Abstract
Epigenetic effects in mammals depend largely on heritable genomic methylation patterns. We describe a computational pattern recognition method that is used to predict the methylation landscape of human brain DNA. This method can be applied both to CpG islands and to non-CpG island regions. It computes the methylation propensity for an 800-bp region centered on a CpG dinucleotide based on specific sequence features within the region. We tested several classifiers for classification performance, including K means clustering, linear discriminant analysis, logistic regression, and support vector machine. The best performing classifier used the support vector machine approach. Our program (called hdfinder) presently has a prediction accuracy of 86%, as validated with CpG regions for which methylation status has been experimentally determined. Using hdfinder, we have depicted the entire genomic methylation patterns for all 22 human autosomes.
Keywords: DNA methylation, epigenomics, methylation prediction, CpG islands
Although progress recently has been made toward whole-genome DNA methylation profiling by using molecular techniques, computational epigenomics is still in its infancy (1). Global analyses of DNA methylation have been focused mainly on two themes: the discovery of methylated CpG islands (CGI) and allele-specific cytosine methylation. Computational prediction of CGIs was introduced in 1987 by Gardiner-Garden et al. (2). They defined CGIs as regions of >200 bp with G+C content of >0.5 and the observed/expected CpG ratio >0.6. Takai and Jones (3) later proposed a more stringent definition that requires CGIs to be >500 bp long, CG content >55%, and the CpG ratio >0.65. This latter method is successful in excluding Alu repeats, many of which were annotated as CGIs when the former criteria were used. Matsuo et al. (4) have provided statistical evidence for erosion of mouse CGIs as compared with human ones. They suggested that an accumulation of TpGs and CpAs observed in mouse, presumably due to the higher rate of deamination of the methylated CpGs, results in a lower CpG ratio in mouse. Antequerra and Bird (5) performed comparative analysis on human and mouse and came to a similar conclusion. Yang et al. (6) proposed a computational method to identify genes with significant differences in gene expression between two parental alleles by searching the UniGene database for the presence of monoallelically expressed (or imprinted) genes in the human genome. Wang et al. (7) compared human and mouse sequences for all known imprinted genes and found 15 motifs that are significantly enriched in the imprinted genes. However, currently there is no algorithm that can predict DNA methylation patterns based on the genomic sequence alone. Because almost nothing is known of the mechanisms that target specific sequences for de novo methylation, a key question that arises is whether there are DNA sequences that are more prone or resistant to methylation.
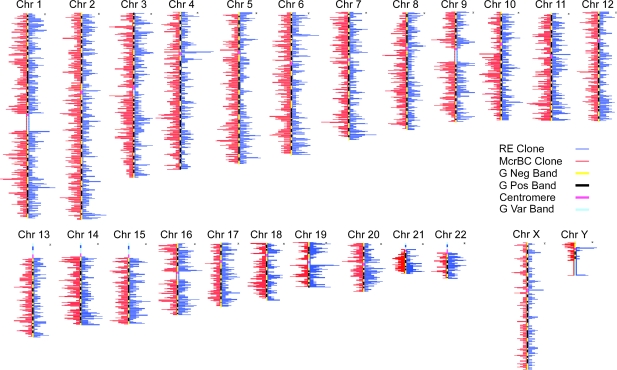
To answer this question, we use data that was generated by enzymatic fractionation of 30 Mb of human brain DNA into nonoverlapping methylated and unmethylated fragments (8) (see Methods). We have 1,948 methylated sequences and 2,386 unmethylated sequences, each sequence is several kilobase pairs long. The distribution of methylated and unmethylated sequences on the chromosome cytogenetic map is given in Fig. 1. The peaks and valleys represent the average number of methylated (M) and unmethylated (U) sequences within a 100-MB window along the map. Interestingly, the M sequences tend to peak at the borders of the pericenteromeric regions corresponding to potential evidence for methylation of satellite repeat elements (heterochromatic chromosomal regions). In contrast, the U sequences tend to peak in euchromatic regions that tend to be gene rich. Mean length of the M sequences is ≈5,400 bp and for U sequences it is 2,700 bp. For further analysis of these sequences, we ignored 250 bp of boundary sequences at both ends to avoid any potential boundary effects that include high density of young Alus transposons (data not shown).
