Abstract
The fungus-growing ants have long provided a spectacular example of coevolutionary integration. Their ecological success is thought to depend largely on the evolutionary alignment of reproductive interests between ants and fungi after vertical transmission and the ancient suppression of fungal sexuality. In the present study we test these assumptions and provide the first evidence of recombination in attine cultivars, contradicting widely held perceptions of obligate clonality. In addition, we document long-distance horizontal transmission of symbionts between leaf-cutter ant species on mainland Central America and South America and those endemic to Cuba, suggesting both lack of pairwise coevolutionary specificity in ant/cultivar interactions and dispersal of symbionts independent of their ant hosts. The coevolution between leaf-cutters and their fungal symbionts is thus not reciprocally pairwise. Rather, a single widespread and sexual fungal symbiont species is engaged in multiple interactions with divergent ant lineages. Strict fungal clonality and vertical transmission evidently have not played a critical role in the long-term evolutionary or ecological success of this well known mutualism.
Keywords: asexuality, Atta, Caribbean
Attine ants evolved agriculture based on the cultivation of fungi some 50–60 million years ago. By allowing the exploitation of food sources unavailable to most other ants, agriculture allowed the fungus gardeners to become a taxonomically diverse group, with various subgroups differing widely in the extent of specialization and nutritional dependency (1–4). The most derived group, the “higher attines,” includes the leaf-cutters, nearly 50 species in the genera Atta and Acromyrmex (5), and they are perhaps the most ecologically dominant organisms in the New World tropics outside of humans (4, 6). Typically, after mating and dispersal from their natal nest, young queens cultivate mycelia brought from parental gardens, a behavior believed to stabilize the mutualism by restricting horizontal cultivar exchange (7). However, horizontal host switches are known to occur in some congeneric taxa (8, 9). For example, in the primitive fungus-gardening ants (the “lower attines”), horizontal transfers occur directly between the cultivar lineages or via free-living fungal populations acting as bridges (8). The higher attine cultivars, by contrast, possess no known free-living relatives, and the presumption has long been that these cultivars are propagated by strictly clonal and vertical inheritance, without an independent means of dispersal (7, 9). Although infrequent observations of fruiting body formation in gardens of higher attines suggest the possibility of a cryptic sexual stage or its vestigial expression, there has been no direct test of these long-standing hypotheses (see ref. 1 for a comprehensive review of the rarity of higher attine cultivar fruiting). Consequently, nearly all of the growing body of work on this complex mutualism has been predicated on the assumption of clonality.
Assuming clonal reproduction as our null hypothesis, we tested for asexuality and admixture in the leaf-cutter cultivars by seeking genetic signatures of clonality. First we characterized protein evolution along fragments of the eukaryote recA homologs DMC1 and RAD51. Both mediate homologous DNA pairing and strand exchange and are necessary for completion of the meiotic cell cycle (10–13). RAD51, the fundamental eukaryotic recombinase, is required for both mitotic and meiotic DNA metabolism, whereas DMC1 is solely expressed during meiosis (14, 15). One prediction of long-term asexuality is that disuse of the meiotic machinery, typically under strong purifying selection in eukaryotes, would be associated with relaxed functional constraints in genes involved in meiosis (16–18). Long-term loss of recombination would thus result in either elevated rates of recombinational protein evolution or perhaps loss-of-function degradation and elimination from genomes not expressing the meiotic cell cycle (17, 18). We also sought evidence of interlocus recombination among RAD51, DMC1, and elongation factor 1-α (EF1-α). Clonality predicts negligible levels of phylogenetic incongruence across genomes, because fully linked genetic markers used to infer taxonomic relationships share an identical evolutionary history (19). Recombination acts to break down linkage and can cause conflicting genealogical relationships between loci.
