa cells traversed normally through the S phase in the presence of 9 μM RO-3306 and effectively accumulated in the G2/M phase (Fig. 2E). The fact that the cells transitioned through the G1 and S phase in the absence of CDK1 activity suggests that CDK1 does not play an essential role outside of mitosis in human cells with normal levels of CDK2/4.
To check the reversibility of cell cycle arrest, HCT116 cells were released from a 18-h RO-3306 block, and the appearance of mitotic cells was followed by time-lapse photography (Movie 1, which is published as supporting information on the PNAS web site) or mitotic index analysis (Fig. 2F). The first mitotic cells appeared as early as 10 min after drug removal and reached a peak number between 30 and 40 min when nearly half of the cell population was in metaphase. Western blot analysis showed changes in phospho-histone H3, a marker of chromosome condensation (15), consistent with the observed synchronous passage through mitosis (Fig. 2H). This rapid entry into mitosis indicates that the cells are arrested at the very border between the G2 and M phases. The cell cycle profile of RO-3306-synchronized HCT116 cells revealed that G2-arrested cells retain their viability after 18 h of exposure to the drug and can transition normally through mitosis and the subsequent G1 phase (Fig. 2G). Four additional cancer cell lines (RKO, SJSA, MDA-MB-435, and DU145) subjected to this treatment also were arrested by RO-3306 and synchronously entered mitosis (data not shown). These results suggest that reversible CDK1 inhibition could provide an effective means for cell synchronization in the late G2 phase and permit the study of the molecular and structural events during the G2/M transition and early mitosis. Large quantities of mitotic cells can be isolated by a mitotic shake-off 30–60 min after release from RO-3306 block or trapped in prometaphase after 2 h of incubation with 100 nM nocodazole immediately after release. Combined RO-3306/nocodazole trap yielded 50–80% mitotic cells in all tested cancer cell lines 2–3 h after release. This short treatment minimizes the toxicity of nocodazole exposure and provides a healthier mitotic cell population.
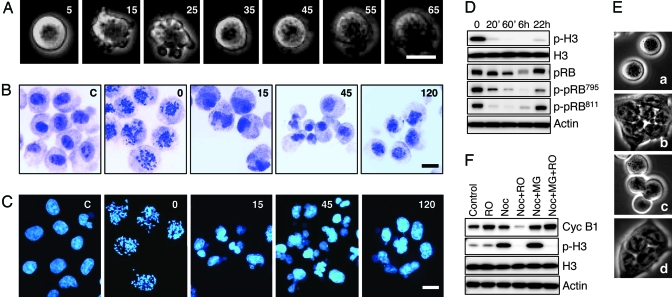
It has been well documented that CDK1 activity is critical for initiation and transition through mitosis, but its multiple functions still are not fully accounted for at the molecular level (8). Using RO-3306 as a specific and reversible probe, we investigated the role of CDK1 activity during mitosis. HeLa cells synchronized in prometaphase by nocodazole were incubated with RO-3306 in the presence of nocodazole and followed by time-lapse photography, mitotic spread analysis, and Western blotting (Fig. 3). In <1 hr, the cells underwent dramatic morphological changes as they transitioned from a rounded prometaphase to a flat interphase morphology. Their surface showed pronounced blebbing 5–20 min after RO-3306 addition (Fig. 3A), accompanied by rapid chromosome decondensation, nuclear reformation (Fig. 3 B and C), loss of phospho-H3, and cyclin B1 (Fig. 3D). Initially, nuclear formation appeared chaotic, but by 2 h, the transformation was completed, and most nuclei appeared normal. The initial loss of PRB phosphorylation on Ser-795 and Ser-811, along with degradation of cyclin B1 and the morphological changes observed, are consistent with mitotic exit into a G1-like state where CDK activities are generally low and APC ubiquitin ligase activity is high as a direct result of loss of CDK1 activity (Fig. 3D).
Fig. 3.