Fig. 1.
Distribution of the methylated (restriction endonucleases, RE) and unmethylated (McrBC) sequences along the chromosome cytogenetic map. The peaks and valleys represent the average number of RE and McrBC sequences within a 100-Mb window along the map. Interestingly, the methylated sequences tend to peak at the borders of the pericenteromeric regions corresponding to potential evidence for methylation of satellite repeat elements (heterochromatic chromosomal regions). In contrast, the unmethylated sequences tend to peak in euchromatic regions that tend to be gene rich.
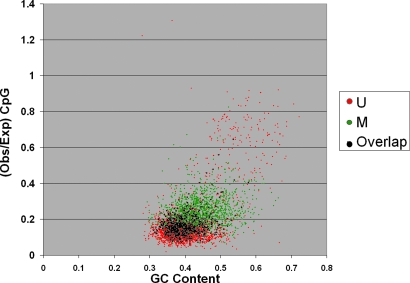
The most marked difference between U and M sets is the distribution of sequences that satisfy the Takai–Jones criteria for CGIs. There is large number of CGIs in the U set (relative to the M set), although the M set is much larger and average sequence length is greater. This difference is evident from Fig. 2, where CpG ratio versus G+C content is plotted for all sequences. The figure shows that in low CpG ratio and G+C content region, there is a large overlap between M and U sequences. On the contrary, in the high CpG ratio region, a majority of the sequences is filled with CGIs (8) and they are mostly unmethylated. Based on this observation, two M-U classifiers were developed, one for CGIs and one for non-CGIs.
Fig. 2.
Distribution of G+C content vs. CpG ratio (observed/expected). For large GC content and CpG ratio values, a majority of sequences are unmethylated.
The second difference between U and M data sets is the distribution of the Alu elements, in particular young and intermediate Alus. Alu elements are primate-specific short interspersed nuclear elements, typically ≈280-nt long (9). These elements account for >10% of the human genome. M sequences are rich in AluY and AluS compared with U sequences. Compared with U sequences, M sequences have 2.5 times more AluY and AluS after length correction.
We also carried out extensive motif discovery within M and U sequences after masking Alu repeats. We scan along the given sequence and evaluate each 500-bp window that is centered on a CpG dinucleotide and test whether it satisfies the Takai–Jones criteria (3). When selected consecutive windows overlap, we merge them to obtain a contiguous sequence. In this manner, we obtain two disjoint sets of sequences: those that satisfy the CGI criteria and those that do not. We used a position weight matrix enumeration program, discriminant matrix enumeration to identify motifs that are most discriminating between U and M sequences (10) and identified the top 10 discriminating hexamer motifs (by using the standard IUPAC codes, see Table 1) for each data set. Many of the motifs that best discriminate between U and M data sets are related to known transcription factor binding sites (see Tables 5–7, which are published as supporting information on the PNAS web site).
Table 1.
Top 10 enriched motifs discovered by Discrete Matrix Enumerator
| Rank | non-CGI |
CGI |
|
|---|---|---|---|
| Enriched in U set | Enriched in M set | Enriched in U set | |
| 1 | AAWGGR | CCDGGV | CCCSGS |
| 2 | AAATKT | BCCCWG | GSCCCS |
| 3 | ATGVAA | GGVCCH | CCGSSC |
| 4 | TGVAAA | CCCWGH | CGSCCS |
| 5 | CWGAMA | GGSCTB | VGCGGG |
| 6 | AATKAA | CCTGMV | GRGCSC |
| 7 | AAATGV | GMCCCN | TCCSSG |
| 8 | TGRAAT | SCCWCR | KCCSGC |
| 9 | GVAAAT | WGCCCH | CTCCSS |
| 10 | TRAATT | CKGSCM | SGMGCC |
Note that standard nucleotide substitution was used.
