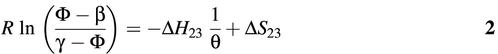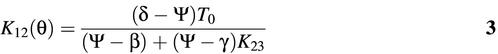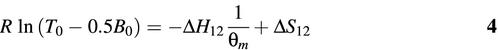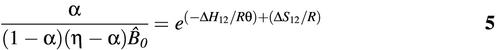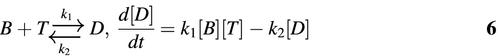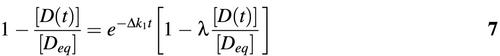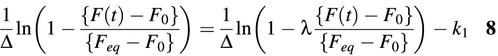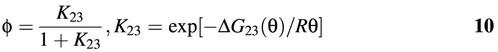Abstract
Molecular beacons are increasingly being used in many applications involving nucleic acid detection and quantification. The stem–loop structure of molecular beacons provides a competing reaction for probe–target hybridization that serves to increase probe specificity, which is particularly useful when single-base discrimination is desired. To fully realize the potential of molecular beacons, it is necessary to optimize their structure. Here we report a systematic study of the thermodynamic and kinetic parameters that describe the molecular beacon structure–function relationship. Both probe and stem lengths are shown to have a significant impact on the binding specificity and hybridization kinetic rates of molecular beacons. Specifically, molecular beacons with longer stem lengths have an improved ability to discriminate between targets over a broader range of temperatures. However, this is accompanied by a decrease in the rate of molecular beacon–target hybridization. Molecular beacons with longer probe lengths tend to have lower dissociation constants, increased kinetic rate constants, and decreased specificity. Molecular beacons with very short stems have a lower signal-to-background ratio than molecular beacons with longer stems. These features have significant implications for the design of molecular beacons for various applications.
INTRODUCTION
Molecular beacons are dual-labeled oligonucleotide probes that have a fluorescent dye (reporter) at one end and a fluorescence quencher (usually Dabcyl) at the opposite end (1). The probe is designed with a target-specific hybridization domain positioned centrally between short sequences that are self-complementary and are usually unrelated to the target sequence (2–4). In the absence of target, the self-complementary domains anneal to form a stem–loop hairpin structure in a unimolecular reaction that serves to bring the fluorescence reporter group into close proximity with the quencher group and results in quenching of the reporter (5,6). In this configuration, the molecular beacon is ‘dark’. Reporter-dye quenchers such as Dabcyl and Black Hole Quencher (BHQ) work based on both fluorescence resonance energy transfer (FRET) and the formation of an exiton complex between the fluorophore and the quencher (7). Thus, Dabcyl quenches the reporter dye only in the hairpin form partly due to its relatively short Förster radius. In the presence of target, the central loop domain will hybridize with the complementary target DNA or RNA in a bimolecular reaction, forcing the molecule to unfold; reporter and quencher are now physically separated and the fluorescence of the reporter dye will be restored upon excitation. In this configuration, the molecular beacon is ‘bright’.
The increase in fluorescence intensity upon hybridization for a given molecular beacon can vary widely, ranging from <10- to >200-fold (1). Several factors influence the signal-to-background (S:B) ratio for a specific molecular beacon, which render proper molecular beacon design critical. For example, hairpins with a stem that is too short can be relatively unstable and result in open (bright) molecular beacons even in the absence of target, leading to a low S:B ratio. Further, probes with a long stem can be too stable and might not hybridize effectively to target, resulting in low S:B ratios as well. The quality of synthesis and purification is also crucial to obtain an optimal S:B ratio, even for a well designed molecular beacon. With proper design and synthesis, high S:B ratio can be achieved which enables molecular beacons to function as probes with very high sensitivity.
Another significant advantage of molecular beacons is that they can recognize target sequences with a greater degree of specificity than linear probes (8). Molecular beacons are readily capable of discriminating between targets that differ by only a single nucleotide. The unimolecular hairpin reaction competes with bimolecular probe–target hybridization and serves to reduce the relative stability of undesired imperfect (mismatch) hybridization events (3,9,10).
The clear advantages of molecular beacons over linear oligonucleotide probes have led to their use in numerous applications ranging from quantitative PCR (11,12), to the study of protein–DNA interactions (13,14), to the visualization of RNA expression in living cells (15,16). However, for any application utilizing molecular beacons it is necessary to carefully design the probe so that their optimal performance is realized. For example, when distinguishing a point mutation [i.e. single nucleotide polymorphism (SNP)] high probe specificity is desired; however, when studying transient RNA expression in living cells in real time, it may be more important to have fast hybridization kinetics. Although some of the thermodynamic features of molecular beacons have been analyzed (8,17), to date only limited quantitative studies of binding kinetics of molecular beacon–target duplexes have been performed (18), and the structure–function relationship of molecular beacons has not been well established. Further, although predictive models have been developed for the hybridization thermodynamics of linear oligonucleotides (19), it is unclear if, and how well, such models apply to the binding stability of molecular beacon–target duplexes.
In this study, we examine molecular beacon design issues relating to target affinity, detection specificity and binding kinetic rates. Specifically, we have designed and synthesized molecular beacons with different probe and stem lengths, performed a systematic study of the thermodynamic and kinetic parameters that control the hybridization of these molecular beacons with perfectly complementary and mismatched targets, and compared experimental results with the prediction of existing models. We have also quantified the S:B ratios of the fluorescence emission from the molecular beacons with different structures. It is demonstrated that the performance of molecular beacons can be very sensitive to its structural features.
MATERIALS AND METHODS
Oligonucleotide synthesis
Oligonucleotide probes and targets were synthesized using standard phosphoramidite chemistry on an Applied Biosystems model 394 automated DNA synthesizer (Foster City, CA). For molecular beacons, a Cy3 fluorophore (Cy3 Amidite, Amersham Pharmacia Biotech, Piscataway, NJ) or a 6-carboxyfluorescein fluorophore (6-FAM Amidite, Applied Biosystems, Inc., Foster City, CA) was attached to the 5′ end of the oligonucleotide and a Dabcyl [4-(4′-dimethylaminophenylazo)benzoic acid] quencher was attached to the 3′ end (Dabcyl-CPG, Glen Research, Sterling, VA). Molecular beacons were purified using dual reverse phase (RP) plus ion-exchange (IE) high performance liquid chromatography (HPLC) on a Waters Model 600E HPLC system (Millipore Corp., Milford, MA). For RP-HPLC purification, oligonucleotides were loaded on a Hamilton PRP-1 column and eluted with a linear 5–50% acetonitrile gradient in 0.1 M triethyl-ammonium acetate pH 7.2 over 40 min. The oligonucleotides were additionally purified by IE-HPLC using a Source™ column (Amersham Pharmacia Biotech, Piscataway, NJ) and eluted with a linear 0–50% 1 M LiCl gradient in 0.1 M Tris pH 8.0 over 40 min. Target oligonucleotides were purified using polyacrylamide gel electrophoresis.