Next we tested the assumption of pairwise congruence between ant and fungal lineages by examining the population genetics of cultivars on two island populations in which the biogeographic histories of the ant hosts are established. The leaf-cutter Acromyrmex octospinosus colonized the French Caribbean islands of Guadeloupe ≈50 years ago (20). Assuming clonality and pairwise coevolutionary specificity between ants and fungi, the A. octospinosus cultivars on Guadeloupe should exhibit evidence of a population bottleneck similar to that of the ants, with mutation acting as the sole source of post-invasion genetic variation. Next we derived the phylogenetic relationship for cultivars from leaf-cutter taxa on Cuba and on the mainland, from Central America and South America (Panama and Brazil). Cuban leaf-cutter ant fauna consists of two endemic species of Atta and an endemic Acromyrmex species (21) (A.S.M., unpublished observations), which is unusual for Caribbean islands, where leaf-cutters are largely absent (22). On both Guadeloupe and Cuba, contrasts between the ants and cultivars in rates of endemism and genetic affinity with mainland populations provide insight into differential dispersal abilities and long-term genealogical incongruence between the two symbiont taxa.
Collectively, these various analyses indicate that the symbiosis between leaf-cutting ants and their cultivars is not one of pairwise specificity between host and symbiont lineages. Different ant genera essentially cultivate the same, sexual fungal symbiont. We discuss the implications of these results for future work on fungus-growing ants and for our understanding of the coevolutionary process.
Results
Conservation of Genes Involved in the Meiotic Cell Cycle.
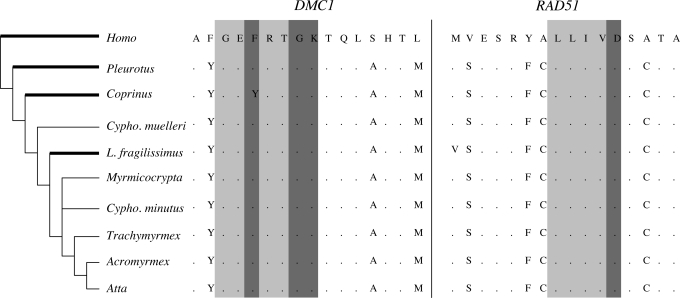
We found that both DMC1 and RAD51 were strongly conserved at the amino acid level and essentially identical to free-living lepiotaceous fungi. Ratios of nonsynonymous-to-synonymous nucleotide changes indicated no evidence of relaxed functional constraints (dN/dS = 0.033 ± 0.012 for DMC1; dN/dS = 0.015 ± 0.006 for RAD51) and no evidence for accelerated rate of evolution in the higher attine clade (P = 0.87 for DMC1; P = 0.75 for RAD51). In particular, no changes were localized to functional regions unambiguously crucial to enzyme activity (e.g., Walker motifs; Fig. 1). In contrast to the conserved coding gene regions, introns in the sequenced fragments of DMC1 (six introns) and RAD51 (two introns) diverged to the point of unalignability, indicating strong purifying selection acting on coding regions of both genes. Whereas conservation of RAD51 could be due to mitotic double-stranded break repair functionality, DMC1 is expressed only during homologous chromosome pairing at meiosis. The strong conservation of DMC1 is not consistent with the long-term arrest of the meiotic chromosome metabolism in the leaf-cutter cultivars.
Fig. 1.
Conservation of ATP-binding Walker motifs in DMC1 and RAD51 at the amino acid level. Essential for gene function, the Walker motifs are shown with lighter gray shading, and their ATP-binding sites are in a darker gray. As elsewhere, cultivar taxa are referred to by the genus of their ant host. Free-living taxa, which include humans and two basidiomycete fungal taxa, are indicated by bold highlighting of branches on the phylogeny. For the entire sequenced fragments, ratios of nonsynonymous-to-synonymous nucleotide changes indicated no evidence of relaxed functional constraints.
Interlocus Recombination Between Nuclear Genes.
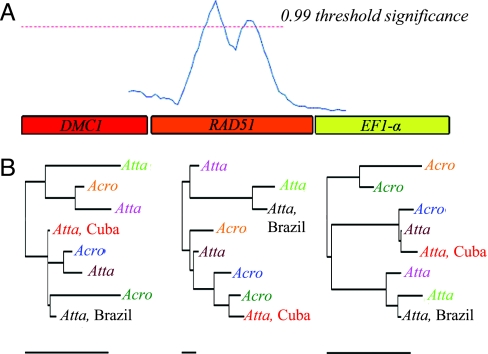
The phylogenetic signals in the DMC1, RAD51, and EF1-α partitions showed a significant lack of homogeneity (P = 0.026) according to a parsimony-based analysis (23). An independent sliding-window analysis (24) of a three-gene alignment also indicated the presence of recombination and automatically partitioned the data into incongruent sections corresponding to boundaries in the three genes (Fig. 2). Although we cannot exclude the possibility of other processes generating conflicting gene-tree topologies [e.g., incomplete lineage sorting or hybridization (25)], these alternatives to recombination are equally at odds with the hypothesis of long-term vertical transmission and clonal reproduction.