CDK1 activity is critical for maintaining the mitotic state. (A) Nocodazole-arrested cells rapidly exit mitosis in the presence of RO-3306. HeLa cells arrested in prometaphase by nocodazole treatment (16 h) were followed by time-lapse photography after the addition of 9 μM RO-3306. (B) Inhibition of CDK1 activity in nocodazole-arrested mitotic cells triggers chromosome decondensation and nuclei reformation. Purified population of nocodazole-arrested metaphase cells was collected by shake-off and treated with RO-3306 (9 μM). Mitotic spreads were prepared at different times and stained with Giemsa. (C) HeLa cells were treated as above but stained for DNA with Hoechst 33258. (D) Western blot analysis of phospho-H3 and phospho-pRB during RO-3306-induced exit from mitosis. (E) Inhibition of cyclin B1 degradation does not prevent mitotic exit in RO-3306-treated HeLa cells. Purified nocodazole-arrested metaphase cells were treated with solvent (a and b) or 10 μM MG132 (c and d) for 30 min followed by the addition of 9 μM RO-3306 for 2 h (b and d). (F) Western blot analysis of cyclin B1 and phospho-H3 in the cells treated as above. (Scale bars: 20 μm.)
It has been well established that exit from mitosis is signaled by the APC-mediated degradation of cyclin B1 (16). Inhibition of cyclin B1 degradation in nocodazole-arrested cells by treatment with the proteasome inhibitor MG132 did not stop HeLa cells from transitioning out of mitosis in response to incubation with RO-3306 (Fig. 3 E and F), suggesting that loss of CDK1 kinase activity but not cyclin B1 function is the cause of the forced mitotic exit. A similar mitotic exit was observed previously in mammalian cells treated with staurosporin, but because of the low specificity of this drug, the role of CDK1 in that experiment was ambiguous (17). Our results with a much more selective inhibitor suggests that mitotic exit is a response solely to the loss of CDK1 activity. This rapid mitotic exit upon CDK1 inhibition is in agreement with the hypothesis that mitosis is an unstable cellular state requiring active maintenance through continuous phosphorylation of multiple protein substrates. The fact that RO-3306 can promote a rapid and complete transition from mitosis to interphase suggests that the catalytic activity of CDK1 is necessary and sufficient for maintaining the mitotic state of human cells and, therefore, serves as a master switch for cell division.
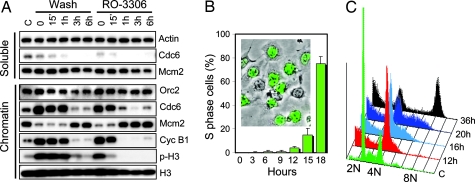
Earlier studies have implicated CDK1 in a mechanism that limits the firing of replication origins to once within a single cell cycle (18, 19). In cycling cells, origin licensing occurs after completion of mitosis (20, 21). We asked whether replication origins could be licensed before completion of cell division by inhibiting CDK1 activity. To this end, purified populations of nocodazole-arrested prometaphase HeLa cells were treated with RO-3306, and the association of three protein members of the replication preinitiation complex, Orc2, Cdc6, and Mcm2, with chromatin was followed for a period of several hours and compared with control cells allowed to undergo normal mitosis. Mcm2, one of six MCM proteins (Mcm2–Mcm7) that form a heterohexamer essential for origin-specific initiation of DNA replication, is released from chromatin during replication and reassociates with DNA at the end of mitosis (22). Inhibition of CDK1 activity by RO-3306 forced cells to exit mitosis without cell division as indicated by the rapid disappearance of phospho-histone H3 (Fig. 3D) and chromatin decondensation (Fig. 3 B and C). Loss of CDK1 activity also was accompanied by a rapid transition of Mcm2 from the soluble fraction to chromatin (Fig. 4A). In the control cells, Mcm2 was loaded onto chromatin only after completion of cell division (Fig. 4A). Cdc6 was found associated with chromatin in the