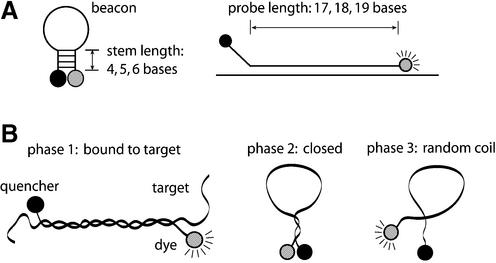
A series of molecular beacons were designed and synthesized, which are in antisense orientation with respect to exon 6 of the human GAPDH gene. Herein, probe length (Lp) is defined as the portion of the molecular beacon that is complementary to the target nucleic acid and stem length (Ls) is defined as the portion of the molecular beacon that is self-complementary. Conventional molecular beacons are designed such that the target-specific probe domain is located between short self-complementary stems that are independent of the target-specific domain. In this study, however, we consider molecular beacons with one stem complementary to the target sequence, as shown in Figure 1A. Specifically, the molecular beacons were designed so that the 5′-stem was entirely derived from the 5′ end of the target-specific probe domain (and therefore was capable of participation in both hairpin formation and target hybridization) while the 3′-stem was created solely to complement the 5′-stem sequence. Our rationale is that this new design, which we call ‘shared-stem’, should lead to an improved FRET efficiency: when a pair of molecular beacons are bound to the same target mRNA in close proximity so that the two dye molecules undergo FRET, hybridization of the fluorophore stem to the target may limit unwanted movement of the dye molecules, thus increasing FRET (20). A systematic comparative study of the structure– function relationships of molecular beacons with the ‘shared-stem’ design was performed, and these molecular beacons were found to possess higher melting temperatures when compared with molecular beacons with the conventional design (21). We intended to synthesize molecular beacons with probe lengths of Lp = 17, 18 and 19 bases and stem lengths of Ls = 0, 4, 5 and 6 bases. However, the ‘share-stem’ design strategy precluded certain stem/probe combinations and consequently, two molecular beacons could not be made, i.e. those with Lp = 19, Ls = 6 and with Lp = 18, Ls = 4.
Figure 1.
(A) The design and structural parameters of molecular beacons considered in the study. (B) Molecular beacons in solution can have three phases: bound to target, closed and random coil.
Probes having Ls = 0 are actually not molecular beacons but are dual-labeled linear probes. Although more commonly employed in assays that involve enzymatic degradation, linear dual-labeled oligos can function as fluorescence-quenched hybridization probes, where the reporter dye remains partially quenched (‘dark’) in the random-coil conformation but is librated from quenching (‘bright’) when the probe is extended in the double-stranded conformation (22). A series of three linear oligonucleotide probes were made using synthesis and purification methods identical to the molecular beacons. Within the range of probe lengths studied, the combination of Cy3 and Dabcyl routinely yields probes with a S:B ratio >5:1 (M.A.Behlke, unpublished data). Dual-labeled linear probes are functionally interchangeable with molecular beacons in the assay formats employed herein.
Five target oligonucleotides were synthesized, one wild-type and four with mismatches at assorted locations (Table 1). All oligonucleotides were synthesized at Integrated DNA Technologies, Inc. (Coralville, IA).
Table 1. The design of probe/target oligonucleotides.
| Name | Sequence (5′–3′) | Notes |
|---|---|---|
| MBa 17-4 | Cy3-GAGTCCTTCCACGATACactc-Dabcyl | Probe 17/stem 4 |
| MB 19-4 | Cy3-GAGTCCTTCCACGATACCActc-Dabcyl | Probe 19/stem 4 |
| MB 20-4 | Cy3-GAGTCCTTCCACGATACCAActc-Dabcyl | Probe 20/stem 4 |
| MB 17-5 | Cy3-GAGTCCTTCCACGATACgactc-Dabcyl | Probe 17/stem 5 |
| MB 18-5 | Cy3-GAGTCCTTCCACGATACCgactc-Dabcyl | Probe 18/stem 5 |
| MB 19-5 | Cy3-GAGTCCTTCCACGATACCAgactc-Dabcyl | Probe 19/stem 5 |
| MB 17-6 | Cy3-GAGTCCTTCCACGATACggactc-Dabcyl | Probe 17/stem 6 |
| MB 18-6 | Cy3-GAGTCCTTCCACGATACCggactc-Dabcyl | Probe 18/stem 6 |
| MB 19-7 | Cy3-GAGTCCTTCCACGATACCAggactc-Dabcyl | Probe 19/stem 7 |
| LPb 17-0 | Cy3-GAGTCCTTCCACGATAC-Dabcyl | Probe 17/stem 0 |
| LP 18-0 | Cy3-GAGTCCTTCCACGATACC-Dabcyl | Probe 18/stem 0 |
| LP 19-0 | Cy3-GAGTCCTTCCACGATACCA-Dabcyl | Probe 19/stem 0 |
| Targc WTd | ACTTTGGTATCGTGGAAGGACTCATGA | Perfect match |
| Targ A | ACTTTGGTATCGTGGAAGGAaTCATGA | Single mismatch |
| Targ B | ACTTTGGTATCGTaGAAGGACTCATGA | Single mismatch |
| Targ C | ACTTTGGTATCGTaGAAGGAaTCATGA | Double mismatch |
| Targ D | ACTTTGGTATCGTaaAAGGACTCATGA | Double mismatch |
aMB, molecular beacons. Lower case, residues added to create stem domains. Upper case, probe target hybridizing domains. Upper case bold, residues participating in both stem hairpin and target binding.
bLP, linear dual-labeled probes.
cTarg, targets. Underscore, 19-base sequence complementary to MB, LP target binding domains. Lower case bold, mismatch bases in targets.
dWT, wild-type.
Equilibrium analysis
Dissociation constants were obtained for all of the molecular beacons and molecular beacon–target pairs as described in Bonnet et al. (8). Briefly, the thermal profile for each molecular beacon and molecular beacon–target pair was obtained by mixing 200 nM of molecular beacons with six different concentrations of target ranging from 0 to 20 µM. Each assay was performed in a 50 µl solution containing 10 mM KCl, 5 mM MgCl2 and 10 mM Tris–HCl pH 7.5 and was excited with a 488 nm laser using a fluorometric thermal cycler (Applied Biosystems Prism 7700). The intensity of fluorescence emission was measured at temperatures ranging from 10 to 80°C. Specifically, the temperature was initially held at 80°C for 10 min, and then decreased by 1°C increments from 80 to 10°C with each step lasting 10 min. The temperature was then raised from 10°C back to 80°C to assure that no hysteresis occurred and equilibrium had been reached. Oligonucleotide probes without Dabcyl quenchers were also tested to correct for the intrinsic variance of fluorescence with temperature.
As shown in Figure 1B, molecular beacons in solution can have at least three distinct conformations: molecular beacon– target duplex (phase 1), stem–loop (phase 2) and random coil (phase 3). Following Bonnet et al. (8), the dissociation constant K23 characterizing the transition between stem–loop conformation and random coils is given by
where Φ is the fluorescence intensity as a function of temperature θ (i.e. the thermal profile), β is the fluorescence emitted by the molecular beacons when they are all in the stem–loop conformation, obtained from the fluorescence intensity measured at 10°C, and γ is the fluorescence emitted by the molecular beacons when they are all randomly coiled, obtained from the fluorescence intensity measured at 80°C. Further,
where R is the gas constant (1.9872 cal mol–1K–1), θ is the temperature in Kelvin, ΔH23 and ΔS23 are, respectively, the changes in enthalpy and entropy of the system.
The dissociation constant K12 corresponding to the transition between molecular beacons bound to target and those that are unbound and still in the stem–loop conformation can be determined from the fluorescence data describing the thermal profile of molecular beacons in the presence of target,
where Ψ is the thermal profile for molecular beacon–target pairs, β is the same as before, δ and γ are, respectively, the fluorescence of bound molecular beacons at 10 and 80°C. The changes in enthalpy and entropy due to this transition are given by
where the melting temperature θm of each molecular beacon– target mixture is defined as the temperature at which half of the molecular beacons are bound to target. With the assumption that T0 > 0.5B0, K12 = T0 – 0.5B0 at θ = θm, where T0 and B0 are the initial concentrations of targets and molecular beacons, respectively.