We rely on a classical pattern recognition framework to develop a methylation predictor. For non-CGIs, we started with 102 features, including G+C content, di- and trinucleotide count, Alu coverage, and 20 hexamers. For CGIs, we used 92 features, including only 10 hexamers. We use recursive feature elimination, which is a backward selection method, and principal component analysis (PCA) for feature subset selection (refs. 11 and 12; see Methods). Once the feature subset was selected, we compared several classifiers to test classification performance including K means clustering, linear discriminant analysis (LDA), logistic regression (LR), and support vector machine (SVM) (13). LDA and LR are representative of linear classification models, whereas SVM is a model that maps the data into a higher dimensional space, where it is possible to apply a linear classification. K means clustering gave completely unpredictable results based on random seed selection (true positive rate was 0.51). LDA gave a 0.84 true positive rate and a 0.25 false positive rate, LR gave a 0.82 true positive and a 0.22 false positive, whereas SVM gave a 0.2 false positive rate and a 0.86 true positive rate. The best performing classifier was the SVM approach (14). We used a sliding window-based prediction approach to determine the methylation propensity. Extensive experiments showed that 800 bp is the optimal window size that best predicts accuracy under SVM (see Methods). The results of 10-fold cross-validation for classification experiments show that SVM can correctly predict methylation status of the unseen non-CGIs regions with 84% accuracy (Table 2). For CGIs, we developed the second classifier, and it has 96.5% accuracy. Our measure for accuracy of prediction is as follows: TP + TN/(TP + FP + TN + FN), where TP means true positives, TN is true negatives, FP is false positives, TN is true negatives and FN is false negatives. To assess the robustness of our method and estimate the significance of our prediction accuracy, we used the standard permutation test, where we randomly shuffled the labels of the known samples and calculated the accuracy of the prediction by following the same procedure. The P value for the prediction accuracy of 84% for non-CGI and 96.5% for CGIs is <10−4. The overall accuracy of our program is 86%, calculated by using the proportion of respective type of CpG regions in the data set.
Table 2.
Methylation prediction accuracy with SVM
| CpG window type | Mean, % | U, % | M, % |
|---|---|---|---|
| CGI | 96.5 | 95 | 98 |
| non-CGI | 84 | 87 | 81 |
| Overall | 86 | 90.7 | 81.3 |
To produce a map of DNA methylation landscape of the human genome, we applied hdmfinder to predict the methylation status of the assembled regions in the 22 autosomes (Build 33). Because of space limitations, it will be available on the UCSC Genome Browser web site. Table 3 shows only the results of our prediction in chromosomes 21 and 22 (we have used a definition of 1-kb upstream of the RefSeq genes as the promoter regions).
Table 3.