Fig. 2.
Evidence for recombination in the alignment of DMC1, RAD51, and EF1-α. (A) A plot of probabilistic divergence measures for the three-gene alignment based on a 500-bp sliding-window analysis. Crossing of the threshold line indicates a significant change in the marginal posterior distribution of topologies, which suggests recombination. (B) Bayesian trees for partitions automatically generated from local divergence scores. Unless otherwise specified, all samples were collected in Panama, and identical samples are color-coded across gene trees and are labeled according to host ant genus. Bars below each tree are scaled to 0.01 substitutions per site.
Genealogical Congruence Between Ants and Their Cultivars on Island Populations.
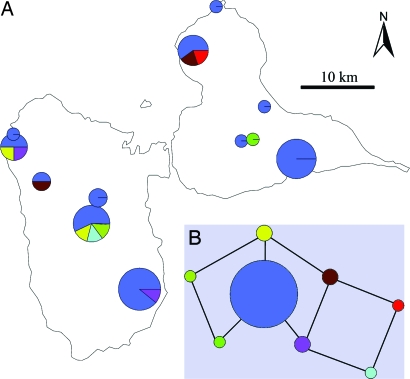
Historical records indicate that the A. octospinosus population on Guadeloupe was founded by a single introduction of a lone female or perhaps a group of sisters (20). We confirmed this by sequencing a highly variable 223-bp fragment of noncoding mtDNA (26), which revealed just one A. octospinosus haplotype from 59 colonies. The fungal cultivar population on Guadeloupe revealed a different pattern. A large fraction of the nests (33/43 genotyped gardens) harbored an identical genotype heterozygous at four microsatellite loci, as expected with a small founding population. However, we detected seven other low-frequency genotypes homozygous at one or more loci, arranged into a loop-like haplotype network, which is a pattern indicative of recombination (Fig. 3). An alternative to recombination is simple mutation, but it is unlikely to have produced the complex network we observed. Moreover, we did not detect a single new allele in the low-frequency cultivars, which would be expected if mutation was sufficient to significantly modify genotype distributions. Genotyping errors are a potential source of artifactual variation (“null alleles” and “allele dropout”) (27). However, we detected genotypes with multiple homozygous loci, independently replicated these results over more than one round of verification, and found no pattern of biased loss of longer alleles, which is typical of allele dropout (28). Thus, it is likely that the loop-like haplotype network is a result of recombination after the colonization of Guadeloupe by A. octospinosus.
Fig. 3.
Frequencies and observed relationships between cultivar genotypes. (A) Distribution of collections and relationships among cultivar genotypes on the islands of Guadeloupe. (B) A parsimony network connecting the observed genotypes. Pie charts and network vertices are scaled according to relative genotype frequencies.
The Cuban population also exhibited a contrasting pattern between ants and cultivars. Whereas the ants have experienced an extensive period of evolutionary isolation (22) (A.S.M., unpublished observations), the cultivars are nearly identical to their mainland counterparts in four intron-rich nuclear genes (DMC1, RAD51, EF1-α, and BiPA) and in the rDNA internal transcribed spacer (ITS) (Fig. 1). Evidently, Cuban and mainland cultivars have not been isolated to the extent of their ant hosts. Although it is conceivable that a now-extinct indigenous cultivar on Cuba was recently replaced (without recombination) by a mainland fungal strain, it is more likely that recurrent long-range dispersal and gene flow between Cuban and mainland populations of leaf-cutter cultivars have prevented the divergence of island and mainland populations. The Cuban cultivars are thus not consistent with the assumption of long-term coevolutionary fidelity between different leaf-cutter ant and fungal lineages, because strictly clonal, vertical propagation would have resulted in the endemic isolation of the fungal cultivars on Cuba.