For all the molecular beacons and molecular beacon–target pairs, the dissociation constants K23 and K12, the corresponding changes in enthalpy and entropy of the system, and the melting temperature θm were quantified using the measured thermal profiles and the above equations. The errors calculated for the thermodynamic parameters signify a 95% confidence interval.
Probe specificity analysis
To quantify the effect of molecular beacon structure on binding specificity, the fraction of molecular beacons bound to target, α, was calculated using the enthalpy change ΔH12 and entropy change ΔS12 obtained from the thermal denaturation profiles for each molecular beacon–target duplex
where η = T0/B0, ![]() 0 = B0/c0, c0 is the unit concentration 1 M (23,24). The value of α was calculated for each molecular beacon–target pair as a function of temperature for samples containing B0 = 200 nM of molecular beacon and T0 = 400 nM of target.
0 = B0/c0, c0 is the unit concentration 1 M (23,24). The value of α was calculated for each molecular beacon–target pair as a function of temperature for samples containing B0 = 200 nM of molecular beacon and T0 = 400 nM of target.
Kinetic analysis
The rate constants that determine the hybridization kinetics for each molecular beacon–target pair were obtained by instantaneously mixing 250 nM of molecular beacons with 2.5 µM of each target using a stopped-flow accessory (Hi-Tech SFA-20) with a SFA-12 temp/trigger module. Fluorescence as a function of time was obtained by exciting the sample at 545 nm and recording the emission at 570 nm using a spectrofluorometer (SPEX fluorolog-2). All tests were conducted in the buffer described above at 37°C and repeated five times. To analyze the hybridization kinetics a second-order reaction was assumed (25),
where [B], [T] and [D] are the concentrations of molecular beacon, target and molecular beacon–target duplex, respectively, k1 is the on-rate constant and k2 the off-rate constant of molecular beacon–target hybridization. Solving the above differential equation gives
where ![]()
![]() and K12 = k2/k1 is the dissociation constant discussed above. Assuming that {F(t) – F0}/{Feq – F0}=[D(t)]/[Deq] where Feq is the fluorescence of the system as t → ∞ and F0 is the initial fluorescence intensity, the rate constants k1 and subsequently k2 = K12·k1 were obtained using two methods. One method used a non-linear least-square method to fit a curve to the normalized fluorescence data {F(t) – F0}/{Feq– F0} directly, using equation 7. A second method fitted the data to an alternative form of equation 7 with a straight line having a slope of k1.
and K12 = k2/k1 is the dissociation constant discussed above. Assuming that {F(t) – F0}/{Feq – F0}=[D(t)]/[Deq] where Feq is the fluorescence of the system as t → ∞ and F0 is the initial fluorescence intensity, the rate constants k1 and subsequently k2 = K12·k1 were obtained using two methods. One method used a non-linear least-square method to fit a curve to the normalized fluorescence data {F(t) – F0}/{Feq– F0} directly, using equation 7. A second method fitted the data to an alternative form of equation 7 with a straight line having a slope of k1.
The exact solution of the second-order kinetic equation given above is valid for probe–target hybridization with any initial probe and target concentrations. Therefore, although the stopped-flow assays were performed with a 10-fold excess of target over molecular beacons, which may warrant the use of a quasi first-order kinetic equation as an approximation, we chose to use equation 7 or equation 8 to obtain the on-rate constant k1 in order to ensure accuracy.
RESULTS
Molecular beacon thermodynamics
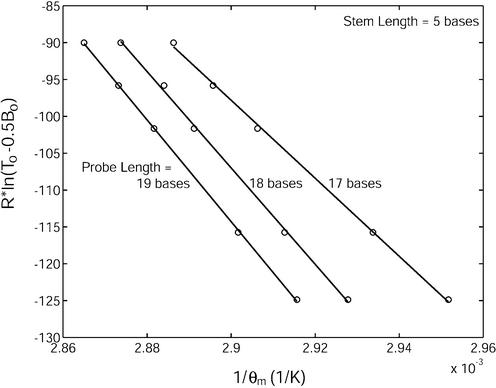
To characterize the thermodynamic properties of molecular beacons, the enthalpy change ΔH12 and entropy change ΔS12, describing the equilibrium state between unbound molecular beacons in the stem–loop conformation and molecular beacon–target duplexes were determined for molecular beacons with various stem–loop structures (see Table 1) using van’t Hoff plots displaying the reciprocal of the melting temperature 1/θm as determined by R·ln(T0 – 0.5B0) shown as the ordinate in Figure 2. Based on equation 4, the slope of the fitted straight line of each curve in Figure 2 represents the enthalpy change –ΔH12 and the y-intercept represents the entropy change ΔS12. To obtain more accurate values of ΔS12, θmRln(T0 – 0.5 B0) versus θm curves were generated and fitted by straight lines. According to equation 4,
Figure 2.
Determination of the changes in enthalpy (slope of the fitted line) and entropy (y-intercept) describing the transition between molecular beacons bound to wild-type target and free in the stem–loop conformation for molecular beacons with stem lengths of five bases and probe lengths of 17, 18 and 19 bases.
θmRln(T0 – 0.5B0) = – ΔH12 + ΔS12 · θm,9
thus, the slope of each fitted straight line gave the corresponding value of ΔS12. As illustrated by Figure 2, small changes in the probe length of a molecular beacon could have a significant impact on its equilibrium state. For example, increasing the probe length Lp from 17 to 18 bases for a molecular beacon with a five-base stem resulted in a more favorable hybridization as ΔH12 increased from 529 ± 27 kJ/mol to 654 ± 39 kJ/mol, and ΔS12 increased from 1437 ± 78 J/mol·K to 1789 ± 113 J/mol·K. When the probe length was further increased from 18 to 19 bases, ΔH12 increased to 690 ± 16 kJ/mol and ΔS12 increased to 1887 ± 46 J/mol·K. Quantitatively, these changes should be influenced by the specific nucleotide that is added when extending the probe length; both the base and the sequence context (nearest neighbor effects) will affect the magnitude of change in thermodynamic stability due to base addition. For the probe sequence considered here, when Lp was increased from 17 to 18 bases a cytosine was added, whereas an adenine was added in extending the probe length from 18 to 19 bases.
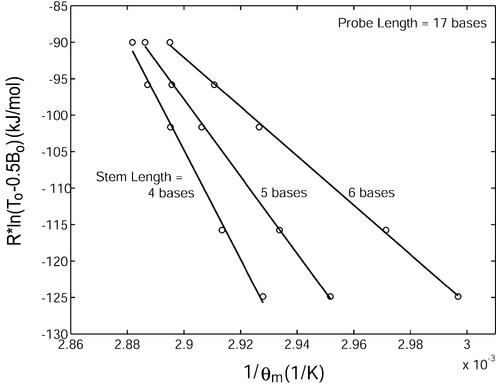
Single-base variation in the stem length of a molecular beacon was found to have a greater impact on its equilibrium state than probe length. As demonstrated in Figure 3, an increase in Ls from four to five bases for a molecular beacon with a probe length of 17 bases resulted in a decrease in ΔH12 from 749 ± 83 kJ/mol to 529 ± 27 kJ/mol and in ΔS12 from 2067 ± 240 J/mol·K to 1437 ± 78 J/mol·K. An increase in stem length from five to six bases further decreased ΔH12 to 338 ± 17 kJ/mol and ΔS12 to 887 ± 49 J/mol·K. This decrease in enthalpy and entropy with stem length reflects the enhanced stability of the molecular beacon in its hairpin conformation. The changes in enthalpy ΔH12 and entropy ΔS12 as indicated in Figures 2 and 3 due to hybridization of molecular beacons with wild-type targets are compiled in Table 2. Note that positive sign of ΔH12 and ΔS12 reflects the definition of the transition from phase 1 (bond to target) to phase 2 (hairpin) of molecular beacons.