mitotic cells but disappeared concomitantly with Mcm2 loading. It was not detectable in the soluble fraction, suggesting that Cdc6 is degraded upon exit from mitosis as has been recently reported (23). The association of Orc2 with the chromatin fraction did not change significantly during the exit from mitosis. Despite their G2 chromosome content, RO-3306-treated cells were fully competent to initiate a new replication cycle and behaved similarly to the control G1 cells exiting an unperturbed mitosis. Although there was an ≈4 h delay compared with control cells, nocodazole-arrested cells entered the S phase fairly synchronously (Fig. 4B). At 18 h after RO-3306 addition, ≈80% of the cells were in the S phase, indicating that RO-3306 treatment permits successful origin licensing in the absence of cell division. The ability of the cells to initiate replication in the presence of RO-3306 also confirms that the compound does not interfere significantly with the function of kinases associated with initiation of DNA replication, CDK2 and Cdc7 (24). Although many cells suffered apoptosis, likely induced by the nocodazole, a large fraction of the tetraploid cell population completed a full round of replication and was arrested by RO-3306 in the G2 phase with 8 N DNA content (Fig. 4C). These results suggest that CDK1 activity is both necessary and sufficient for preventing replication origin licensing before completion of mitosis in human cells.
Fig. 4.
CDK1 activity is essential for prevention of replication origin licensing and cytokinesis before completion of mitosis. (A) CDK1 inhibition in nocodazole-arrested mitotic cells resets them for a new replication cycle without cell division. Prometaphase HeLa cells purified from nocodazole-arrested population by a shake-off were divided into two fractions and either washed and incubated in drug-free medium to allow transition through the G1 phase or incubated in the presence of nocodazole (100 nM) and RO-3306 (9 μM). Cells were harvested at different times, lysed, fractionated into chromatin and soluble fractions, and analyzed for Cdc6, Orc2, Mcm2, cyclin B1, and phospho-histone H3 by Western blotting. (B) HeLa cells were treated as above in the presence of nocodazole and RO-3306, and the S phase cells were labeled by BrdU-immunostaining. Inset shows a representative image (green BrdU-labeled cells) taken at 18 h under fluorescence (FITC filter) and phase contrast. (C) Cell cycle analysis of HeLa cells treated as in B at different times after the addition of 9 μM RO-3306 in the presence of 100 nM nocodazole.
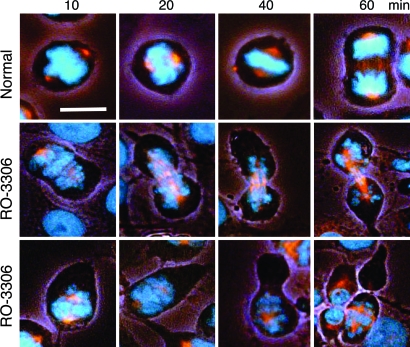
Next, we studied the effect of CDK1 inhibition on cells undergoing normal mitosis. Proliferating HeLa cells, synchronized in mitosis by release from RO-3306 block, were retreated with RO-3306 and followed for morphological markers of mitotic progression (Fig. 5). Control cells proceeded normally through mitosis with a majority reaching anaphase at 60 min and undergoing cytokinesis within the next half hour (Fig. 5 and data not shown). Cells in which CDK1 activity was inhibited started to exhibit extended morphology after 10 min, and the majority initiated premature cytokinesis within 20–40 min. At 1 h, most of the cells underwent cytokinesis-like changes. However, the unscheduled RO-3306-induced cytokinesis appeared chaotic because it began before chromosome alignment and chromatid segregation and was accompanied by chromatin decondensation. In some cases, a cleavage furrow-like structure appeared near the equatorial plain of the spindle (Fig. 5). In others, it formed outside of the spindle zone and led to unequal cleavage and formation of daughter-like cells without chromosomes (Fig. 5). These data suggest that one of the critical functions of CDK1 is to inhibit cytokinesis until chromosome segregation is complete in telophase.