Discussion
Collectively, these data reveal that a single biological species of fungus, previously named Leucoagaricus gongylophorus (1, 29, 30), coevolves with at least two different genera of leaf-cutting ants. Acting over relatively short periods of time, recombination within L. gongylophorus homogenizes genetic variation over great geographic distances and taxonomic divergence between ant hosts. This conclusion is consistent with previous findings that Brazilian Acromyrmex and Atta cultivars show little or no differentiation in ribosomal ITS regions over thousands of kilometers (31). Thus, the evolutionary dynamics between ants and the cultivar fungus may be qualitatively different from what is currently believed. Rather than a one-to-one correspondence between ant and cultivar lineages, a many-to-one relationship exists, in which a single general-purpose fungal mutualist acts as a nexus for indirect interactions between divergent ant lineages.
These data do not directly reveal the mechanisms of dispersal and recombination in the cultivars. In addition to sexual fruiting and meiosis, most fungal groups express a variety of irregular reproductive modes. Various “parasexual” processes are common in filamentous fungi, involving fusion and recombination between somatic cells (32–34). However, to date, outside of laboratory settings, parasexuality has not been described from natural populations of basidiomycete fungi (34–41). Vegetative incompatibility probably limits opportunities for hyphal fusion and thus reduces the natural frequency of parasexual mechanisms that depend on anastomosis (39). Generally, the rareness of parasexuality in basidiomycetes, the conservation of the meiotically expressed DMC1, and the occasional observations of sexual fruiting bodies (basidiocarps) in the fungus gardens (1) imply that meiotic sporulation is a principle cause of recombinant cultivars. However, it may well be that both meiotic and mitotic processes contribute to admixture in attine cultivars. We envision a scenario similar to the following: (i) meiosis occurs during spore production in the occasional basidiocarp in attine cultivars, and (ii) the spores then travel to ant-tended gardens and undergo heterokaryosis via mitotic recombination (42). Consistent with the existence of nuclear admixture in the attine symbiosis, like other mitotic recombinants leaf-cutter cultivars exhibit polyploidy (43) (J. Scott, M. Kweskin, and U.G.M., unpublished observations; and A.S.M., U.G.M., and J. Boomsma, unpublished observations). Alternatively, recombination may conceivably occur via a free-living population of higher attine cultivars, as with the lower attines, but such a population has not yet been detected despite intensive surveys (9) (T. Vo, A.S.M., and U.G.M., unpublished observations). Whether sexual or parasexual mechanisms are at work, the population-genetic and evolutionary consequences are indistinguishable: recombination shuffles genetic variation in attine cultivars and results in nonclonal, reticulate propagation.
Initially, long-term clonal propagation was proposed as a central mechanism promoting ant–fungus coevolution (3). Atta and Acromyrmex, for example, exhibit behaviors such as vertical transmission and garden monocultures thought to function in the alignment of reproductive interests (1). This interpretation was revised, at least in the lower fungus-gardening ants, whose cultivars were discovered to exist as free-living forms (9). Subsequently, the presumed long-term asexuality of the higher attine cultivars, predicted to reduce fungal fitness by accumulation of deleterious mutations, has been invoked to explain a number of phenomena occurring at the transition between lower and higher attines, such as greater virulence of the specialized garden parasite Escovopsis (44, 45) and the evolution of multiple mating by queens of the leaf-cutting ants (46, 47). Our data require a reevaluation of hypotheses such as these predicated on fungal clonality. Recombinant attine cultivars also require reassessment of the contrasts typically drawn between attines and other insect farmers (48, 49). The termites and beetles farm sexual fungal crops, which presumably express effective pathogen resistance. The ants rely on antibiotics from mutualistic actinomycete bacteria that rapidly coevolve with garden pathogens, a mechanism hypothesized to compensate for fungal asexuality in the evolutionary arms race with parasites (50). The discovery of ongoing sexual reproduction suggests that ant cultivars require no novel explanations for the maintenance of immune function (although such explanations are not prohibited). However, because clonal propagation was a major point of difference between attines and other insect farmers (48, 50), closing this gap suggests an opportunity for the development of a more unitary understanding of insect agriculture.