Figure 3.
Determination of the changes in enthalpy (slope of the fitted line) and entropy (y-intercept) describing the transition between molecular beacons bound to wild-type target and free in the stem–loop conformation for molecular beacons with probe lengths of 17 bases and stem lengths of 4, 5 and 6 bases.
Table 2. Melting temperature, thermodynamic parameters and on-rate constant.
| Name | Predicted θm (°C) | Measured θm (°C) | Measuredb H12 (kJ/mol) | Measuredb S12 (J/mol K) | Measuredb k1 (M–1s–1) | ||||
|---|---|---|---|---|---|---|---|---|---|
| MB alone | Duplexa WT | Duplexa Mut B | MB alone | Duplexa WT | Duplexa Mut B | ||||
| MB 17-4 | 26.2 | 59.5 | 49.1 | 44.4 | 68.5 | 56.3 | 749 | 2067 | 4540 |
| MB 19-4 | 26.2 | 63.3 | 54.2 | 41.0 | 71.2 | 61.2 | 863 | 2383 | 7420 |
| MB 20-4 | 29.2 | 63.8 | 55.2 | 39.9 | 71.4 | 61.8 | 928 | 2570 | 5760 |
| MB 17-5 | 43.5 | 59.5 | 49.1 | 58.2 | 65.8 | 52.1 | 529 | 1437 | 416 |
| MB 18-5 | 42.6 | 61.3 | 51.7 | 56.7 | 68.6 | 56.7 | 654 | 1789 | 1050 |
| MB 19-5 | 45.2 | 62.9 | 53.5 | 57.4 | 70.0 | 57.6 | 690 | 1887 | 1470 |
| FamMBc 19-5 | 45.2 | 62.9 | 53.5 | 53.9 | 66.3 | 52.1 | 522 | 1413 | 1580 |
| MB 17-6 | 56.6 | 59.5 | 49.1 | 67.0 | 60.7 | 43.4 | 338 | 887 | 39 |
| MB 18-6 | 55.8 | 61.3 | 51.7 | 65.5 | 66.9 | 51.2 | 550 | 1494 | 342 |
| MB 19-7 | 59.7 | 62.9 | 53.5 | 67.0 | 65.5 | 51.8 | 413 | 1096 | 506 |
| LP 17-0 | - | 58.9 | 48.4 | - | 66.5 | 55.3 | 528 | 1429 | 10200 |
| LP 18-0 | - | 61.6 | 52.1 | - | 69.2 | 58.6 | 638 | 1740 | 12200 |
| LP 19-0 | - | 63.1 | 53.8 | - | 70.1 | 60.6 | 705 | 1930 | 14000 |
aDuplex, formation of probe–target duplex due to hybridization, with wild-type (WT) for 200 nM probe with 400 nM wild-type target; Mut B for 200 nM probe with 400 nM mutant target B (Targ B in Table 1).
bThe measured H12, S12 and k1 values were obtained using wild-type targets.
cFamMB 19-5, molecular beacon with the same design as MB 19-5 shown in Table 1 except that the Cy3 fluorophore at the 5′ end was replaced by a 6-FAM fluorophore.
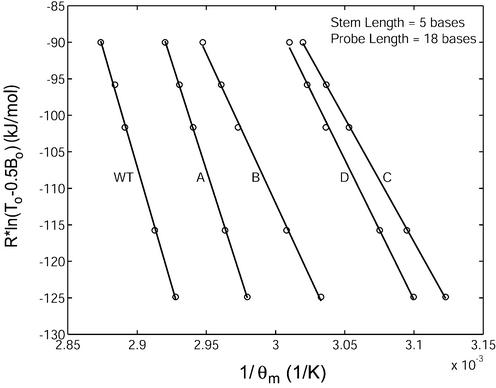
The thermodynamic parameters describing the equilibrium state between molecular beacons in the unbound, stem–loop conformation and those bound to targets containing mismatches were also calculated (see Table 1 for target sequences). As expected, the presence of single-base mismatches in the target resulted in a less favorable binding between molecular beacon and targets (Fig. 4). Moreover, it was found that a mismatch at the center of the molecular beacon binding region (target B) has a more profound effect on the equilibrium state than a mismatch near the end of the molecular beacon probe segment (target A). Specifically, compared with wild-type target, binding with target A resulted in decrease in ΔH12 from 654 ± 39 kJ/mol to 592 ± 15 kJ/mol and in ΔS12 from 1788 ± 113 J/mol·K to 1638 ± 44 J/mol·K. However, with target B, ΔH12 decreased to 409 ± 28 kJ/mol and ΔS12 decreased to 1115 ± 84 J/mol·K. The presence of two mismatches caused an even larger decrease in enthalpy and entropy, as demonstrated by the curves in Figure 4 describing the equilibrium state of the molecular beacon– target C duplex (ΔH12 = 339 ± 8 kJ/mol, ΔS12 = 934 ± 23 J/mol·K) and that of the molecular beacon–target D duplex (ΔH12 = 384 ± 26 kJ/mol, ΔS12 = 1065 ± 79 J/mol·K).
Figure 4.
Determination of the changes in enthalpy (slope of the fitted line) and entropy (y-intercept) describing the transition between molecular beacons bound to target and free in the stem–loop conformation for molecular beacons with a stem length of five bases and a probe length 18 bases in the presence of wild-type (WT) and mutant targets (target A – D).
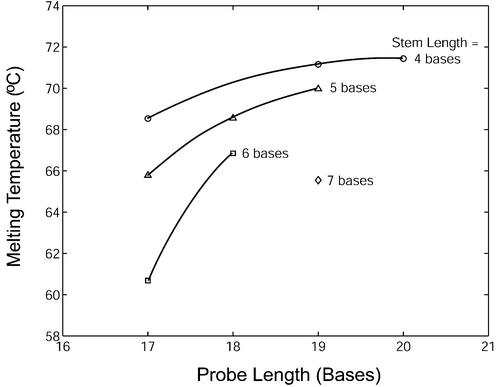
Melting temperature
To illustrate the effect of molecular beacon structure on its melting behavior, the melting temperatures θm (i.e. temperature at which α = 0.5) were determined for molecular beacons with various stem–loop structures under the condition B0 = 200 nM and T0 = 400 nM (Fig. 5). Melting temperature increased with probe length. Stem length also had a significant effect on the melting temperature of molecular beacon–target duplexes. For example, within the set of three molecular beacons having a probe length of 17 bases, when the stem length was increased from four to five bases the melting temperature was reduced by 2.7°C. Further increasing the stem length from five to six bases resulted in an additional 5.1°C reduction in duplex melting temperature. The relative effect of stem length on melting temperature is greater for shorter probes. The observed melting temperatures for probes hybridized with wild-type and mutant-B targets are summarized in Table 2.
Figure 5.
Melting temperatures for molecular beacons with various structures in the presence of wild-type target. The melting temperature was determined for samples containing 200 nM of molecular beacons and 400 nM of targets.
In Table 2 the experimental results were compared with melting temperatures θm predicted using nearest neighbor methods for unimolecular hybridization (hairpin) and bimolecular hybridization (duplex). First, the program ‘DNA mfold’, developed by Michael Zuker (26) (available at http://www.bioinfo.rpi.edu/applications/mfold/old/dna/) was used to predict θm of molecular beacon hairpin under conditions of 10 mM KCl and 5.0 mM MgCl2. In all cases the observed hairpin θm was significantly higher than the predicted θm, with the magnitude of discrepancy being greater for hairpins with shorter stems. The mfold algorithm does not take into account any stabilizing interactions that might occur between reporter dye and quencher, which may account for some of the observed differences. Marras et al. recently studied the effect of interactions between various reporter dyes and quenchers on θm in the setting of 20mer blunt-ended hybrids (7) and reported an increase of 3°C for the fluorescein–Dabcyl pair and 5°C for the Cy3–Dabcyl pair. The dye–quencher interaction might have an even greater impact on the stability of the short hairpin stems present in the molecular beacons studied here.