Fig. 5.
Inhibition of CDK1 during mitosis causes premature cytokinesis. HeLa cells were enriched in mitotic cells by release from RO-3306 block (9 μM for 18 h) and followed for morphological changes in the absence (Top) or presence of 9 μM RO-3306 (Middle and Bottom). Cells were fixed and immunostained for β-tubulin (red) and DNA by Hoechst 33258 (blue). Images were captured by using a Nikon Eclipse TE2000-U inverted microscope with a CF1 DAPI/FITC/tetramethylrhodamine B isothiocyanate fluorescence filter cube and phase contrast under low light. (Scale bar: 20 μm.)
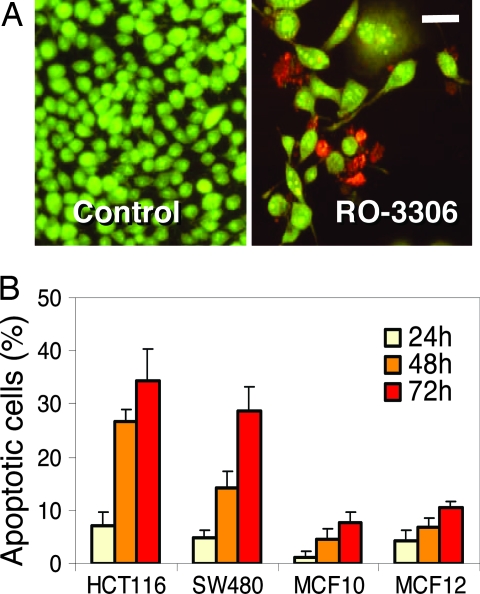
Our experiments indicated that CDK1 inhibition by RO-3306 for up to 20 h is fully reversible and does not noticeably alter cell viability. We next tested the tolerability of CDK1 inhibition for extended periods of time. Two colon cancer cell lines, HCT116 and SW480, were treated with RO-3306 at concentrations that effectively arrest cell cycle progression for up to 96 h, and their cell cycle distribution and viability were analyzed. At 48 h, cells accumulated a substantial sub-G1 fraction, and by 72 h, the majority had lost viability (Fig. 6A and data not shown). We then compared the response of cancer cells and nontumorigenic cells to the cytotoxicity of RO-3306. Exponentially growing HCT116 and SW480 cells were incubated with 9 μM RO-3306 alongside of MCF10A and MCF12A, immortalized nontumorigenic breast epithelial cell lines. Annexin V staining, an early marker of apoptosis, revealed a substantially larger apoptotic fraction in the cancer cell lines (30–40%) compared with nontransformed cells (≈10%) after 72 h of drug exposure (Fig. 6B). Although proliferation of both cancer and normal cells is effectively blocked by RO-3306 (data not shown), extended CDK1 inhibition appears to be more proapoptotic in cancer cells. Therefore, selective CDK1 inhibitors may have utility as anticancer agents. Further optimization of the RO-3306-class of compounds may help to address the therapeutic potential of CDK1 inhibition.
Fig. 6.
RO-3306 induces apoptosis in cancer cells. (A) Exponentially growing SW480 cells were treated with 4 μM RO-3306 for 48 h and stained with propidium iodide (red, dead cells) and acridine orange (green, live cells). (Scale bar: 50 μm.) (B) Proliferating HCT116, SW480, MCF10A, and MCF12A cells were treated with 9 μM RO-3306, and the percentage of Annexin V-positive (apoptotic) cells minus background was determined by flow cytometry.
Methods
Kinase Assays.
