Abstract
Background:
For most vertebrate organs and tissues, the majority of development occurs during embryogenesis, and postnatal changes are primarily concerned with growth. The central nervous system is unusual in that a considerable amount of morphological development, cell differentiation and acquisition of function, takes place during postnatal development. As yet, the molecular mechanisms underlying these complex developmental processes are not well understood. In order to identify markers for these developmental processes, we have analyzed the expression profiles, during postnatal murine brain development, of approximately 25,000 transcripts. This analysis, performed at day 1, day 10, day 20 and day 42 of postnatal development, identified a large number of developmentally regulated genes which we have assigned into three broad expression categories.
Results:
Expression levels at four timepoints during postnatal murine brain development were established for approximately 25,000 gene transcripts. Approximately 1% of the genes examined displayed a developmentally regulated pattern of expression and we provide all the necessary information required to easily obtain molecular markers for a subset of these developmentally regulated transcripts. Of this subset, 61 showed increasing expression during development, 61 showed decreasing expression during development, and 9 exhibited a peak of expression during this period.
Conclusions:
A small percentage of the genes expressed in the postnatal developing brain show changes in expression level during the newborn to adult phase of development. It is likely that these developmentally regulated transcripts represent molecular markers for the complex developmental process occurring in the postnatal brain.
Background
For most vertebrate organs and tissues, the majority of development occurs during embryogenesis, and postnatal changes are primarily concerned with growth. The central nervous system (CNS) is unusual in that a considerable amount of morphological development, cell differentiation and acquisition of function takes place during postnatal development [1,2]. As yet, the molecular mechanisms underlying these complex developmental processes are not well understood.
We have recently completed a differential display (DDRT-PCR) [3] analysis of genes expressed in the murine postnatal developing brain. This analysis formed the baseline for an ongoing study and produced a series of RNA fingerprints representing genes transcribed at specific stages of brain development. For our analysis we compared RNA fingerprints at four different timepoints during postnatal development: newborn (day 1), day 10, day 20 and adult (day 42). Incidental to our experimental objective, we also identified a large number of genes which were clearly developmentally regulated in the wild-type mouse brain. We submit this data principally to allow others to isolate specific transcripts with developmentally regulated expression in the postnatal brain without the necessity of performing extensive screening procedures. From a total of approximately 25,000 transcripts displayed, around 200-300 (approximately 1%) exhibited developmentally regulated expression profiles. Here we present groups of RNA fingerprints displaying a subset of these developmentally regulated genes, along with sufficient technical information to replicate specific fingerprints and recover selected cDNAs.
The particular fingerprints presented here were selected on the basis that identical DDRT-PCR profiles were obtained with at least two separate batches of RNA, and that each fingerprint displayed at least three developmentally regulated transcripts. On each group of fingerprints we have indicated the position of at least three bands representing the more obviously developmentally regulated genes. We have assigned these marked transcripts into three broad categories as follows: genes for which mRNA levels increase during brain development; genes for which mRNA levels decrease during development; and genes exhibiting a peak in mRNA levels during this developmental period.
To ensure that changes in DDRT-PCR profiles represent genuine changes in expression levels, two cDNA fragments were recovered and used in downstream expression analyses. The overall procedure followed is illustrated in Figure 1 (and see [4]). The northern blot expression profiles of both transcripts accurately replicated the original DDRT-PCR expression profiles, confirming the validity of our approach.
Figure 1.

Differential display procedures; mSSCP: modified single-stranded conformational polymorphism [4].