The program ‘HyTher’ developed by John SantaLucia (19,24,27–32) (available at http://ozone2.chem.wayne.edu/Hyther/hythermenu.html) was used to predict the θm of probe–target duplexes under the same conditions. The fluorescein-labeled molecular beacon (FamMB) had a θm that was close to the predicted value for duplexes with both the perfect-match (wild-type) and Mutant-B targets. The Cy3-labeled molecular beacons, however, had measured duplex stabilities that were generally higher than predicted. Members of the cyanine class of dyes are well known for their capacity to form stable complexes with DNA via intercalation or minor-groove binding (33–35). While Cy3 is not normally considered a DNA-binding dye, the high duplex stabilities of Cy3-labeled molecular beacons might reflect a thermodynamic contribution from interactions between the cyanine dye and the target nucleic acid.
Molecular beacon specificity
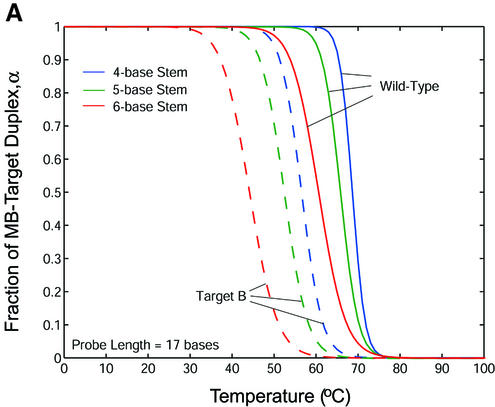
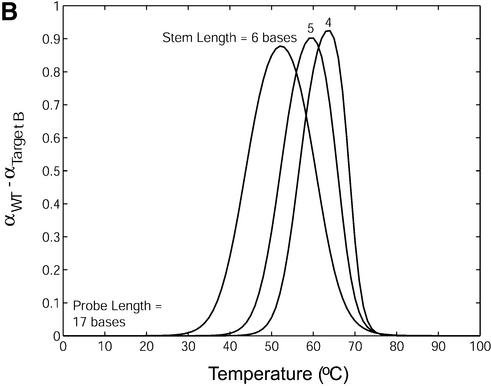
Melting curves that display the fraction of molecular beacons bound to target α as a function of temperature were created for each molecular beacon design and molecular beacon–target pair utilizing their respective thermodynamic parameters ΔH12 and ΔS12 obtained from the thermal equilibrium analysis. The melting temperature θm (°C) in these diagrams is taken as the temperature at which α = 0.5 (i.e. half of the molecular beacons are bound to target), consistent with the previous definition. In general, the specificity of a probe can be characterized by calculating the difference in melting temperature Δθm between the duplex of probe/wild-type target and that of probe/mutant targets. It was found that Δθm increased as the stem length of a molecular beacon increased. However, the transition between the bound and unbound states also became larger, as shown in Figure 6A. Therefore, a larger difference in melting temperature may not accurately reflect the increase in the fraction of molecular beacons bound to wild-type targets compared with that bound to mutant targets. To better evaluate the ability of molecular beacons to discriminate between wild-type and mutant targets, melting curves quantifying the fraction of molecular beacons bound to wild-type and mutant targets were overlaid and the difference in α was calculated. As displayed in Figure 6B, molecular beacons with a four-base stem indeed benefit from the sharper transition in their melting curve as demonstrated by their higher peak value. However, as the stem length increases so does the molecular beacon’s ability to discriminate targets over a broader range of temperatures.
Figure 6.
(A) Melting curves for molecular beacons hybridizing to wild-type target (solid line) and target B (dashed line) and (B) the difference in the fraction of molecular beacons bound to wild-type target and that to mutant target B. The molecular beacons have a probe length of 17 bases and stem lengths of 4, 5 and 6 bases.
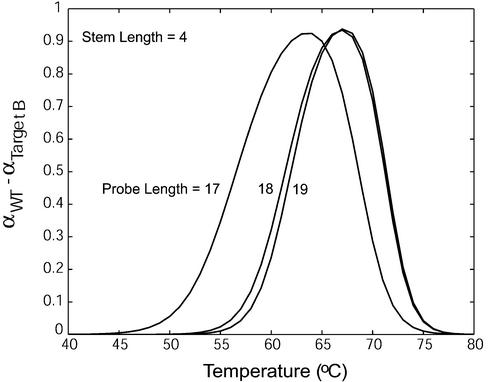
The effect of probe length on molecular beacon–target binding specificity is illustrated in Figure 7 in which the difference in the fraction of bound beacons Δα = αWT– αtarget B was plotted as a function of temperature for molecular beacons with Lp = 17, 18, 19 and Ls = 4. It was found that changing the probe length did not have a large influence on the maximum value of Δα. However, the range of temperatures over which a molecular beacon can discriminate between wild-type and mutant targets increased as probe length decreased. Therefore, the specificity of molecular beacons increases as probe length decreases, similar to linear probes.
Figure 7.
The difference in the fraction of molecular beacons bound to wild-type target and that to mutant target B for molecular beacons with a stem length of four bases and probe lengths of 17, 18 and 19 bases.
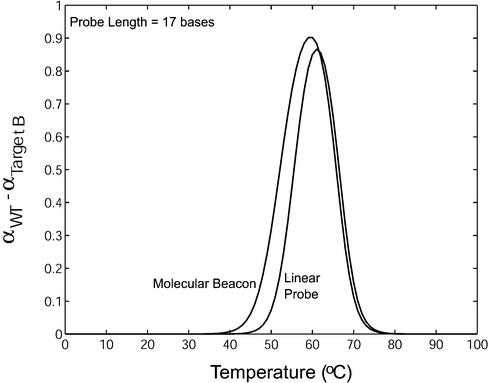
To evaluate the specificity of molecular beacons as compared with that of linear probes, Δα = αWT – αtarget B was plotted as function of temperature in Figure 8 for both linear and hairpin probes. It was found that for all the stem length and probe length combinations considered, molecular beacons had an improved ability to detect mismatches compared with linear probes. Specifically, molecular beacons not only demonstrated the ability to discriminate between wild-type and mutant targets over a broader range of temperatures but also maintained a larger differential between the fraction of molecular beacons bound to wild-type targets and that to mutant targets. Although in Figure 8 the comparison between molecular beacons (Ls = 4) and dual-labeled linear probes is given only for probe length of 17 bases, the general trend is the same for probe lengths of 18 and 19 bases.
Figure 8.
A comparison between the difference in the fraction of molecular beacons and that of linear probes bound to wild-type target and mutant target B.
It should be noted, however, that the behavior of the dual-labeled linear probes uncovered in this study might be influenced by the dye–quencher, dye–target or quencher– target interactions. In particular, the close proximity of the quencher to the target may have limited probe–target hybridization. In contrast, when molecular beacons hybridized to a target, the quencher was separated from the target by the free ‘stem’, as shown in Figure 1A.