DNA methylation prediction results for chromosomes 21 and 22
| Chromosome | Total no. of CpG dinucleotides | Predicted in non-CGIs |
Predicted in CGIs |
RefSeq genes with predicted methylated promoters | ||
|---|---|---|---|---|---|---|
| U | M | U | M | |||
| 21 (218 known genes, 41% G+C content) | 367,215 | 24,656 | 319,438 | 22,522 | 599 | NM_001685, NM_002040, NM_006447, NM_005806, NM_003024, NM_005111, NM_145311, NM_145858, NM_005069, NM_009586, NM_018964, NM_018669, NM_021075, NM_033661, NM_033662 |
| 22 (341 known genes, 48% G+C content) | 541,955 | 17,773 | 474,495 | 48,381 | 1,306 | NM_053006, NM_022719, NM_001835, NM_007098, NM_012143, NM_030758, NM_153044, NM_000967, NM_005008, NM_006071, NM_014246, NM_014177 |
An earlier study by Feltus et al. (15) had indicated similar sequence dependence on the epigenetic state of some selected CGIs. However, our study differs at least on two counts. First, their study was conducted on a limited set of preselected CGIs, whereas no prior selection of sequences was involved in the present study. Second, the CGIs used in their analysis were methylated by overexpression of methyltransferase (DNMT1) in vitro; whereas in our case, the data comes from normal human adult brain DNA. Our method shows that the sequence dependence of CpG methylation can be generalized to all CpGs in the genome even though the nature of this dependence is still an open issue. It is known that there are tissue-specific differences between the methylation profiles, but basic patterns are very similar for many tissues (16). Although our prediction is based on the human brain DNA, we hope people could compare it with DNA methylation result for other tissues to study variation. The current estimate on tissue-specific CpG methylation in mouse CGIs is ranging from 5% to 16% (17, 18), and further in-depth studies of tissue-specific CpG methylation variations (regardless of whether it is in CGI) and epigenetic polymorphisms within the human population undoubtedly will be extremely valuable.
Methods
Data Source.
Here we describe the method briefly and refer to Rollins et al. (8) for the experimental details. During enzymatic fractionation, McrBC digestion removes methylated sequences resulting in 11.6 Mb of unmethylated sequence domains. Similarly, five methylation-sensitive restriction endonucleases digestion removes unmethylated sequences, resulting in 18.6-Mb methylated sequence domains covering all 22 autosomes. Specifically, methylated sequence libraries were created by digestion with the methylation-sensitive restriction endonucleases Tail (ACGT), BstUI (CGCG), HhaI (GCGC), HpaII (CCGG) and AciI (CCGC and GCGG). Although the methylation status in these libraries is only experimentally known for the listed sites, for the purposes of this computational work, all other CpG sequences (e.g., TCGA and ACGC) in the library are assumed to have the same methylation status as surrounding sites. Unmethylated sequence libraries were created by digestion with McrBC, which cleaves at Rm5CG(N)40–500m5CGR sites. Interior CpGs not flanked by a purine are assumed to have the same methylation status as surrounding purine-flanked CpGs. All sequence data are available upon request.
Feature Selection by Using PCA and Recursive Feature Elimination.
We performed PCA and set a threshold of 0.2 for the coefficient value of the principal components to find the features that contribute significantly to the first four principal components. We also used SVM with recursive feature elimination (RFE) for the feature selection and compared the result with PCA-based features (for further details, see, which are published as supporting information on the PNAS web site). It has been shown that feature selection can improve classification results for DNA methylation (19). RFE is a feature selection method that ranks the features by the change in objective function when one feature is removed (11). The classifier is trained initially by using all of the features. The method is based on backward sequential selection of features. The ranking criterion is computed for all features. At each iteration the feature with smallest ranking criterion is removed. The features are iteratively removed in a greedy fashion until the largest margin of separation is reached. Fig. 5, which is published as supporting information on the PNAS web site, presents the result of RFE, which shows the change in prediction accuracy with respect to the change in the number of features. Classification accuracy increases exponentially with the number of features until one uses 17 features, after which the accuracy does not improve significantly with the increase in the number. In the order of best to worst, these features are AAWGGR, TGRAAT, AAT, ATGVAA, ACG, CG, GCG, AC, Alu-coverage, CGG, GAA, CAC, CKGSCM, SCCWCR, ATG, TGC, and CCG. There is a significant overlap between the features between the PCA results and RFE method. Alu, hexamers, and some of the trimers are shown to be important by both methods: Alu, AAWGGR, TGRAAT, ATGVAA, CG, GCG, CGG, and GAA for non-CGI. The CG is very prominent in top features selected by RFE and significant both in the second and fourth principal component for the CGI set. Hence, based on these results, we selected 17 and 16 features for classifiers of non-CGI and CGI (Table 4), respectively. It should be noted here that because we are using an SVM, which is a nonlinear classifier, differences in the mean values of the variables do not directly correspond to their discriminability.