Results
In this study, we have observed a large number of transcripts which apparently increase and/or decrease in expression level during postnatal murine brain development. The period of development under study spanned from day 1 to day 42 post-partum. Figure 2 shows a selection of RNA fingerprints containing bands representing such developmentally regulated transcripts. Thirty-four groups of RNA fingerprints were selected on the basis that identical DDRT-PCR profiles were obtained with at least two separate batches of RNA, and that each fingerprint displayed at least three developmentally regulated transcripts. The gel positions of a subset of developmentally regulated transcripts are indicated in Figure 2 and individual transcripts are assigned into one of three broad expression categories: A (+), genes for which mRNA levels increase during brain development; B (-), genes for which mRNA levels decrease during development; and C (^), genes exhibiting a peak in mRNA levels during this developmental period. Of a total of 131 transcripts indicated, 61 are assigned to category A, 61 to category B, and 9 to category C. The sequence details of the primers used to produce the RNA fingerprints shown in Figure 2 are contained in Table 1. A detailed description of the various procedures employed and the animals used in this study are presented in the Materials and methods section.
Figure 2.

Gene expression profiles in the postnatal developing mouse brain. RNA fingerprints produced by specific DDRT-PCR primer combinations at four stages of brain development run in parallel. Anchored primer/arbitrary primer combinations are shown at the top for each group of RNA fingerprints and stages of development are indicated in days (1, 10, 20 and 42). Gel positions of developmentally regulated transcripts in expression category A (+), expression category B (-) and expression category C (^) are highlighted.
Table 1.
Oligonucleotide sequences of primers used
| Anchored primer |
| dT12AA 5'-d(TTTTTTTTTTTTAA)-3' |
| dT12GA 5'-d(TTTTTTTTTTTTGA)-3' |
| dT12CA 5'-d(TTTTTTTTTTTTCA)-3' |
| dT12AG 5'-d(TTTTTTTTTTTTAG)-3' |
| dT12GG 5'-d(TTTTTTTTTTTTGG)-3' |
| dT12CG 5'-d(TTTTTTTTTTTTCG)-3' |
| dT12AC 5'-d(TTTTTTTTTTTTAC)-3' |
| dT12GC 5'-d(TTTTTTTTTTTTGC)-3' |
| dT12CC 5'-d(TTTTTTTTTTTTCC)-3' |
| dT12AT 5'-d(TTTTTTTTTTTTAT)-3' |
| dT12GT 5'-d(TTTTTTTTTTTTGT)-3' |
| dT12CT 5'-d(TTTTTTTTTTTTCT)-3' |
| Random primer |
| (arbitrary but defined sequence) |
| P3 5'-d(GCCGTTCCAT)-3' |
| R1 5'-d(GGAACTCCGT)-3' |
| R2 5'-d(GGCAAGTCAC)-3' |
| R4 5'-d(AGGACCGCTA)-3' |
| R5 5'-d(CGGACCCCGG)-3' |
| R7 5'-d(TACAACGAGG)-3' |
| R8 5'-d(TGGATTGGTC)-3' |
| R9 5'-d(TGGTAAAGGG)-3' |
| R10 5'-d(TCGGTCATAG)-3' |
| R11 5'-d(TACCTAAGCG)-3' |
| R12 5'-d(CTGCTTGATG)-3' |
| R14 5'-d(GATCGCATTG)-3' |
| R18 5'-d(GGAACCAATC)-3' |
| M2 5'-d(CACAGTTTGC)-3' |
| M3 5'-d(CCACAGAGTA)-3' |
| S 5'-d(GCGACCCATG)-3' |
| A1 5'-d(ACAGAGCACA)-3' |
| A2 5'-d(ACGTATCCAG)-3' |
| Oligo(dT) primer mix |
| (poly(A) anchored) |
| MA Equal mix of dT12AA, dT12GA and dT12CA |
| MG Equal mix of dT12AG, dT12GG and dT12CG |
| MC Equal mix of dT12AC, dT12GC and dT12CC |
| MT Equal mix of dT12AT, dT12GT and dT12CT |
| Extended random primer |
| 5'-d(GTCAGAATTC-random primer)-3' |
| Extended oligo(dT) primer |
| 5'-d(GTCAGAATTCT12)-3' |
To ensure that developmentally dependent changes in intensity of individual DDRT-PCR bands reflected changes in levels of individual mRNAs, two apparently up-regulated transcripts were chosen for further study. The cDNA fragments represented by these bands were recovered, purified from co-migrating DNA species by mSSCP [4], cloned and used in northern hybridization studies (Figure 3). For northern analyses, whole brain RNA was isolated from a separate batch of animals to those used in the original DDRT-PCR. As can be seen from Figure 3, for both transcripts northern analyses produced expression profiles similar to those observed by DDRT-PCR, thereby validating this approach to identifying developmentally regulated transcripts.