Molecular beacon kinetics
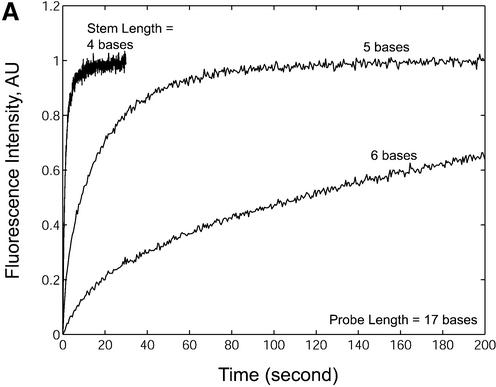
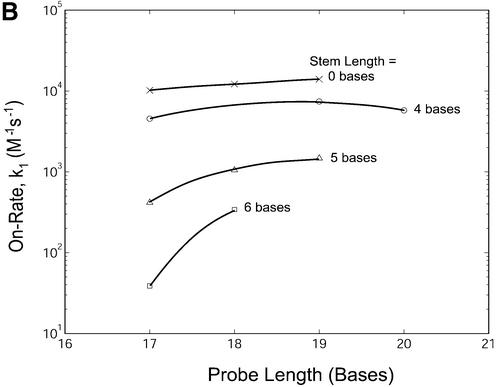
In addition to the studies of thermodynamic behavior of molecular beacon hairpins and duplexes at equilibrium as described above, the effects of probe length and stem length on hybridization kinetics were also examined. The normalized fluorescence restoration as a function of time due to probe– target hybridization for molecular beacons with probe lengths of 17 bases and stem lengths of four, five and six bases are shown in Figure 9A. The measured kinetic on-rate constants for all probes are summarized in Figure 9B and Table 2. It is clear that changes in stem length resulted in significant variations in hybridization on-rate constants. For example, molecular beacons with a four-base stem had an on-rate constant up to 100 times greater than molecular beacons with a six-base stem. Dual-labeled linear probes (stem = 0) hybridized to target only slightly faster (approximately twice) than molecular beacons with a four-base stem. Hybridization kinetics were studied at 37°C, which is close to the melting temperature for the four-base stem hairpins (40–44°C). It is possible that greater differences in hybridization rate constants between linear and four-base stem probes would be seen if the reaction kinetic rates were studied at a lower temperature.
Figure 9.
Hybridization kinetics of molecular beacons in the presence of wild-type targets. (A) The normalized restoration in fluorescence that accompanies target hybridization for molecular beacons with probe lengths of 17 bases and stem lengths of 4, 5 and 6 bases. (B) The rate of hybridization k1 (on-rate) for molecular beacons with various probe and stem lengths hybridized to their complementary targets.
Changing the probe length of a molecular beacon also influenced the on-rate constant of hybridization. In general, molecular beacons with longer probe domains bound to their targets more quickly than those with shorter probe domains. In fact, with a stem length of six bases, the addition of only a single base to the probe length of the molecular beacon resulted in a 10-fold increase of the on-rate constant.
Hybridization between molecular beacon and target involves both the opening of the hairpin structure and formation of probe–target duplex; in most cases they occur simultaneously, since during hybridization probe–target binding drives the opening of molecular beacons. However, insight may be gained by imagining the hybridization process as consisting of the following steps: (i) the target ‘wraps’ onto part of the probe domain of the molecular beacon which remains closed; (ii) as the number of ‘zipped’ base pairs becomes large, the hairpin opens; (iii) zippering of the remaining base pairs. Of the three steps, the rate-limiting steps are step (i) and step (ii), since it takes time for the target to conform to the loop geometry of the hairpin, and for the duplex to overcome the energy barrier induced by the stem. It is reasonable to assume that compared with steps (i) and (ii), step (iii) (zippering of a short duplex after the hairpin opens) is much faster. Indeed, our results indicated that, with linear probes, the formation of a short duplex (17–19 bp) is very fast (with an on-rate constant of 10 000–14 000 M–1s–1), as shown in Table 2.
For molecular beacons with short stems, the time required for step (i) is shorter compared with long-stem beacons: with a longer stem more base pairs must zip up to overcome the energy barrier encountered in unzipping the stem and this is the reason why increasing the stem length at constant probe length will give rise to slower binding kinetics as indicated by the results shown in Figure 9B. For sufficiently long stems, the binding kinetics may become dominated by the rate constant of step (i), and the number of the ‘first forming’ base pairs in step (i) could become close to the number of bases in the loop. In fact, for a molecular beacon with a six-base stem and 17-base probe, the loop length is just 11 bases and the hybridization between the loop and target might be barely enough to cause the hairpin to open. This may also be the reason why, for long-stem/short-probe molecular beacons, the kinetic on-rate constant appears to depend on the probe length.
It should be noted that the stopped-flow experiments only measured the rate constants for steps (i) and (ii) because during step (iii), the change in fluorescence is small: once a molecular beacon is open, the direct quenching disappears and the FRET-induced quenching is minimal. However, the two-state model used in this study as well as the results given in Figure 9B and Table 2 for the apparent on-rate constant are still valid, since step (iii) is much faster compared with steps (i) and (ii) as discussed above.
The choice of fluorophore used in a molecular beacon does not seem to affect the rate of hybridization. For example, a molecular beacon with a fluorescein dye molecule at the 5′ end hybridized at a similar rate to an identical molecular beacon with a Cy3 dye molecule at the 5′ end, as shown in Table 2.
In general, the on-rate constant k1 of beacon–target hybridization is buffer sensitive, as indicated by Kuhn et al. (18). Thus, it is likely that the values of k1 given in Figure 9B, which are consistent with those obtained by Kuhn et al. (18), may reflect the specific ionic conditions of the buffer used in the present study. However, it is believed that the general trend revealed in Figure 9B is valid as far as the effect of molecular beacon structure is concerned.
Background fluorescence
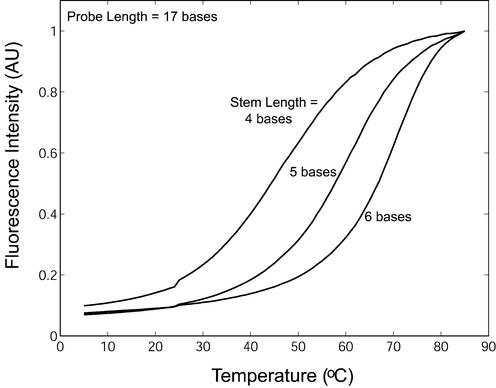
Our results show that molecular beacons with a four-base stem have a large on-rate constant and can still recognize their targets with a higher specificity than linear probes. However, these beacons may have higher background fluorescence at 37°C and could become unsuitable for in vivo applications. Figure 10 shows the normalized background fluorescence emission as a function of temperature for molecular beacons with stem lengths of four, five and six bases and a probe length of 17 bases in the absence of target. It is clear that a molecular beacon with a short stem has background fluorescence higher than that from molecular beacons with longer stems. For example, at 37°C, the background fluorescence of molecular beacons with a four-base stem is about four times greater than molecular beacons with five- or six-base stems, consistent with the prediction that compared with beacons with longer stems, a significantly larger fraction of molecular beacons with a four-base stem will be in an open random coil conformation (bright) as opposed to the closed hairpin structure (dark) at this temperature. In contrast, for molecular beacons with a fixed stem length of four bases, it was found that as the probe length increased from 17 to 20 bases there was only a small increase in the level of background fluorescence (data not shown), which is consistent with that reported by Goddard et al. (36).
Figure 10.
Normalized background fluorescence as a function of temperature for molecular beacons with a probe length of 17 bases and stem lengths of 4, 5 and 6 bases. Similar results were obtained for molecular beacons with probe lengths of 18 and 19 bases (data not shown).
