Abstract
Familial essential tremor (ET), the most common inherited movement disorder, is generally transmitted as an autosomal dominant trait. A genome-wide scan for ET revealed one major locus on chromosome 3q13. Here, we report that the Ser9Gly variant in the dopamine D3 receptor gene (DRD3), localized on 3q13.3, is associated and cosegregates with familial ET in 23 out of 30 French families. Sequencing revealed no other nonsynonymous variants in the DRD3-coding sequence and in the first 871 bp of the 5’ flanking region. Moreover, Gly-9 homozygous patients presented with more severe and/or earlier onset forms of the disease than heterozygotes. A replication study comparing 276 patients with ET and 184 normal controls confirmed the association of the Gly-9 variant with risk and age-at-onset of ET. In human embryonic kidney (HEK) 293-transfected cells, the Gly-9 variant did not differ from the Ser-9 variant with respect to glycosylation and to anterograde and retrograde trafficking, but dopamine had an affinity that was four to five times higher. With the Gly-9 variant, the dopamine-mediated cAMP response was increased, and the mitogen-associated protein kinase (MAPK) signal was prolonged, as compared with the Ser-9 variant. The gain-of-function produced by the Gly-9 variant may explain why drugs active against tremor in Parkinson's disease (PD) are usually not effective in the treatment of ET and suggests that DRD3 partial agonists or antagonists should be considered as novel therapeutic options for patients with ET.
Keywords: dopamine receptor, gain-of-function, gene dosage, movement disorder, polymorphism
Essential tremor (ET) is a slowly progressive disorder most frequently characterized by an action (kinetic or postural) tremor of the arms and hands, and less frequently of the head and voice; legs are rarely affected (1). Severe cases cause substantial disability, especially in the elderly, and, if medications such as propranolol, primidone, and topiramate fail, patients may require surgical treatment with deep brain stimulation (2, 3).
The etiology and pathogenesis of ET are unknown, but clinical studies suggest a dysfunction of the cerebello-thalamo-cortical pathway (4). ET is a genetically transmitted neurological disorder, in which the frequency of positive family history ranges from 17.4 to 100%, according to various studies (1, 5). ET prevalence estimates worldwide range from 0.3% to 4.0% in the general population (1). There is a definite increase in prevalence of ET with age; prevalence has thus been reported as high as 22% in elderly populations (1). Previous genome-wide scans have identified two ET loci: a first study of 16 Icelandic families (6) found only one ET locus (ETM1, also called FET1) on chromosome 3q13; a second locus (ETM2) on chromosome 2p22–25 was subsequently identified in a large family of Czech descent (7). Although an A265G substitution in the HS1-binding protein 3 gene (HS1-BP3) was reported in ET families (8), a study of a larger series of affected families and controls found no evidence of cosegregation of the A265G polymorphism with ET (9).
The Gly-9 variant of dopamine D3 receptor (DRD3) (10, 11) has been associated with tardive dyskinesia, a frequent motor side effect of antipsychotic drugs (12). DRD3 expression is reduced in Parkinson's disease (PD) (13), and a study in a monkey model of PD indicates that DRD3 hyperfunction is responsible for dyskinesia, a motor complication of PD treatment (14). Largest DRD3 densities occur in the basal ganglia, but cerebellum also expresses DRD3 at a lower density (15). In the rat cerebellum, DRD3 is present in Purkinje cells (16), and cerebellum has been implicated in the pathogenesis of ET (17). The DRD3 gene is located on chromosome 3q13.3 between markers D3S1278 and D3S1267, which gave highest logarithm of odds (LOD) scores at the ETM1 locus (6). Therefore, the implication of the DRD3 in several movement disorders, brain DRD3 distribution, and its chromosomal localization made the DRD3 gene as a candidate for familial ET.