Table 4.
Selected features of non-CGI and CGI
| Rank | Features | Mean (U) | SE (U) | Mean (M) | SE (M) |
|---|---|---|---|---|---|
| non-CGI | |||||
| 1 | AAWGGR | 1.4 | 2.7 | 1.1 | 1.3 |
| 2 | TGRAAT | 0.85 | 2.1 | 0.69 | 1.2 |
| 3 | AAT | 17 | 7.9 | 15 | 7.6 |
| 4 | ATGVAA | 1.1 | 2.3 | 0.88 | 1.3 |
| 5 | ACG | 1.5 | 1.5 | 2.7 | 2.1 |
| 6 | CG | 6.3 | 5.6 | 12 | 7.4 |
| 7 | GCG | 1.3 | 2 | 3 | 2.7 |
| 8 | AC | 1.9 | 2.4 | 3.5 | 3.4 |
| 9 | ALU-COVER | 25 | 87 | 120 | 170 |
| 10 | CGG | 34 | 8 | 39 | 8.2 |
| 11 | GAA | 1.2 | 1.3 | 1.8 | 1.8 |
| 12 | CAC | 9.2 | 4 | 12 | 4.6 |
| 13 | CKGSCM | 14 | 6.7 | 14 | 6.2 |
| 14 | SCCWCR | 1 | 1.1 | 1.6 | 1.5 |
| 15 | ATG | 13 | 6.1 | 13 | 5.1 |
| 16 | TGC | 10 | 4 | 13 | 4.7 |
| 17 | CCG | 1.9 | 2.4 | 3.6 | 3.2 |
| CGI | |||||
| 1 | CGG | 31 | 11 | 17 | 6.5 |
| 2 | CAT | 4.9 | 2.8 | 8.4 | 5.6 |
| 3 | TCCSSG | 3 | 1.9 | 0.99 | 1.7 |
| 4 | CCG | 29 | 10 | 16 | 8.5 |
| 5 | CCA | 14 | 4.5 | 18 | 9.1 |
| 6 | TTC | 9 | 4 | 7.7 | 4.4 |
| 7 | GCC | 34 | 10 | 22 | 12 |
| 8 | TAT | 2.1 | 2.1 | 5.3 | 5.8 |
| 9 | SGMGCC | 4.7 | 2.8 | 2.1 | 2.1 |
| 10 | TCG | 9.7 | 3.5 | 5.9 | 3.7 |
| 11 | ACG | 7.7 | 3.3 | 13 | 11 |
| 12 | CCC | 24 | 9.2 | 16 | 9.2 |
| 13 | CCGSSC | 6.2 | 3.9 | 2.1 | 2.7 |
| 14 | CG | 79 | 20 | 61 | 25 |
| 15 | CGC | 26 | 9.1 | 18 | 8.7 |
| 16 | ATG | 4.8 | 2.8 | 8.8 | 8 |
Model Selection for SVM.
The SVM algorithm (13) applies a kernel function to fit a maximum-margin hyperplane in the transformed feature space. The transformation may be nonlinear (e.g., polynomial or radial basis function), and the transformed space is usually high dimensional. Although the classifier is a hyperplane in the high-dimensional feature space, it may be nonlinear in the original input space. If the kernel used is a radial basis function, the corresponding feature space is a Hilbert space of infinite dimension. We trained a two-class SVM by using a radial basis kernel. The SVM is computationally expensive, but it is compensated for its higher prediction accuracy when we compared it to other classifiers. There are two parameters associated with SVM training. One is regularization of the cost parameter C and kernel parameter γ, which determines the RBF width. We performed extensive grid search (Fig. 6, which is published as supporting information on the PNAS web site) to select the optimal parameter values of 10 for C and 0.5 for γ.
Window Length Dependency.