The discovery of horizontal exchange and sexual reproduction in attine cultivars mirrors an ongoing and fundamental change in our understanding of coevolution. Unlike parasitic interactions, one-to-one specificity in mutualistic interactions is evidently not common, as once thought, even in exemplary systems where specificity was once the rule (51). Such specificity rarely occurs in any but the most integrated and intimate interactions (52, 53). Rather, molecular studies of virtually all other well characterized mutualisms, from lichens (54) to plant–pollinator mutualisms (55–57), reveal evolutionary diffuseness, asymmetric specialization, and variability to a degree only now beginning to be understood (51–60) Leaf-cutters were counterexamples and exemplified specificity and pairwise reciprocity between mutualist lineages. However, it seems that even in the leaf-cutters, with their distinctive form of multiguild mutualism, the general principle at work is one of diffuse coevolution between participants.
Methods
Amplification of ITS, DMC1, RAD51, EF1-α, and BiPA.
Except for ITS4 and ITS5 primers (61), commonly used for fungal ribosomal ITS amplification, primer sequences were developed from conserved regions of gene homologs in the GenBank database (Table 1). DNA was extracted according to Kweskin (43) or by incubating a single cluster of fungal gongylydia (structures used to feed ant larvae) in 100-μl 5% aqueous Chelex resin for 1.5 h at 60°C and then at 99°C for 10 min.
Table 1.
Primer sequences used for amplifying fungus-gardener cultivar genes
| Primer name | Sequence (5′→3′) |
|---|---|
| BIP3-F | GAT GTY AGC AAG AAC CTY CG |
| BIP3-R | TTG TCC TTG GTG AGS GRA CG |
| EF1-α-F | GTT GCT GTC AAC AAG ATG GAC ACT AC |
| EF1-α-R | GCC TTG ATG ATA CCA GTC TCG ACA CG |
| RAD51-F | GGC AAA TGT TTG TAT ATA GAT ACT G |
| RAD51-R | CAC CGA TAG GTT TCT TCT CAT TAC C |
| DMC1-FB | GGT ATG TCG GAA GCC AAA G |
| DMC1-RF | TCG GCC CAA CCT CCT TCG TC |
| DMC1-F | AAG CTG CAC ACA AAA TCT TGG TTA G |
| DMC1-R | GTC AAT GTC AAG AGA TCG GAT ACA C |
One microliter of the extract was used as template in 10-μl reaction volumes. The PCR contained 1× reaction buffer, 1 mM dNTPs, 0.5 μM primers, and 2.5 mM MgCl2 with 0.1 units of Bioline Taq polymerase. Average reaction conditions involved an initial denaturing step of 94°C for 2 min followed by 30 cycles of 94°C for 10 s, 60°C for 20 s, and 72°C for 30 s. PCR products were ≈500 bp long, except for the DMC1-FB/RF primer combination, which produced fragments slightly more than 1 kb in size. The annealing temperature was varied slightly to correspond with primer Tm. Two microliters of the products was run on 1.5% agarose gels and visualized by staining with ethidium bromide. Reactions that yielded strong bands were cleaned by polyethylene glycol precipitation (a 1:1 PCR product/20% PEG mixture was incubated for 15 min at 37°C followed by a 10-min centrifugation at 2,688 × g and two washes with 80% ethanol). Purified products were cycle-sequenced by using the ABI BigDye Terminator Kit (version 3.1) and sequenced on a PRISM 3100 genetic analyzer (Applied Biosystems) according to the manufacturer’s instructions. To ensure the accuracy of sequence information, both the forward and reverse sequences were generated.
Population Genetics of the Guadeloupe Invasion.
As above, fungal DNA was extracted by boiling gongylydia in Chelex resin. Ant DNA was extracted by crushing legs of workers in 50 μl of extraction buffer [50 g of guanidinium isothiocyanate, 50 ml of PCR-grade water, 5.3 ml of 1 M Tris·HCl (pH 7.6), 5.3 ml of 0.2 M EDTA, 10.6 ml of 20% sodium lauryl sarcosinate, and 1 ml of 2-mercaptoethanol] and incubating for 1 h at 70°C with occasional vortexing. The extractions were briefly centrifuged, and the supernatant was collected. The extraction was chilled for at least 2 h at −20°C after addition of 50 μl of isopropanol. Afterward the extraction was centrifuged at 2,688 × g for 20 min, and the supernatant was discarded. The pellet was washed with 150 μl of cold ethanol, dried, and redissolved in 50 μl of PCR-grade water.