Figure 3.
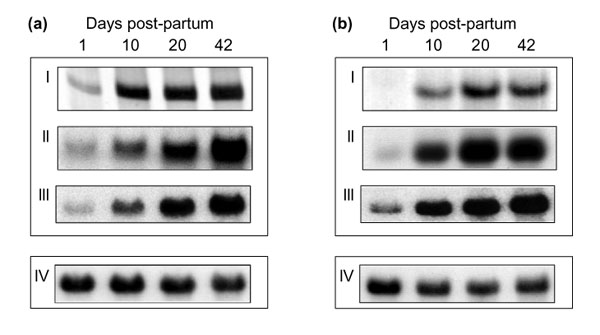
Verification of expression profiles. Downstream analysis of two developmentally regulated transcripts - (a) and (b) are the two transcripts. Candidate cDNAs were selected on the basis of the original DDRT-PCR profile (I), purified from co-migrating sequences by mSSCP (II) and used as probes in northern analysis (III). To correct for variation in RNA loading, northern blot membranes were re-probed for expression of the mouse beta actin gene (IV).
Sequence analysis revealed that these cDNA fragments represented portions of the prion Protein (PrP) and of the Thy-1.2 gene transcripts [5,6]. The cloned DNA used as a probe in Figure 3a is identical to nucleotides 588-1,414 of the PrP transcript while that used in Figure 3b is identical to nucleotides 1,599-1,700 of the Thy-1.2 transcript (Figure 4). This shows that the original DDRT-PCR reactions have amplified an internal fragment of the PrP transcript and the terminal 3' untranslated region (UTR) of the Thy-1.2 transcript. The PrP fragment was amplified by a combination of arbitrary primer R4 and anchored primer T12CC while the Thy-1.2 fragment was amplified by a combination of arbitrary primer P3 and the anchored primer T12CA. For the PrP fragment, the arbitrary primer showed a 100% match while the anchored primer showed 86% homology (12/14) with the published sequence. In contrast, for the Thy 1.2 fragment, while the anchored primer matched with 100% homology, the arbitrary primer showed only 70% homology (7/10) with the corresponding region of the published sequence.
Figure 4.
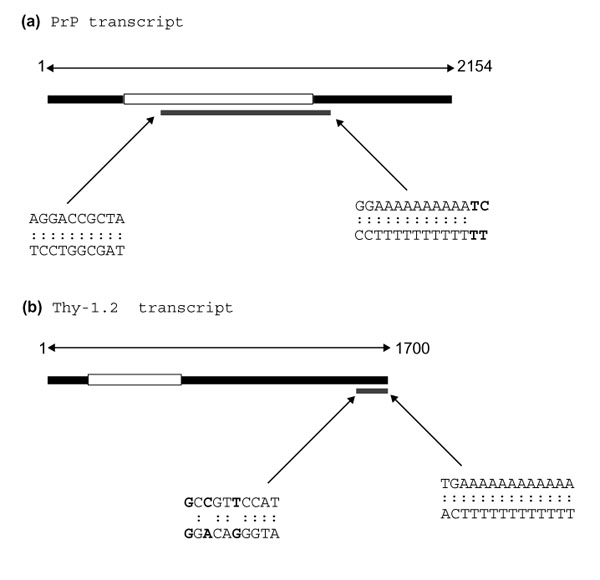
Origin of DDRT-PCR fragments. Schematic illustrating the position of DDRT-PCR cDNA fragments in relation to full-length transcripts of (a) PrP and (b) Thy-1.2. Black boxes represent transcripts, with open reading frames in white. Areas of transcripts amplified by DDRT-PCR are shown as gray boxes. Sequences of regions matching arbitrary and anchored primers are detailed with mismatched pairs in bold.