The CDK assays were run by using recombinant human CDK/cyclin complexes (CDK1/cyclin B1, CDK1/cyclin A, CDK2/cyclin E, and CDK4/cyclin D) expressed and isolated from Hi5 insect cells. The expression constructs, GST-cyclin B1, CDK1, GST-cyclin-E, CDK2, GST-CDK4, and cyclin D, were provided by W. Harper (Baylor College of Medicine, Houston). The GST-tagged proteins were coexpressed and purified in complex with their partners as described in ref. 25. All assays used a His-6-tagged fragment of pRB (amino acids 385–928) as a substrate. The protein was expressed from a construct provided by Veronica Sullivan (Roche Research Center, Welwyn, U.K.). It was expressed in M15 Escherichia coli cells and bound on a Ni-chalated agarose column pretreated with 1 mM imidazole and eluted with 500 mM imidazole. The eluted protein was dialyzed against 20 mM Hepes, pH 7/6.25 mM MgCl2/1.5 mM DTT, aliquoted, and stored at −80°C.
The activity of CDK1/cyclin B1, CDK1/cyclin A, CDK2/cyclin E, and CDK4/cyclin D was measured by a homogeneous time-resolved fluorescence assay in a 96-well format. The assay buffer contained 25 mM Hepes, 6.25 mM MgCl2, 0.003% Tween 20, 0.3 mg/ml BSA, 1.5 mM DTT, and ATP as follows: 162 μM (CDK1), 90 μM (CDK2), or 135 μM (CDK4). CDK1 and CDK2 buffer contained 10 mM MgCl2. Test compounds were diluted in assay buffer to 3-fold their final concentration in 20 μl, and the reaction was started by the addition of a 40-μl assay buffer containing the pRB substrate (0.185 μM). The plates were incubated at 37°C for 30 min with constant agitation, and the reaction was terminated by the addition of 15 μl of 1.6 μM anti-phospho pRB antibody (Ser-780; Cell Signaling Technology, Beverly, MA) in 25 mM Hepes/24 mM EDTA/0.2 mg/ml BSA. After an additional 30 min of incubation with shaking, 15 μl of 3 nM Lance-Eu-W1024-labeled anti-rabbit IgG and 60 nM Alophycocyanin-conjugated anti-His-6 antibody (PerkinElmer Life Sciences) in 25 mM Hepes/0.5 mg/ml BSA was added and incubated for 1 h. The plates were read in the Victor-V multilabel reader (PerkinElmer Life Sciences) at excitation 340 nm and emission 615 nm and 665 nm. The IC50 values were calculated from the readings at 665 nm and normalized for Europium readings at 615 nm. Ki values were calculated according to the equation: Ki = IC50/(1 + S/Km), where S is the ATP concentration in the assay and Km is the Michaelis-Menten constant for ATP. The inhibitory activity against the panel of kinases was determined by the IMAP assay technology (Molecular Devices) as described in ref. 26.
Molecular Docking.
Docking of RO-3306 to the ATP-binding site of CDK2 was carried out manually by using modeling package moe from Chemical Computing Group, Inc. (Montreal). Energy minimization was performed to RO-3306 first in vacuum and then in the environment of the ligand-binding site of CDK2 with fixed CDK2 atoms. After unfixing the CDK2 atoms, a tethering force was applied to all heavy atoms in the system. The tethering force was removed gradually, and the CDK2/RO-3306 complex was subjected to energy minimization until the rms gradient is <0.05 Å. The force field used for energy minimization was MMFF94x.
Cells and Drug Treatment.
All cell lines used in this study were purchased from American Type Culture Collection (ATCC, Manassas, VA) and grown in the recommended media supplemented with 10% heat-inactivated FBS (Invitrogen) in a humidified environment with 5% CO2. Drugs were dissolved in DMSO and kept at −20°C as 10 mM stock solutions. For cell cycle analysis, 106 cells were grown in 75 cm2 tissue-culture flasks. They were harvested and analyzed as described in ref. 27.
BrdU Labeling and Detection.