The intergenic spacer between cytochrome oxidase subunits I and II was amplified by using primers (COI and COII) and conditions previously published by Sumner et al. (26). PCR products were purified and sequenced by using the conditions described for the fungal genes.
Cultivars were genotyped by using M13-tailed primers B0150B, A0460, C606, and B0358 (J. Scott, M. Kweskin, and U.G.M., unpublished observations). PCR reagent concentrations were the same as those used for the fungal genes, except that forward and reverse primers were added up to 0.01 and 0.15 μM, respectively. Additionally, 0.18 μM fluorescently labeled M13 tail was included in the reaction. PCR conditions involved an initial denaturing step of 94°C for 2 min followed by 20 cycles of 94°C for 10 s, 60°C for 20 s, and 72°C for 10 s. Subsequently, 15 more cycles were performed with the annealing temperature at 53°C. The length of the amplified microsatellite loci was read by using a PRISM 3100 genetic analyzer.
Statistical Analyses: Positive Selection Analysis.
HyPhy (62) was used to test for relaxed selection on DMC1 and RAD51 and to calculate dN/dS values and 95% confidence intervals. First, an appropriate codon model was selected independently for both genes (HKY85 was the best model for both). Subsequently, we tested whether DMC1 and RAD51 evolved under a different selection regime in the putatively asexual clade of higher attine cultivars. The analysis was carried out by using a complete model of site-to-site rate variation with four rate classes, a polarity/charge/hydrophobicity-based amino acid model.
Recombination Between Genes.
Recombination analyses were carried out on a concatenated alignment of DMC1, RAD51, and EF1-α sequences (1,504 bp) for eight Atta and Acromyrmex cultivars. The paup Homopart test (23) used 18 parsimony-informative characters of 37 total variable characters. The presence of recombination was tested independently by using topali (24), with default settings and a 500-bp window sliding at 10 bp per step. The significance threshold was calculated by using 100 bootstrap replicates.
Acknowledgments
We thank J. Scott for developing the microsatellite library and sharing his expertise, Vikram Chhatre for assisting with molecular bench work, and Nancy Moran for helpful suggestions on previous drafts. This work was supported by National Science Foundation Grant DBI-0102094 and funds from Vanderbilt University (to P.A.), National Science Foundation/Integrated Research Challenges in Environmental Biology Grant DEB-0110073 (to U.G.M.), and National Science Foundation Dissertation Improvement Grant DEB-0508613 (to A.S.M. and U.G.M.). Additional funds were provided by fellowships from the U.S. Environmental Protection Agency, the Dolores Zohrab Liebmann Fund, and research grants from the University of Texas Section of Integrative Biology (to A.S.M.).
Abbreviations
- ITS
internal transcribed spacer
- EF1-α
elongation factor 1-α.