The fraction, φ, of molecular beacons that may become open at temperature θ in the absence of target can be predicted by
where the free energy change ΔG23 resulted from the melting of the stem as well as the dye–quencher interactions and/or their interactions with the oligonucleotide. If these interactions are neglected, ΔG23 = ΔH23 – θΔS23 can be obtained using the mfold program (http://www.bioinfo.rpi.edu/ applications/mfold/old/dna/). For example, for the specific design of molecular beacon with a 17-base probe length and a five-base stem length as shown in Table 1, the program gives ΔH23 = 34 kcal/mol, ΔS23 = 107.4 cal/mol·K. The predicted values for φ, however, are higher than the experimental results shown in Figure 10 over a wide range of temperatures, presumably due to the omission of the dye–quencher and dye/quencher–oligonucleotide interactions. This is consistent with the lower melting temperature predicted using mfold. Note that the normalized fluorescence intensity Φ(θ) given in Figure 10 is related to the fraction of opened molecular beacons φ by
where β ≈ 0.07 is the normalized fluorescence intensity at 5°C.
DISCUSSION
It has long been recognized that, compared with linear oligonucleotide probes, stem–loop hairpin probes can have better specificity in gene detection (37). Molecular beacons are not only stem–loop hairpin probes but also have the ability to ‘switch on’ fluorescence upon binding to a complementary target, and are therefore becoming a powerful tool for many applications (38–43). However, as with any biochemical sensor/transducer system, it is necessary to understand the structure–function relationships of molecular beacons to fully exploit their potential. In many applications, the choices of the sequence of a probe are limited by target-specific considerations, such as the sequence context surrounding a SNP of interest. Probe features that can be adjusted independent of target choice include probe length and stem length. To directly test these aspects of probe design, we systematically studied the relative effects of varying probe and stem lengths on target affinity, mismatch discrimination and hybridization kinetics.
Insight into the structure–function relationship of molecular beacons can be gained by considering the free energy differences between unbound molecular beacons and molecular beacon–target duplexes. The formation of the stem– loop structure between self-complementary sequences at the ends of a molecular beacon is driven by a favorable free energy difference ΔGs that depends on the stem length Ls, stem sequence, ionic conditions, and the temperature θ (8,44). A longer Ls leads to a larger ΔGs and results in a more stable stem–loop structure against thermal fluctuations. Likewise, the free energy difference ΔGp due to the binding of the probe to its complementary target depends on the probe length, probe sequence and temperature. Since ΔGp is larger than ΔGs for a typical molecular beacon design, the stem–loop opens upon molecular beacon–target binding. Competition between ΔGp and ΔGs largely determines the conformational state of a molecular beacon at a given temperature with a given ionic environment and thus dictates the stability, specificity and hybridization kinetic rates. Although these general trends can be understood qualitatively from an energy point of view, it is still necessary to quantify the structure–function relationship of molecular beacons.
Here we have performed a systematic study of molecular beacons with Lp = 17, 18, 19 and 20 bases and Ls = 4, 5 and 6 bases and show how these structural differences affect function. It was found that molecular beacons with longer (more stable) stem lengths and shorter probe lengths have an improved ability to discriminate between targets over a wider range of temperatures; however, this comes at the cost of reduced molecular beacon–target duplex stability and a decreased rate of hybridization. Molecular beacons with longer stems also generated melting curves with a broader transition between unbound molecular beacons in the stem– loop conformation and molecular beacon–target duplexes. Consequently, there was a reduction in the optimal ratio of molecular beacons bound to wild-type targets to those bound to mutant targets. These findings can be explained by the effect of molecular beacon structure on ΔGp and ΔGs. As the probe length is decreased (lower ΔGp) or as the stem length is increased (higher ΔGs) the difference between ΔGp and ΔGs becomes smaller and the preference of target hybridization becomes less favorable. Any further reduction in this free energy difference, possibly due to a point mutation in the target, will subsequently have an amplified effect on the binding of molecular beacons to these targets.
Increasing ΔGs by lengthening the stem of a molecular beacon also lowered the rate of hybridization due to the larger energy barrier that must be overcome for target binding to occur. It is interesting to note, however, that even the kinetic rate constants for the dual-labeled linear probes were slightly lower than previously reported for unmodified oligonucleotides (18,45,46). Although it is not clear what caused the lower rates of hybridization it could be a consequence of the experimental set-up and the buffer conditions. Another possibility is that there was some interaction between the fluorophore and the quencher, or between the end-labels and the single-stranded target that is not present with unmodified oligonucleotides (47).
In determining the binding specificity of a molecular beacon–target duplex, we used melting curves to establish the difference in the fraction of molecular beacon bound to wild-type target and that to mutant target. Therefore, the average width of each curve shown in Figures 6B, 7 and 8 is an indication of the range of temperatures at which the molecular beacon can differentiate between wild-type and mutant targets and the height reflects how well the molecular beacon can differentiate between targets. The melting curves, however, are dependent on the initial concentrations of molecular beacons and targets. This implies that in order to realize the maximum specificity in living cells the concentration of molecular beacons must be tailored such that the maximum differential between the fraction of molecular beacon bound to wild-type targets and that to mutant targets occurs at physiological temperature. This is very critical in applications in which detection of point mutations of the target is desired.
Using van’t Hoff plots, the changes in enthalpy ΔH12 and entropy ΔS12 associated with beacon/target binding were obtained for molecular beacons with different probe and stem lengths hybridized to wild-type and mutant targets. In general, the measured changes in ΔH12 and ΔS12 owing to structural variations and probe/target mismatches are in good agreement with those reported in the literature (19,24,27–31). For example, for a molecular beacon with a five-base stem and an 18-base probe hybridized to mutant target B, the changes of ΔH12 and ΔS12 due to a single-base mismatch (G·A) were, respectively, 58.8 kcal/mol and 161 cal/mol K (entropy unit, eu), while the reported values are in the range of 50.5– 75.2 kcal/mol for ΔH12 and 138.4–215.0 eu for ΔS12 (19). Further, for a molecular beacon with a five-base stem, increasing the probe length Lp from 18 to 19 bases resulted in a change in ΔH12 by 8.64 kcal/mol and in ΔS12 by 23.5 eu, respectively, which are comparable to the average values of the unified NN parameters ΔH0 = –8.36 kcal/mol and ΔS0 = –22.4 eu (27). Unexpectedly, however, when the probe length Lp of the molecular beacon changed from 17 to 18 bases, both ΔH12 and ΔS12 changed about 3.7 times the average values of ΔH0 and ΔS0. One possible reason is that, due to the competition between ΔGp and ΔGs, the effect of increasing Lp from 17 to 18 bases is ‘amplified’, i.e. the dissociation constant K12 at θ = θm is more sensitive to certain changes in probe length. Additional experiments and analysis need to be carried out to study the underlying reasons of this intriguing difference.
The thermodynamic studies present here were conducted in a buffer containing 10 mM monovalent cation and 5 mM divalent cation. For in vivo applications of molecular beacons, the ionic milieu will change, which should affect both melting temperature and kinetic rates. Preliminary studies indicated that kinetic on-rate constants in phosphate buffered saline are significantly slower than the on-rate constants reported here using a buffer that has a relatively high Mg++ ion concentration (data not shown).
In calculating the rate constants for the formation of molecular beacon–target duplexes, we assumed that hairpin unfolding and probe–target binding are linked events and therefore can be analyzed using a two-state approach (25). However, this assumption might not apply to all scenarios and a multi-step reaction might need to be considered.