Conclusions
In the course of analyzing gene expression in the postnatal developing murine brain we have observed that approximately 1% of genes transcribed are developmentally regulated. The objective of this report is to permit those interested in such transcripts to selectively isolate specific developmentally regulated transcripts without the necessity of performing an extensive screen. Here we present the RNA fingerprints containing a subset of these developmentally regulated transcripts and we include all the necessary information to permit individual transcripts to be isolated and identified. We have highlighted a total of 131 developmentally regulated transcripts in three broad categories of expression profiles. Of these, approximately 7% (9) fell into category C while the remainder were equally divided between categories A (61) and B (61). We also present confirmatory evidence that changing DDRT-PCR expression profiles represent genuine alterations in expression levels during brain development. It should be noted that changes in expression profile reflect changes in RNA level per microgram total RNA in whole brain and, given that the postnatal brain is not a uniform structure, expression profiles should be interpreted accordingly.
Materials and methods
Collection of tissues
Male animals from an inbred line of 129/Ola mice were used. Animals were humanely sacrificed and whole brains from six individual animals at P1, P10, P20 and P42 stages of postnatal development were collected, flash frozen in liquid nitrogen and stored at -80°C.
RNA extraction
Total RNA was extracted using the RNAzol™ B method (AMS Biotechnology, Oxon, UK), based on the guanidinium thiocyanate/phenol/chloroform extraction method of Chomczynski and Sacchi [7]. Following the initial precipitation step, the RNA pellet was washed once with 85% ethanol, resuspended in water, and 0.1 volumes of 3M sodium acetate (pH 5.2) and 2.5 volumes ethanol were added, the tubes mixed briefly and RNA stored as ethanol precipitates at -80°C. RNA from six individuals at each timepoint was pooled for DDRT-PCR analysis. For quantitation of total RNA, a small aliquot was removed, pelleted by centrifugation, resuspended in water and the concentration estimated by spectrophotometry. Stock RNA solutions were adjusted to a concentration of approximately 1 μg/μl in ethanol suspension. The use of RNAzol B and homogenization of tissue with a Polytron homogenizer resulted in the isolation of total RNA with OD260/280 ratios consistently greater than 1.95.
Synthesis of first-strand cDNA
For each sample, a volume of RNA stock solution containing approximately 8 μg RNA was transferred to a fresh tube. Samples were centrifuged at 13,000 rpm for 25 minutes and the resulting RNA pellet was washed with 85% ethanol, dried at 45°C for 2 minutes and resuspended in 7 μl RNAse-free water. A small portion (2 μl) of each sample was re-quantitated by spectrophotometery and 5 μg total RNA (in 5 μl volume) was used to synthesize first-strand cDNA (First-strand cDNA synthesis kit; Amersham Pharmacia Biotech, Hertfordshire, UK). Reactions contained 1 μl DTT (200 mM), 5 μl of bulk first-strand mix (containing Moloney murine leukemia virus reverse transcriptase) and 4 μl of either d(T)12MA, d(T)12MG, d(T)12MC, d(T)12MT primer (24 μM; M = A, G or C). Reactions were mixed and incubated at 37°C for 1 hour and then heated to 95°C for 10 minutes to inactivate reverse-transcriptase. The synthesized cDNA was then dispensed into 1 μl aliquots and stored at -20°C.