Footnotes
References
- 1.Mueller U. G. Am. Nat. 2002;160:S67–S98. doi: 10.1086/342084. [DOI] [PubMed] [Google Scholar]
- 2.Mueller U. G., Schultz T. R., Currie C. R., Adams R. M. M., Malloch D. Q. Rev. Biol. 2001;76:169–197. doi: 10.1086/393867. [DOI] [PubMed] [Google Scholar]
- 3.Chapela I. H., Rehner S. A., Schultz T. R., Mueller U. G. Science. 1994;266:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler W. M. Bull. Am. Mus. Nat. Hist. 1907;23:669–807. [Google Scholar]
- 5.Schultz T. R., Meier R. Syst. Entomol. 1995;20:337–370. [Google Scholar]
- 6.Hölldobler B., Wilson E. O. The Ants. Cambridge, MA: Harvard Univ. Press; 1990. [Google Scholar]
- 7.Bot A. N. M., Rehner S. A., Boomsma J. J. Evolution (Lawrence, Kans.) 2001;55:1980–1991. doi: 10.1111/j.0014-3820.2001.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 8.Green A. M., Adams R. M. M., Mueller U. G. Mol. Ecol. 2002;11:191–195. doi: 10.1046/j.1365-294x.2002.01433.x. [DOI] [PubMed] [Google Scholar]
- 9.Mueller U. G., Rehner S. A., Schultz T. R. Science. 1998;281:2034–2038. doi: 10.1126/science.281.5385.2034. [DOI] [PubMed] [Google Scholar]
- 10.Sung P., Krejci L., Van Komen S., Sehorn S. G. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 11.West S. C. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 12.Bishop D. K., Park D., Xu L., Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A., Ogawa H., Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 14.Mikosch T. S., Sonnenberg A. S. M., Van Griensven L. J. L. D. Fungal Genet. Biol. 2001;33:59–66. doi: 10.1006/fgbi.2001.1265. [DOI] [PubMed] [Google Scholar]
- 15.Nara T., Saka T., Sawado T., Takase H., Ito Y., Hotta Y., Sakaguchi K. Mol. Gen. Genet. 1999;262:781–789. doi: 10.1007/s004380051141. [DOI] [PubMed] [Google Scholar]
- 16.Normark B. B., Judson O., Moran N. A. Biol. J. Linn. Soc. 2003;79:69–84. [Google Scholar]
- 17.Paoletti M., Rydholm C., Schwier E. U., Anderson M. J., Szakacs G., Lutzoni F., Debeaupuis J. P., Latge J. P., Denning D. W., Dyer P. S. Curr. Biol. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 18.Ramesh M. A., Malik S. B., Logsdon J. M. Curr. Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Smith J. M., Smith N. H. Mol. Biol. Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 20.Malato G., Kermarrec A., Troup J. M. Nouv. Agron. Antilles-Guyane. 1977;3:473–484. [Google Scholar]
- 21.Fontenla J. L. Avicennia. 1995;3:77–86. [Google Scholar]
- 22.Cherrett J. M. Proc. Am. Soc. Hort. Sci.; 1968. pp. 295–310. [Google Scholar]
- 23.Swofford D. L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 24.Milne I., Wright F., Rowe G., Marshall D. F., Husmeier D., McGuire G. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- 25.Maddison W. Syst. Biol. 1997;46:523–536. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 26.Sumner S., Aanen D. K., Delabie J., Boomsma J. J. Insectes Soc. 2004;51:37–42. [Google Scholar]
- 27.Chakraborty R., De Andrade M., Daiger S. P., Budowle B. Mol. Ecol. 1992;5:453–455. [Google Scholar]
- 28.Wattier R., Engel C. R., Saumitou-Laprade P., Valero M. Mol. Ecol. 1998;7:1569–1573. [Google Scholar]
- 29.Möller A. Die Pilzga Ërten Einiger su Ëdamerikanischer Ameisen. Jena, Germany: Fisher; 1893. [Google Scholar]
- 30.Pagnocca F. C., Bacci M., Fungaro M. H., Bueno O. C., Hebling M. J., Santanna A., Capelari M. Mycol. Res. 2001;105:173–176. [Google Scholar]
- 31.Silva-Pinhati A. C. O., Bacci M., Hinkle G., Sogin M. L., Pagnocca F. C., Martins V. G., Bueno O. C., Hebling M. J. A. Braz. J. Med. Biol. Res. 2004;37:1463–1472. doi: 10.1590/s0100-879x2004001000004. [DOI] [PubMed] [Google Scholar]
- 32.Buller A. H. R. Researches on Fungi. Vol. 4. London: Longmans Green; 1931. [Google Scholar]