Here, we examined association and linkage of DRD3 in a set of 30 families with familial ET and subsequently tested for association in a large independent case-control study. Then, we investigated the functional consequences of the Ser9Gly variant on receptor signaling, glycosylation, and trafficking in a heterologous expression system. Our genetic investigations found linkage and association of the DRD3 Gly-9 variant with ET, and functional studies demonstrated an increased affinity for dopamine of the Gly-9 variant, which elicited increased response to dopaminergic stimulation. Thus, the Gly-9 variant may confer susceptibility to ET.
Results
The Gly-9 Variant Is Associated and Cosegregates with ET in a Set of 30 Families.
In a first exploratory approach to assess the role of the DRD3 Ser9Gly variant in ET, we performed a case-control association study with probands of 30 French families with ET (Fig. 1). Patients were matched for gender and age, with 50 control individuals originating from the same geographical area. Genotype counts in patients were significantly different from the controls' genotypes (χ2 = 10.55, df = 2, P = 0.005; Table 1). The Gly-9 allele appeared more frequent in patients (46.7%) than in controls (26.0%; Pearson's χ2 = 7.16, P = 0.007).
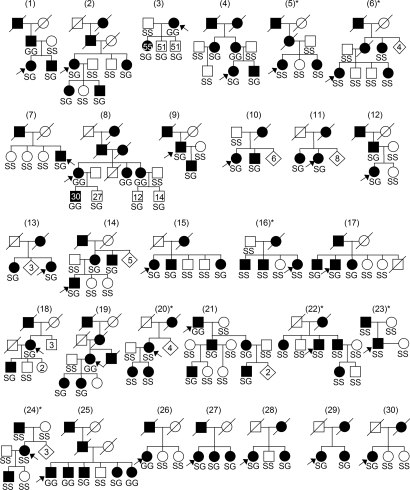
Fig. 1.
Pedigrees of affected families used in this study with the genotypes (S, Ser-9; G, Gly-9) of individuals (arrows, probands). Age at assessment is depicted in members of families 3 and 8. Families in which Gly-9 allele did not cosegregate with ET are marked with an asterisk. [Reproduced with permission from Clinical Genetics, ref. 40 (Copyright 2006, Blackwell Publishing).]
Table 1.
Genotype counts and allele frequencies in the two samples
| Subjects | Genotype frequencies |
P value (χ2) | Allele frequencies |
P value (χ2) | |||
|---|---|---|---|---|---|---|---|
| AA (Ser9/Ser9) | AG (Ser9/Gly9) | GG (Gly9/Gly9) | A (Ser9) | G (Gly9) | |||
| French sample | |||||||
| ET patients | 0.233 (7) | 0.600 (18) | 0.167 (5) | 0.533 (32) | 0.467 (28) | ||
| Controls | 0.600 (30) | 0.280 (14) | 0.120 (6) | 0.005 (10.55) | 0.740 (74) | 0.260 (26) | 0.007 (7.16) |
| American sample | |||||||
| ET patients | 0.449 (124) | 0.424 (117) | 0.127 (35) | 0.661 (365) | 0.339 (187) | ||
| Controls | 0.506 (93) | 0.440 (81) | 0.054 (10) | 0.035 (6.732) | 0.726 (267) | 0.274 (101) | 0.039 (4.247) |
The number in parentheses in the genotype column is the number of individuals; the number in parentheses in the allele column is the number of chromosomes. P values show the comparison results between patients and normal controls.
Then, the affected and unaffected members of 30 families were genotyped for the DRD3 Ser9Gly variant. The Gly-9 allele cosegregated with ET in the 23 of 30 families (Fig. 1): in these families, all affected members were Gly-9 carriers. In families 3 and 8, unaffected Gly-9 carriers were younger at clinical assessment than affected members. This observation is in agreement with the increasing incidence of ET with age (1, 5). In the 7 remaining families (families 5, 6, 16, 20, 22, 23, and 24), all members were homozygous for the Ser-9 allele. In the latter families, ET may be linked to another locus, because it is known that the disorder is genetically heterogeneous (1).