Differential display PCR
For differential display PCR reactions, 1 μl aliquoted cDNA was diluted to 133 μl with water and 10 μl of this solution was used for each display PCR (equivalent to the amount of cDNA produced from 25 ng RNA). To each 10 μl cDNA on ice, 2 μl arbitrary 10-mer primer (5 μM) was added and the solution overlaid with 30 μl mineral oil (Sigma, Dorset, UK). Master mix (8 μl) containing 2 μl 10X PCR buffer (Boehringer Mannheim, East Sussex, UK), 2 μl dNTPs (20 μM; Amersham Pharmacia Biotech, Hertfordshire, UK), 0.3 μl Taq polymerase (1.5 U, Boehringer Mannheim, East Sussex, UK), 2 μl (dT)12MN (25 μM), 1 μl [α-35S]dATP (1000 Ci/mmol) and 0.7 μl dH2O was added to each tube. Tubes were centrifuged briefly and incubated in a Biometra Unoblock PCR machine at 94°C (2 minutes), followed by 40 cycles of 94°C denaturation (30 seconds), 40°C annealing (2 minutes) and 72°C extension (30 seconds), followed by a final extension step at 72°C for 5 minutes. Type III [8] loading dye (4 μl) was added to each tube and 8 μl of each sample loaded onto a 6% non-denaturing HR-1000 GenomyxLR polyacrylamide gel (Beckman Instruments Ltd., Buckinghamshire, UK). Samples were run for 2 hours 15 minutes at 2,700V (50°C) on a GenomyxLR DNA analyzer (Beckman Instruments Ltd., Bucks, UK). The gels were transferred to 3 MM blotting paper, dried and exposed to BiomaxMR autoradiography film for 16 hours.
Recovery of cDNA from dried polyacrylamide gels
Gel regions corresponding to bands representing candidate cDNAs were excised from all four lanes using sterile scalpels and transferred to sterile 0.5 ml Eppendorf tubes. Glogos™ autoradiograph markers (Stratagene, Cambridgeshire, UK) were used to align the gel with the autoradiograph and identical regions were excised from all lanes. The gel fragments were rehydrated at room temperature for 15 minutes in 100 μl water, and cDNA eluted at 99°C for 15 minutes before transfer of the liquid phase to fresh 0.5 ml tubes. cDNA was precipitated by the addition of 2.5 volumes ethanol, 0.1 volumes of 3 M sodium acetate (pH 5.2) and 1 μl See-DNA (Amersham Pharmacia Biotech, Buckinghamshire, UK), and stored on dry ice for 1 hour. Following centrifugation (13,000 rpm, 25 minutes, 4°C) the pellet was washed in 85% ethanol and resuspended in 4 μl of water.
Modified single-strand conformation polymorphism (mSSCP)
For the mSSCP-PCR reaction, 4 μl 10X PCR buffer (Boehringer Mannheim, East Sussex, UK), 3.2 μl dNTPs (2.5 mM dGTP, dCTP, dTTP; 0.025 mM dATP; Amersham Pharmacia Biotech, Buckinghamshire, UK), 2.5 μl anchored primer (20 μM), 2.5 μl arbitrary primer (20 μM), 0.3 μl Taq polymerase (1.5 U; Boehringer Mannheim, East Sussex, UK) and 0.5 μl [α-33P] dATP (3,000 Ci/mmol) was added to the DNA and the reaction volume adjusted to 40 μl with water. PCR conditions were similar to those used for display-PCR, with the exception that only five cycles were performed. After removal of mineral oil, PCR products were purified by phenol/chloroform/isoamyl alcohol extraction and precipitated for 1 hour on dry ice. Pellets were washed, resuspended in 8 μl of mSSCP loading buffer (80% deionized formamide, 0.01% bromophenol blue, 0.01% xylene cyanol, 1 mM EDTA, 10 mM NaOH) and denatured at 95°C for 10 minutes before loading onto a 0.5X MDE gel (Flowgen, Staffordshire, UK). Samples were electrophoresed typically for 18 hours at 8W (25°C) in 0.6X TBE buffer using the Genomyx LR system. Following autoradiography, areas of the gel corresponding to candidate cDNAs were excised and the cDNA eluted and precipitated as described above.