- 33.Pontecorvo G. Annu. Rev. Microbiol. 1956;10:393–400. doi: 10.1146/annurev.mi.10.100156.002141. [DOI] [PubMed] [Google Scholar]
- 34.Taylor J. W., Jacobson D. J., Fisher M. C. Annu. Rev. Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 35.Xu J., Horgen P. A., Anderson J. B. Mycol. Res. 1996;100:188–192. [Google Scholar]
- 36.Johannesson H., Stenlid J. Fungal Genet. Biol. 2004;41:563–570. doi: 10.1016/j.fgb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Abomo-Ndongo S., Tourvieille J., Guillaumin J. J. Cryptogam. Mycol. 2002;23:335–347. [Google Scholar]
- 38.Carvalho D. B., Smith M. L., Anderson J. B. Mycol. Res. 1995;99:641–647. [Google Scholar]
- 39.Anderson J. B., Kohn L. M. Trends Ecol. Evol. 1998;13:444–449. doi: 10.1016/s0169-5347(98)01462-1. [DOI] [PubMed] [Google Scholar]
- 40.Polak E., Hermann R., Kuees U., Aebi M. Fungal Genet. Biol. 1997;22:112–126. doi: 10.1006/fgbi.1997.1010. [DOI] [PubMed] [Google Scholar]
- 41.Selosse M. A. New Phytol. 2001;149:159–162. doi: 10.1046/j.1469-8137.2001.00047.x. [DOI] [PubMed] [Google Scholar]
- 42.Hallenberg N., Kuffer N. Nordic J. Bot. 2001;21:431–436. [Google Scholar]
- 43.Kweskin M. Molecular and Behavioral Ecology of Fungus-Growing Ants and Their Fungi. Austin: Univ. of Texas; 2003. p. 79. [Google Scholar]
- 44.Currie C. R. Annu. Rev. Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 45.Currie C. R., Mueller U. G., Malloch D. Proc. Natl. Acad. Sci. USA. 1999;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villesen P., Gertsch P. J., Frydenberg J., Mueller U. G., Boomsma J. J. Mol. Ecol. 1999;8:1819–1825. doi: 10.1046/j.1365-294x.1999.00767.x. [DOI] [PubMed] [Google Scholar]
- 47.Villesen P., Murakami T., Schultz T. R., Boomsma J. J. Proc. R. Soc. London B; 2002. pp. 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aanen D. K. Proc. Natl. Acad. Sci. USA. 2002;99:14887–14892. doi: 10.1073/pnas.222313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller U. G., Gerardo N. M., Aanen D. K., Six D. L., Schultz T. R. Annu. Rev. Ecol. Syst. 2005;36:563–595. [Google Scholar]
- 50.Mueller U. G., Gerardo N. M. Proc. Natl. Acad. Sci. USA. 2002;99:15247–15249. doi: 10.1073/pnas.242594799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machado C. A., Robbins N., Gilbert M. T. P., Herre E. A. Proc. Natl. Acad. Sci. USA. 2005;102:6558–6565. doi: 10.1073/pnas.0501840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark M. A., Moran N. A., Baumann P., Wernegreen J. J. Evolution (Lawrence, Kans.) 2000;54:517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 53.Taylor D. L., Bruns T. W. Proc. Natl. Acad. Sci. USA. 1997;94:4510–4515. doi: 10.1073/pnas.94.9.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DePriest T. Annu. Rev. Microbiol. 2004;58:273–301. doi: 10.1146/annurev.micro.58.030603.123730. [DOI] [PubMed] [Google Scholar]
- 55.Hoeksema J. D., Bruna E. M. Oecologia. 2000;125:321–330. doi: 10.1007/s004420000496. [DOI] [PubMed] [Google Scholar]
- 56.Stanton M. L. Am. Nat. 2003;162:S10–S23. doi: 10.1086/378646. [DOI] [PubMed] [Google Scholar]
- 57.Strauss S. Y., Irwin R. E. Annu. Rev. Ecol. Syst. 2004;35:435–466. [Google Scholar]
- 58.Knowlton N., Rohwer F. Am. Nat. 2003;162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- 59.Vazquez D. P., Aizen M. A. Ecology. 2004;85:1251–1257. [Google Scholar]
- 60.Thompson J. N. The Geographic Mosaic of Coevolution (Interspecific Interactions) Chicago: Univ. of Chicago Press; 2005. [Google Scholar]
- 61.White T. J., Bruns T., Lee S., Taylor J. W. In: PCR Protocols: A Guide to Methods and Applications. Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., editors. New York: Academic; 1990. pp. 315–322. [Google Scholar]
- 62.Kosakovsky Pond S. L., Frost S. D. W., Muse S. V. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]