The basic features and structure–function relationships revealed in this study have important implications for the design of molecular beacons. This study has demonstrated clearly that in designing molecular beacons for a specific application, both stem and probe lengths must be carefully chosen. For example, when high probe specificity is required, as in the case of detecting point mutations or polymorphisms, molecular beacons will offer improved discrimination when relatively more stable (longer) stems are matched with shorter probe domains. Conversely, when studying RNA expression in living cells in real-time, it may be more important to have fast hybridization kinetics. In this case, molecular beacons with less stable (shorter) stems and longer probe domains would be preferred. Mismatch discrimination is further improved if the mutation is positioned centrally within the probe domain. Finally, since molecular beacons with a four-base stem length may have high background fluorescence, the use of longer stems would be preferred in most applications. In summary, the quantitative studies of structure–function relationships of molecular beacons can provide guidance to the design of molecular beacons to achieve an optimal balance among specificity, S:B ratio and kinetic rates desirable for a specific application.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. M. Nickerson for his help in conducting the stopped-flow experiments, A. Laikhter and E. Edgar for assistance with oligonucleotide synthesis, and R. Owczarzy and L. Huang for helpful discussions. This work was supported in part by NSF grant BSE-0222211, a seed grant from the Wallace H. Coulter Foundation, and a Cutting Edge Research Award from Georgia Institute of Technology.
REFERENCES
- 1.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 3.Kostrikis L.G., Tyagi,S., Mhlanga,M.M., Ho,D.D. and Kramer,F.R. (1998) Spectral genotyping of human alleles. Science, 279, 1228–1229. [DOI] [PubMed] [Google Scholar]
- 4.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 5.Mullah B. and Livak,K. (1999) Efficient automated synthesis of molecular beacons. Nucl. Nucl., 18, 1311–1312. [Google Scholar]
- 6.Lakowicz J.R. (1999) Principles of Fluorescence Spectroscopy, 2nd edn, Kluwer Academic/Plenum Publishers, New York, NY.
- 7.Marras S.A., Kramer,F.R. and Tyagi,S. (2002) Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res., 30, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet G., Tyagi,S., Libchaber,A. and Kramer,F.R. (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA, 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 10.Dubertret B., Calame,M. and Libchaber,A. (2001) Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol., 19, 365–370. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B. and Kinzler,K.W. (1999) Digital PCR. Proc. Natl Acad. Sci. USA, 96, 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W., Martinez,G. and Mulchandani,A. (2000) Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal. Biochem., 280, 166–172. [DOI] [PubMed] [Google Scholar]
- 13.Fang X., Li,J.J. and Tan,W. (2000) Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem., 72, 3280–3285. [DOI] [PubMed] [Google Scholar]
- 14.Li J.J., Geyer,R. and Tan,W. (2000) Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res., 28, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuo T. (1998) In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta, 1379, 178–184. [DOI] [PubMed] [Google Scholar]
- 17.Bonnet G., Krichevsky,O. and Libchaber,A. (1998) Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl Acad. Sci. USA, 95, 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn H., Demidov,V.V., Coull,J.M., Fiandaca,M.J., Gildea,B.D. and Frank-Kamenetskii,M.D. (2002) Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc., 124, 1097–1103. [DOI] [PubMed] [Google Scholar]
- 19.SantaLucia J. Jr (1998) A unified view of polymer, dumbbell and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsourkas A. and Bao,G. (2001) Detecting mRNA transcripts using FRET-enhanced molecular beacons. In Proceedings of the 2001 Bioengineering Conference, ASME BED-Vol. 50, pp. 169–170.
- 21.Tsourkas A., Behlke,M. and Bao,G. (2002) Structure–function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res., 30, 4208–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak K.J., Flood,S.J.A., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 23.Ratilainen T., Holmen,A., Tuite,E., Haaima,G., Christensen,L., Nielsen,P.E. and Norden,B. (1998) Hybridization of peptide nucleic acid. Biochemistry, 37, 12331–12342. [DOI] [PubMed] [Google Scholar]
- 24.Xia T., SantaLucia,J.,Jr, Burkard,M.E., Kierzek,R., Schroeder,S.J., Jiao,X., Cox,C. and Turner,D.H. (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry, 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- 25.Lauffenburger D.A. and Linderman,J.J. (1993) Receptors: Models for Binding, Trafficking and Signaling. Oxford University Press, New York, NY.
- 26.Zuker M. (2000) Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol., 10, 303–310. [DOI] [PubMed] [Google Scholar]
- 27.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 28.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 29.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 31.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 32.Bommarito S., Peyret,N. and SantaLucia,J.,Jr (2000) Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res., 28, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikheikin A.L., Zhuze,A.L. and Zasedatelev,A.S. (2000) Binding of symmetrical cyanine dyes into the DNA minor groove. J. Biomol. Struct. Dyn., 18, 59–72. [DOI] [PubMed] [Google Scholar]
- 34.Yarmoluk S.M., Lukashov,S.S., Ogul’Chansky,T.Y., Losytskyy,M.Y. and Kornyushyna,O.S. (2000) Interaction of cyanine dyes with nucleic acids. XXI. Arguments for half-intercalation model of interaction. Biopolymers, 62, 219–227. [DOI] [PubMed] [Google Scholar]
- 35.Thompson M. and Woodbury,N.W. (2001) Thermodynamics of specific and nonspecific DNA binding by two DNA-binding domains conjugated to fluorescent probes. Biophys. J., 81, 1793–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goddard N.L., Bonnet,G., Krichevsky,O. and Libchaber,A. (2000) Sequence dependent rigidity of single stranded DNA. Phys. Rev. Lett., 85, 2400–2403. [DOI] [PubMed] [Google Scholar]
- 37.Roberts R.W. and Crothers,D.M. (1991) Specificity and stringency in DNA triplex formation. Proc. Natl Acad. Sci. USA, 88, 9397–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leone G., van Schijndel,H., van Gemen,B., Kramer,F.R. and Schoen,C.D. (1998) Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res., 26, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vet J.A., Majithia,A.R., Marras,S.A., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao G., Chicas,A., Olivier,M., Taya,Y., Tyagi,S., Kramer,F.R. and Bargonetti,J. (2000) A DNA damage signal is required for p53 to activate gadd45. Cancer Res., 60, 1711–1719. [PubMed] [Google Scholar]
- 41.Tan W., Fang,X., Li,J. and Liu,X. (2000) Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chemistry, 6, 1107–1111. [DOI] [PubMed] [Google Scholar]
- 42.Steemers F.J., Ferguson,J.A. and Walt,D.R. (2000) Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat. Biotechnol., 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 43.Tung C.H., Mahmood,U., Bredow,S. and Weissleder,R. (2000) In vivo imaging of proteolytic enzyme activity using a novel molecular reporter. Cancer Res., 60, 4953–4958. [PubMed] [Google Scholar]
- 44.Ying L., Wallace,M.I. and Klenerman,D. (2001) Two-state model of conformational fluctuation in a DNA hairpin-loop. Chem. Phys. Lett., 334, 145–150. [Google Scholar]
- 45.Kambhampati D., Nielsen,P.E. and Knoll,W. (2001) Investigating the kinetics of DNA–DNA and PNA–DNA interactions using surface plasmon resonance-enhanced fluorescence spectroscopy. Biosens. Bioelectron., 16, 1109–1118. [DOI] [PubMed] [Google Scholar]
- 46.Mazumder A., Majlessi,M. and Becker,M. (1998) A high throughput method to investigate oligodeoxyribonucleotide hybridization kinetics and thermodynamics. Nucleic Acids Res., 26, 1996–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkhurst K.M. and Parkhurst,L.J. (1995) Kinetic studies by fluorescence resonance energy transfer employing a double-labeled oligonucleotide: hybridization to the oligonucleotide complement and to single-stranded DNA. Biochemistry, 34, 285–292. [DOI] [PubMed] [Google Scholar]