Mitotic cells isolated by a gentle shake-off of nocodazole-treated cell population (100 nM for 15 h) were seeded in 35 mm2 dishes (2 × 105 cells per dish) and incubated in the presence or absence of nocodazole and or RO-3306. BrdU (20 μM; Sigma) was added for 30 min, and the cells were fixed in 70% ice-cold ethanol for 5 min and left to air dry. Cells were washed once with PBS, and DNA was denatured by 10- to 15-sec treatment with 0.07 M NaOH and neutralized with PBS, pH 7.5. Anti-BrdU FITC-conjugated antibody (Becton Dickinson; catalog no. 347583) was used as recommended by the manufacturer. Labeled cells were mounted with coverslips by using Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and examined under fluorescence microscope. S phase cells were determined by counting at least 500 cells and were expressed as a percentage of the total population.
Immunostaining.
Cell were seeded in 35 mm2 dishes (2 × 105 cells per dish) 24 h before treatment with RO-3306 (9 μM) for 18 h. The cell population was released from the G2 block by washing twice in prewarmed drug-free media and incubated in fresh media for 30 min to enrich in mitotic cells. Cells were fixed in 4% methanol-free paraformaldehyde for 20 min, washed, and kept in PBS. They were permeabilized with 0.01% Triton X-100 in PBS for 1 min, washed with PBS, blocked with 5% Blotto, and incubated with Cy3-conjugated anti-β-tubulin antibody (Sigma; catalog no. c-4585) overnight at room temperature in a humidified chamber (1:100 dilution). DNA was stained with Hoechst 33258 (Sigma) for 5 min, and the cells were observed under combined phase contrast and fluorescence to reveal both cell shape and the fluorescently labeled structures.
Cellular Fractionation.
HeLa cells were lysed and fractionated into cytoplasm, soluble nuclear material, and chromatin after the procedure of Mendez and Stillman (28). Cells were resuspended (4 × 106 cells/ml) in buffer A (10 mM Hepes, pH 7.9/10 mM KCl/1.5 mM MgCl2/0.34 M sucrose/10% glycerol/1 mM DTT/Roche protease inhibitors/0.1% Triton X-100) and incubated on ice for 5 min. Nuclei were collected (P1) by low-speed centrifugation (1,300 × g for 5 min at 4°C). P1 fraction was washed once again in buffer A and then lysed in buffer B (3 mM EDTA/0.2 mM EGTA/1 mM DTT/Roche protease inhibitors) and incubated on ice for 30 min. Nuclei were separated into soluble fraction (S3) and insoluble chromatin by centrifugation (1,700 × g for 5 min at 4°C). The final chromatin fraction (P3) was washed again in buffer B and centrifuged under the same conditions. P3 fraction was resuspended further in Laemmli buffer and sonicated for 10 sec.
Western Blotting.
Control asynchronous or purified mitotic cells were grown in 75 cm2 flasks (106 cells per flask), lysed in RIPA buffer, and Western blotting was performed as described in ref. 29. Antibodies used are as follows: phospho-histone H3 (Upstate Biotechnology, Lake Placid, NY; catalog no. 06-570), histone H3 (Cell Signaling Technology; catalog no. 9715), cyclin B1 (Santa Cruz Biotechnology; catalog no. SC-245), β-actin (Sigma; catalog no. 5361), Orc2 (Santa Cruz Biotechnology; catalog no. SC-13238), Cdc6 (Santa Cruz Biotechnology; catalog no. SC-9964), pRB (Cell Signaling Technology; catalog no. 9309), phospho-pRB795 (Cell Signaling Technology; catalog no. 9301s), and phospho-pRB811 (Cell Signaling Technology; catalog no. 9308s), and the rabbit anti-human MCM2 antibody was a gift from I. Todorov (Beckman Institute, Duarte, CA) and used at a 20,000-fold dilution.
Apoptosis Assays.