Linkage in the presence of association was positively demonstrated by the use of the Transmission Disequilibrium Test (TDT) in families with both parents available and at least one heterozygous parent (families 2, 9, 12, 14, 19, 21) or the Sib-TDT (S-TDT) in other informative sibships (families 7, 8, 15, 17, 18, 25, 26, 28, 30) (18). TDT χ2 value was 7.364 (P = 0.0067), and S-TDT z’ score was 4.306 (P < 0.00001). Combined TDT-STDT z’ score was 4.951 (P < 0.000001), indicating a strong linkage disequilibrium between the Gly-9 variant and ET in these families. To compare our results with the previously published linkage studies, we also performed a parametric linkage analysis of the Ser9Gly variant in the 30 families, assuming a penetrance of 0.90, which was similar to previous reports and compatible with the observed transmission in our families. Analyses assuming a phenocopy rate ranging from 0.01 to 0.10 and a mutation frequency from 0.01 to 0.50 yielded logarithm of odds (LOD) scores ranging from 5.38 to 7.99 at θ = 0 (Table 2, which is published as supporting information on the PNAS web site).
No Other Candidate Variants in DRD3 Coding Sequence and Promoter Region.
We examined HapMap data (www.hapmap.org) to delineate the haplotype block of strong linkage disequilibrium containing the Ser9Gly variant. The latter was mapped to a block of ≈50 kb containing the three first DRD3 exons and introns, and ≈ 30 kb of noncoding sequence upstream from the DRD3 promoter region (Table 3, which is published as supporting information on the PNAS web site). We amplified and sequenced a total of 5,098 bp of the DRD3 gene in nine probands (n = 3 with each genotype) and found no nonsynonymous variants in the entire coding sequence, except the Ser9Gly, and no variants in the splicing junctions or in 871 bp of the promoter region (Table 4, which is published as supporting information on the PNAS web site).
Gly-9 Homozygotes Have More Severe ET Symptoms.
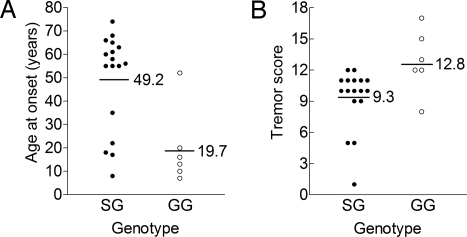
Among probands, age at onset (AAO) was significantly younger (two-tailed P = 0.006 by the Mann–Whitney U test) in Gly-9 homozygotes than in heterozygotes (Fig. 2A). Gly-9 homozygotes also had more severe tremor scores than heterozygotes. For instance, forearm tremor scores were higher (two-tailed P = 0.02 by the Mann–Whitney U test; see Fig. 2B). Tremor severity increases with age (1, 5), but an older age at assessment could not account for increased severity in Gly-9 homozygotes in our sample (57.2 ± 11.3 y and 64.5 ± 3.5 y in homozygotes and heterozygotes, respectively, P = 0.81). Therefore, these results suggest a gene dosage effect.
Fig. 2.
Gly-9 homozygotes were affected with more severe ET than heterozygotes. (A) Age at onset was lower in Gly-9 homozygotes (GG) than in heterozygotes (SG). (B) Forearm tremor score was higher in homozygotes (GG) than in heterozygotes (SG). [Modified from ref. 40.]
Association of the Ser9Gly Variant with Risk and AAO of ET in an Independent Case-Control Study.