We tested the effect of window size on classification performance by applying the same method but on data that were calculated based on varying window size. Fig. 7, which is published as supporting information on the PNAS web site, shows how the prediction accuracy depends on the window size. Classification accuracy improves with increases in window size and reaches its maximum at a window size of 800 bp (one explanation is that the methylated set is rich in AluY and its effect on prediction becomes prominent at longer window size).
Overall Prediction: hdmfinder.
We designed the algorithm for the genomewide prediction (see Fig. 8, which is published as supporting information on the PNAS web site). For each window centered around a CpG, we test whether it satisfies the Takai–Jones CGI criteria. Next, for all of the windows, we apply the SVM classifier to predict their methylation status. Using our predictor function, we calculate two posterior probability P(Class|data) for both the “+” strand and “−” strand. In our case, “data” is either CGI or non-CGI. SVM-based methods do not generate any probability measure directly. However, one can use a logistic link function to generate a class probability. Methylation status of the sequence is determined by the strand that has higher posterior probability. After obtaining the probabilities, we apply a Gaussian smoothing function with a window length of 5 to remove fluctuations. hdmfinder is available upon request.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants (to J.J., T.H.B., and M.Q.Z.), a Fellowship from the Leukemia and Lymphoma Society (to R.A.R.), NIH Grant HG002915-01A1 (to F.H.), and National Institute of Mental Health Grant MH074118-01.
Abbreviations
- CPI
CpG island
- M
methylated
- PCA
principal component analysis
- RFE
recursive feature elimination
- SVM
support vector machine
- U
unmethylated.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Fazzari M. J., Greally J. M. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 2.Gardiner-Garden M., Frommer M. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 3.Takai D., Jones P. A. Proc. Natl. Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuo K., Clay O., Takahashi T., Silke J., Schaffner W. Somatic Cell Mol. Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- 5.Antequera F., Bird A. Proc. Natl. Acad. Sci. USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H. H., Lee M. P. Ann. N.Y. Acad. Sci. 2004;1020:67–76. doi: 10.1196/annals.1310.008. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Fan H., Yang H. H., Hu Y., Buetow K. H., Lee M. P. Genomics. 2004;83:395–401. doi: 10.1016/j.ygeno.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Rollins R. A., Haghighi F., Edwards J. R., Das R., Zhang M. Q., Ju J., Bestor T. H. Genome Res. 2005;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mighell A. J., Markham A.F., Robinson P.A. FEBS Lett. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 10.Smith A. D., Sumazin P., Zhang M. Q. Proc. Natl. Acad. Sci. USA. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guyon I., Weston J., Barnhill S., Vapnik V. Mach. Learn. 2002;46:389–422. [Google Scholar]
- 12.Ambroise C., McLachlan G. J. Proc. Natl. Acad. Sci. USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortes C., Vapnik V. Mach. Learn. 1995;20:273–297. [Google Scholar]
- 14.Vapnik V. The Nature of Statistical Learning Theory. New York: Springer; 1995. [Google Scholar]
- 15.Feltus F. A., Lee E. K., Costello J. F., Plass C., Vertino P. M. Proc. Natl. Acad. Sci. USA. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunau C., Hindermann W., Rosenthal A. Hum. Mol. Genet. 2000;9:2651–2663. doi: 10.1093/hmg/9.18.2651. [DOI] [PubMed] [Google Scholar]
- 17.Shiota K., Kogo Y., Ohgane J., Imamura T., Urano A., Nishino K., Tanaka S., Hattori N. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- 18.Song F., Smith J. F., Kimura M. T., Morrow A. D., Matsuyama T., Nagase H., Held W. A. Proc. Natl. Acad. Sci. USA. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Model F., Adorjan P., Olek A., Piepenbrock C. Bioinformatics. 2001;17(Suppl. 1):S157–S164. doi: 10.1093/bioinformatics/17.suppl_1.s157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.