Cells were seeded in six-well tissue culture plates (2 × 105 cells per well) 24 h before drug treatment. For staining of live/dead cells, acridine orange and propidium iodide (Sigma) were dissolved in PBS at 0.3 mg/ml and added to culture media at 1 μg/ml. After 5 min of incubation at 37°C, cells were examined under fluorescence microscope. Acridine orange stains live cells green, and propidium iodide stains dead cells red. Annexin V-positive cells were determined by using the GuavaNexin kit (Guava Technologies, Hayward, CA) and the Guava personal cell analyzer as described in ref. 30. Percent apoptosis is calculated as the percentage of both live and dead Annexin V-positive cells in the population at the time of measurement.
Supplementary Material
Acknowledgments
We thank D. Carvajal, N. Le, and T. Simcox for experimental help; I. Todorov for providing the Mcm2 antibody; and B Graves, N. Fotouhi, and L. Babiss for helpful suggestions during the course of this work.
Abbreviation
- CDK
cyclin-dependent kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Norbury C., Nurse P. Annu. Rev. Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 2.Morgan D. O. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 3.Murray A. W. Chem. Biol. 1994;1:191–195. doi: 10.1016/1074-5521(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 4.Tetsu O., McCormick F. Cancer Cell. 2003;3:233–245. doi: 10.1016/s1535-6108(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 5.Ortega S., Prieto I., Odajima J., Martin A., Dubus P., Sotillo R., Barbero J. L., Malumbres M., Barbacid M. Nat. Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 6.Sherr C. J., Roberts J. M. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 7.Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 8.Murray A. W. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 9.Knockaert M., Greengard P., L. Meijer L. Trends Pharmacol. Sci. 2002;23:417–425. doi: 10.1016/s0165-6147(02)02071-0. [DOI] [PubMed] [Google Scholar]
- 10.Hirai H., Kawanishi N., Iwasawa Y. Curr. Top. Med. Chem. 2005;5:167–179. doi: 10.2174/1568026053507688. [DOI] [PubMed] [Google Scholar]
- 11.Canduri F., Uchoa H. B., de Azevedo W. F., Jr. Biochim. Biophys. Res. Comm. 2004;324:661–666. doi: 10.1016/j.bbrc.2004.09.109. [DOI] [PubMed] [Google Scholar]
- 12.Nurse P. Eur. J. Cancer. 1997;33:1002–1004. doi: 10.1016/s0959-8049(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 13.Nigg E. A. Nat. Rev. Mol. Cell. Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa M., Higashi H., Jung H. K., Suzuki-Takahashi I., Ikeda M., Tamai K., Kato J., Segawa K., Yoshida E., Nishimura S., Taya Y. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak S. J., Corces V. G. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 16.King R. W., Deshaies R. J., Peters J. M., Kirschner M. W. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 17.Hall L. L., Th’ng J. P., Guo X. W., Teplitz R. L., Bradbury E. M. Cancer Res. 1996;56:3551–3559. [PubMed] [Google Scholar]
- 18.Diffley J. F. Curr. Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Machida Y. J., Hamlin J. L., Dutta A. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 20.DePamphilis M. L. Gene. 2003;310:1–15. doi: 10.1016/s0378-1119(03)00546-8. [DOI] [PubMed] [Google Scholar]
- 21.Stillman B. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Todorov I. T., Attaran A., Kearsey S. E. J. Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen B. O., Wagener C., Marinoni F., Kramer E. R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J. M., Helin K. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 25.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 26.Gaudet E. A., Huang K. S., Zhang Y., Huang W., Mark D., Sportsman J. R. J. Biomol. Screen. 2003;8:164–175. doi: 10.1177/1087057103252309. [DOI] [PubMed] [Google Scholar]
- 27.Carvajal D., Tovar C., Yang H., Vu B. T., Heimbrook D. C., Vassilev L. T. Cancer Res. 2005;65:1918–1924. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- 28.Mendez J., Stillman B. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Fillipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 30.Thompson T., Tovar C., Yang H., Carvajal D., Vu B. T., Xu Q., Wahl G. M., Heimbrook D. C., Vassilev L. T. J. Biol. Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.