To confirm the association of the Gly-9 variant with risk of ET, we genotyped the Ser9Gly variant in 276 American patients with ET and 184 American normal controls with no history of familial movement disorders. Genotype counts in patients were significantly different from the controls' genotypes (χ2 = 6.73, df = 2, P = 0.035; Table 1) Distribution of the genotypes in control group was in Hardy–Weinberg equilibrium. The Gly-9 allele was more frequent in patients than in controls (Pearson's χ2 = 4.25, P = 0.039; Table 1). Mean AAO of ET was 42.7 y in Ser-9 homozygotes, 39.2 y in heterozygotes, and 32.8 y in Gly-9 homozygotes. In addition, 91.4% (32 of 35) of Gly-9 homozygotes had onset before age 55, compared with 74.3% (87 of 117) of heterozygotes and 66.9% (83 of 124) of Ser-9 homozygotes. AAO was indeed significantly younger (two-tailed P = 0.02 by the Mann–Whitney U test) in Gly-9 homozygotes than in the other patients. As expected, when considering only patients with AAO <55 years, the association between ET and the Gly-9 variant appeared stronger (χ2 = 11.49, df = 2, P = 0.003), and the Gly-9 allele frequency increased to 37.4% in the patients' group (χ2 = 8.64, P = 0.003).
Glycosylation, Localization, and Trafficking of DRD3 Variants.
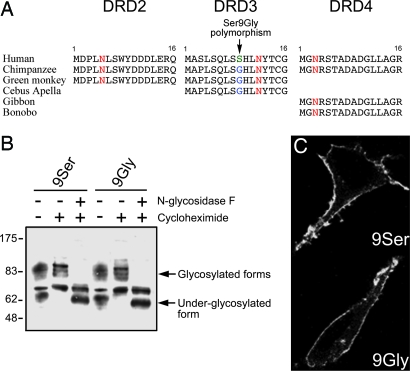
The Ser9Gly variation is located in the extracellular N terminus of the DRD3 (Fig. 3A), a region that is glycosylated and predicted to play a role in receptor conformation, maturation, and trafficking. We investigated the possibility that the Ser9Gly variation affected DRD3 glycosylation and trafficking, using yellow fluorescent protein (YFP)-tagged DRD3 variants transfected to human embryonic kidney (HEK) 293 cells (Fig. 3B). Treatment with cycloheximide, a protein synthesis inhibitor, was used to avoid nonmaturated forms of the receptor, either alone (to detect higher molecular weight glycosylated forms) or in combination with N-glycosidase F, a deglycosylating enzyme (to reveal lower molecular weight underglycosylated forms). There were no differences in the presence and the intensity of the glycosylated and underglycosylated forms of the two DRD3 variants, suggesting that the Ser9Gly variation did not alter the glycosylation process.
Fig. 3.
Glycosylation and trafficking of DRD3 variants. (A) The Ser9Gly variant is located in the extracellular N terminus of the DRD3, a region conserved among human and nonhuman primates. The highly conserved putative glycosylation site is shown in red, whereas Ser-9 and Gly-9 alleles are shown in green and blue, respectively. (B) Glycosylation status of each DRD3 variants was compared by Western blotting. HEK293 cells transiently transfected by either YFP-tagged variant were treated with cycloheximide for 3 h to stop synthesis of new DRD3. Membrane proteins were subjected to deglycosylation by N-glycosidase F enzyme before resolution in 10% SDS/PAGE and immunoblotting by using YFP antibody. Blot is representative of two independent experiments. (C) Subcellular localization of both YFP-tagged variants in transiently transfected HEK293 cells after treatment with cycloheximide for 3 h to reduce intracellular fluorescence and assess targeting to the cell surface.
In addition, both DRD3 variants displayed the same subcellular localization, mainly at the plasma membrane of transfected HEK293 cells (Fig. 3C). In striatal primary neurons, characterized by a marker of medium spiny neurons, DARPP32 (Fig. 5B, which is published as supporting information on the PNAS web site), both variants localized mostly in the soma and to a lesser extent in projections where they partially colocalized with a synaptic marker, synaptophysin (Fig. 5C). No apparent discrepant subcellular distribution between both DRD3 variants was observed.