PCR re-amplification of mSSCP-purified candidate cDNAs
A final PCR reamplification of the recovered cDNA was performed using modified versions of the arbitrary and anchored primers used in the DDRT-PCR and mSSCP steps. For both the anchored and arbitrary primers, oligonucleotides were synthesized with an additional 5' sequence containing an EcoRI restriction site to aid cloning (referred to as extended primers [9]; see Table 1). Reactions contained 4 μl mSSCP purified cDNA, 4 μl 10X buffer, 3.2 μl dNTPs (10 mM each dATP, dCTP, dTTP, dGTP), 2.5 μl extended anchored primer (20 μM), 2.5 μl extended arbitrary primer (20 μM) and 0.5 μl Taq polymerase (Boehringer Mannheim, East Sussex, UK) in a 40 μl volume.
Samples were denatured at 94°C for 2 minutes and a single round of PCR performed at 94°C for 30 seconds, 40°C for 2 minutes and 72°C for 30 seconds. A further five cycles of PCR were then performed at 94°C for 30 seconds, 58°C for 1 minute and 72°C for 30 seconds, followed by a final extension step of 72°C for 5 minutes.
PCR products were phenol/chloroform extracted, precipitated and then subjected to EcoRI restriction endonuclease digestion, prior to further purification and cloning into the EcoRI site of pBluescript SKII+ or KSII+ (Stratagene, Cambridgeshire, UK).
Cloned cDNAs were sequenced and the sequences compared to non-redundant GENBANK/EMBL protein and nucleotide databases, using either the GCG [10] or NCBI worldwide web [11] implementation of the BLAST algorithm [12]. Under standard procedures, sequences would also be compared with the GENBANK/EMBL expressed sequence tags (EST) databases and matching I.M.A.G.E. ESTs obtained from the Human Genome Mapping (HGMP) Resource Centre, Cambridge [13]. The I.M.A.G.E. clones are then fully sequenced in both directions and the resulting sequence re-checked against the databases to confirm their identity. The expression profiles of transcripts represented by isolated clones were confirmed by standard northern procedures [13].
Acknowledgments
Acknowledgements
This work was supported by a BBSRC programme grant and a MAFF commission.
References
- Noback CR, Strominger NL, Demarest RJ. The Human Nervous System. Philadelphia: Williams and Wilkins, 1995.
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kagiyama R. A mammalian helix-loop-helix structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Miele G, MacRae L, McBride D, Manson J, Clinton M. Elimination of false positives generated through PCR-reamplification of differential display cDNA. Biotechniques. 1998;25:138–144. doi: 10.2144/98251rr02. [DOI] [PubMed] [Google Scholar]
- Lee IY, Westaway D, Smit AFA, Wang K, Seto J, Chen L, Acharya C, Ankener M, Baskin D, Cooper C, et al. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res. 1998;8:1022–1037. doi: 10.1101/gr.8.10.1022. [DOI] [PubMed] [Google Scholar]
- Giguere V, Isobe K-I, Grosveld F. Structure of the murine Thy-1 gene. EMBO J. 1985;4:2017–2024. doi: 10.1002/j.1460-2075.1985.tb03886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocynate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch J, Sambrook EF. Molecular Cloning: A Laboratory Manual. . New York: Cold Spring Harbor Laboratory Press, 1982.
- Miele G, Slee R, Manson J, Clinton M. A rapid protocol for the authentication of isolated differential display RT-PCR cDNAs. Prep Biochem Biotech. 1999;29:245–255. doi: 10.1080/10826069908544927. [DOI] [PubMed] [Google Scholar]
- Wisconsin Package Version 10.0 http://www.oxmol.co.uk/software/wisconsin/
- National Centre for Biotechnology Information http://www.ncbi.nlm.nih.gov/BLAST/
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- UK Human Genome Mapping Project Resource Centre http://www.hgmp.mrc.ac.uk/Biology/descriptions/image.html