To compare anterograde trafficking of DRD3 variants, we used a biochemical assay of receptor insertion at the plasma membrane (19). Cells were first blocked from biotinylation by using sulfo-NHS-acetate and then incubated at 37°C for the indicated times (Fig. 6, which is published as supporting information on the PNAS web site). Cells were then biotinylated with sulfo-NHS-LC-biotin to purify surface-labeled proteins with streptavidin-agarose. Biotinylated DRD3 newly inserted at the cell surface was similar for the Ser-9 and Gly-9 variants, with an apparent rate of 0.0783 ± 0.0076 and 0.0786 ± 0.0098 fold per hour, respectively (Fig. 6), suggesting that the Ser9Gly variant did not affect receptor anterograde trafficking.
To compare retrograde trafficking of DRD3 variants, we used a biochemical internalization assay based upon the use of a cleavable sulfo-NHS-SS-biotin (20). After endocytosis, receptors biotinylated at the plasma membrane became inaccessible to cleavage with a membrane-nonpermeant reducing agent (glutathione). Constitutive and dopamine-induced internalization was measured for a 90-min interval in the absence or the presence of 10 μM dopamine, respectively (Fig. 7, which is published as supporting information on the PNAS web site). DRD2 and DRD3 subtypes displayed similar constitutive retrograde trafficking (14 ± 5.5% vs. 11.3 ± 2.7%). In contrast, DRD2 undertook dopamine-induced internalization more prominently than DRD3 (54.6 ± 7.5% vs. 22 ± 4.3% after 90-min stimulation; Fig. 7B). Endogenous transferrin receptor internalization was not regulated by dopamine and was similar in cells transfected either by DRD2 or DRD3, thus confirming the specificity of the test. Then, we compared DRD3 variants retrograde trafficking (Fig. 7C). No significant discrepancies were observed between the Ser-9 and Gly-9 variant during constitutive (11.3 ± 2.7% vs. 10 ± 5%) and dopamine-induced (22 ± 4.3% vs. 23 ± 6.1%) trafficking. These results suggest that the Ser9Gly variant did not alter retrograde trafficking.
Dopamine Has a Higher Affinity and Efficacy at the Gly-9 Variant.
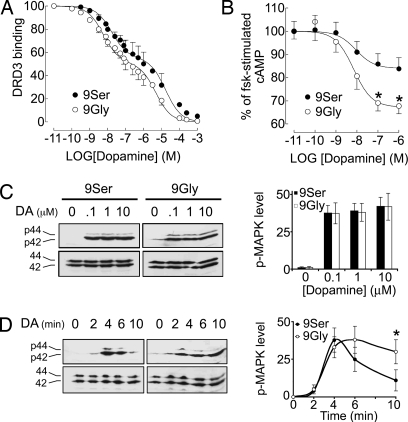
To further assess the functional properties of the Gly-9 variant, we stably transfected each of the two DRD3 variants in HEK293 cells and obtained a set of clonal cell lines with similar expression levels (2.1 ± 0.3 pmol·mg protein−1, n = 8 clones, and 2.2 ± 0.25 pmol·mg protein−1, n = 9 clones, for Ser-9 and Gly-9 variants, respectively). We measured the ability of dopamine to inhibit the binding to surface DRD3 receptors, measured on whole cells obtained from independent clonal cell lines. Dopamine at 40 nM inhibited 38 ± 2% and 58 ± 2% of binding to the Ser-9 (n = 8 clones) and Gly-9 (n = 9 clones) variant, respectively (P < 0.0001). Using a representative clonal cell line of each DRD3 variant, complete inhibition curves (Fig. 4A) show that dopamine inhibited surface DRD3 binding in a biphasic manner, a feature common to G protein-coupled receptors (GPCRs). The Ser9Gly variation affected similarly each of the two affinity sites, with an increased dopamine affinity by 4–5 times (P < 0.01), but did not change the proportion of each affinity site (45 ± 2% and 49 ± 2% for the Ser-9 and Gly-9 variants, respectively). This result is in agreement with a previous study, in which binding of dopamine to the high- and low-affinity sites has not been examined (21).
Fig. 4.
Gain-of-function of the DRD3 Gly-9 variant. (A) Dopamine inhibited surface DRD3 binding in a biphasic manner, as measured on live HEK293 cells stably transfected; eight and nine independent clones for the Ser-9 and Gly-9 variants, respectively, were compared. Results are mean ± SEM of data obtained in three independent experiments, expressed as percentage of total specific binding. (B) The maximal inhibition by dopamine of forskolin (FSK)-stimulated cAMP accumulation in HEK293 cells stably transfected with either variant (two independent clones for each variant). Results are mean ± SEM of data obtained in four independent experiments, expressed as percentage of control cAMP accumulation measured in the absence of dopamine. ∗, P < 0.01 by the two-tailed t test. (C) Dopamine-induced activation of the MAPK (p42 and p44) was assessed in HEK293 cells transiently transfected with either variant. Levels of activated MAPK were detected by a monoclonal antibody raised against the phosphorylated forms of MAPK. Membranes were stripped and then reprobed with an anti-MAPK antibody to detect total MAPK loaded in gel. Serum-starved cells were stimulated by the indicated doses of dopamine for 5 min. (D) Serum-starved cells were stimulated by 1 μM dopamine for the indicated times. Blots are representative of four independent experiments, and quantifications were performed by densitometric analysis. ∗, P < 0.05 by the two-tailed t test.
Then, we compared dopamine-mediated inhibition of forskolin-induced cAMP accumulation, a typical DRD3-mediated response (22), through each DRD3 variant in two independent clones for each variant. The response to dopamine in HEK293 cells transfected with the DRD3 alone was weak (<15% and 7% for the Gly-9 and Ser-9 variants, respectively; data not shown) but increased by prior transient transfection with the adenylate cyclase type V, which preferentially confers responsiveness to human DRD3 in these cells (23). In these latter conditions, the dopamine response was twice as high when elicited by stimulation of the Gly-9 as the Ser-9 variant (33 ± 3% vs. 16 ± 4%, P < 0.01; see Fig. 4B). This finding indicates that the Gly-9 variant more robustly increased cAMP inhibition than the Ser-9 in response to dopamine.
To further support the possibility that the Ser9Gly variation alters normal DRD3 signaling properties, we compared dopamine-induced activation of the mitogen-associated protein kinase (MAPK), another DRD3-mediated response (24). Dopamine activated the MAPK optimally at 0.1, 1, and 10 μM without apparent dose effect (Fig. 4C), a response that was blocked by the DRD3 antagonist, haloperidol (data not shown). Interestingly, the levels of phospho-MAPK detected after Gly-9 variant activation were similar to those after Ser-9 variant activation (Fig. 4C). Activation of the MAPK initiated rapidly (≈2 min) after dopamine addition and peaked at ≈5 min whether the Ser-9 or the Gly-9 variant was stimulated. However, the Gly-9 variant promoted a longer-lasting elevation of phospho-MAPK levels (>10–15 min) than the Ser-9 variant (<10 min; Fig. 4D). This result shows that the MAPK signaling mediated by the Gly-9 variant is prolonged compared with the Ser-9 variant.
Discussion
Our results, demonstrating cosegregation of the Gly-9 variant with ET in a set of 30 French families and association of the same variant with risk and AAO of ET in a large American case-control study, suggest that this variant is either causal or in strong linkage disequilibrium with the causal mutation. The haplotype block of strong linkage disequilibrium containing the Ser9Gly variant is close to a previously identified marker of the ETM1 locus (6). There is no other coding sequence but three DRD3 exons in this block. Except the Ser9Gly variant, the identified variants are intronic or upstream from DRD3 promoter region. The variant seems to cause a gain-of-function of the receptor, as assessed by increased dopamine affinity and signaling responses. Future studies of DRD3 expression in normal and ET brains may provide insights into the relevance of DRD3 to the pathogenesis of ET.
From a genetic point of view, the results of our familial study replicate the first linkage study of ET, which found a disease locus on 3q13.3; in addition, they suggest a gene dosage effect for the Gly-9 allele. In the case-control study, we confirmed the association of this variant with ET. Here again, Gly-9 homozygosity seemed to have the most important effect on disease risk and AAO. The effect of a single Gly-9 allele seemed much more limited, perhaps because of the relatively young age of numerous Ser9Gly heterozygotes in the control group (median age of heterozygotes, 51 y; range, 16–88 y). Some of them could indeed develop ET when advancing in age. Conversely, we noted that, among the ten Gly-9 homozygous controls, none was older than 64 years of age.
Large variations in ET prevalence estimates across various studies are accounted for by the varying severity of the disease. Without a physiological or other marker for ET, the diagnosis is based on a set of diagnostic criteria. The consensus classification defines ET as a bilateral action tremor of the hands and forearms (but not rest tremor) present for at least 3 y as a core criterion (25), and all patients in our samples meet these criteria. Because mild or early symptoms of ET may escape the diagnosis, it would be of great interest to search for an association between the DRD3 Gly-9 variant and a broadly defined tremor phenotype in a large sample of aged adults to determine the prevalence of ET related to this genetic abnormality.
Most small-molecule transmitters or hormones acting at GPCR interact within the transmembrane region of the receptor (26). The Ser9Gly variant is located in the extracellular N terminus of the DRD3 and is not predicted to take part in receptor ligand binding. However, the Ser9Gly variant affected dopamine affinity at two sites, which correspond to the high-affinity state, coupled to the G protein that transduces the signal to the effector, and to the low-affinity, uncoupled state, respectively. Because the proportion of each site is conserved in each variant, the Ser9Gly variant does not affect the equilibrium between the G protein-coupled and uncoupled forms of the receptor. We predict that the variant could affect the overall conformation of the receptor. In agreement with this hypothesis, several mutations responsible for autosomal dominant retinitis pigmentosa were identified in the extracellular N terminus of rhodopsin, a prototypical GPCR, producing misfolded rhodopsins that are defective in normal intracellular transport (27). Posttranslational modifications like glycosylation are critical to efficiently fold and sort dopamine receptors through the anterograde biosynthetic pathway (28). Although the Ser9Gly variant is located close to a highly conserved glycosylation site, both variants displayed similar glycosylation and anterograde trafficking. Indeed, both variants reached efficiently the plasma membrane and internalized in response to dopamine, but to a limited extent as previously reported (29). Nevertheless, the Gly-9 variant was different from the Ser-9 by its ability to signal more robust dopamine responses. Indeed, both cAMP inhibition and MAPK signal duration were higher with the Gly-9 than the Ser-9 variant. The N-terminal region may therefore provide an unexpected structural determinant to DRD3 function. We hypothesize that the Gly-9 variant could increase dopamine affinity and efficacy by altering an interaction with a protein that affects the conformation of the receptor, which may occur as homo- or heterooligomers (30). For example, pharmacological modulation of class II GPCR through its extracellular N terminus by receptor activity-modifying proteins has emerged as a new phenotype-modifying determinant of GPCR function (31).
Our genetic and functional data, together with the gene dosage effect, are therefore consistent with the hypothesis that the DRD3 Gly-9 variant confers inheritable susceptibility to ET in our sample, with a penetrance that increases with age and is almost complete in the elderly population, but with highly variable expression. The relationship between DRD3 and motor control is supported by the finding that DRD3 overexpression produces in nonhuman primates levodopa-induced dyskinesia (14). Although DRD3 receptor overexpression in the basal ganglia has been implicated in levodopa-induced dyskinesia, abnormalities in the cerebellar DRD3 receptors could be responsible for ET. Although dopaminergic drugs are ineffective in ET, olanzapine and clozapine, which are nonselective dopamine D2-like receptor blockers (acting at both DRD2 and DRD3 receptors), have been found to improve ET (32, 33). Therefore, we propose DRD3-selective partial agonists or antagonists (14, 34) as a possible pharmacotherapy of ET.
Methods
Patients.
For the family study, 30 unrelated families with familial ET were recruited in France at La Pitié-Salpétrière Hospital in Paris and Hospital Center in Charleville-Mézières. All patients fulfilled the diagnostic criteria of classic hereditary ET as defined by the Consensus Statement of the Movement Disorder Society (25), and only patients with probable and definite familial ET were included in the study (35). The screening procedure for signs of ET consisted of the drawing of an Archimedes spiral with the dominant hand within preprinted lines (36) and a clinical examination that included neurological examination, tremor type and its topography, search for associated signs or symptoms, drugs in use, and paraclinical investigations to define the diagnosis. Forearm tremor scores were measured as described (37). For the case-control replication study, 276 patients with ET, all of Caucasian origin (age range, 14–93 y; age-at-onset range, 3–83 y; male/female ratio, 126/150), diagnosed according to common diagnostic criteria (25), and 184 normal controls with no family history of movement disorders (age range, 9–88 y; male/female, 83/101) were recruited by the Department of Neurology, Baylor College of Medicine, and the study was approved by Institutional Review Board. All participating individuals gave their informed consent.
DRD3 Genotyping and Analysis.
Genomic DNA was prepared from whole blood by using standard techniques. The DRD3 Ser9Gly variant was genotyped as described (10) or by using a TaqMan SNP genotyping assay (see Table 3). The program tdt/s-tdt 1.1 (http://genomics.med.upenn.edu/spielman/TDT.htm) was used to perform the nonparametric analysis of linkage in the presence of association (38). To perform the parametric linkage analysis, we used the genehunter program (18). To identify other DRD3 mutations, genomic DNA from nine probands was amplified by PCR and screened for sequence variations by direct sequencing on a Li-Cor 4000L automated DNA sequencer (see Table 4).
Plasmid Constructs.
A plasmid pRc/CMV bearing a cDNA encoding the Ser-9 variant (39) was used to generate cDNA encoding the Gly-9 variant by site-directed mutagenesis using the QuikChange multi Site-directed Mutagenesis Kit (Stratagene). DRD3 stop codon of both Ser-9 and Gly-9 variants was deleted by site-directed mutagenesis, and subsequent sequences were subcloned into pEYFPN1 (BD Biosciences) upstream of the enhanced yellow fluorescent protein (E-YFP). Constructions were checked by nucleotide sequencing (Licor).
Cell Culture and Transfections.
HEK293 cells were cultured in a cell culture medium made with a 1:1 mixture of DMEM and F-12 (Life Technologies, Grand Island, NY), supplemented with 10% FCS and 100 μg·ml−1 penicillin/streptomycin in humidified atmosphere of 5% CO2, 95% air. Cells were seeded in 10-cm dishes at 50–80% confluency, and cDNA encoding the Ser-9 or the Gly-9 variant was transfected (10 μg of DNA for 106 cells) by using Superfect (Qiagen, Valencia, CA). Transfectants were selected with G418 (Invitrogen) at the concentration of 800 μg·ml−1.
Deglycosylation Assays.
Membranes were incubated for 45 min at 4°C in 500 μl of 50 mM Na/Na2PO4 (pH 7.4), 300 mM NaCl, 40 mg/ml digitonin, 10 mg/ml deoxycholate, 10 mM DTT plus protein inhibitor mixture (Sigma). Cleared membranes (50 μg) were incubated with recombinant N-glycosidase F (10 units) for 16 h at 37°C according to the supplier's instructions (Roche Biochemicals). Controls were incubated without enzyme. Analysis was carried out by SDS/PAGE under reducing conditions, and immunoblots were revealed by anti-YFP antibody (1:7,000; BD Biosciences).
Fluorescence Microscopy.
Cells were grown on coverslips in the presence of cycloheximide (20 μg·ml−1 for 3 h) to reduce intracellular fluorescence. Cells were stimulated or not by 10 μM dopamine for various times and fixed in 2% paraformaldehyde for 20 min at room temperature, and YFP fluorescence was captured by using a confocal microscope (Leica, Deerfield, IL).
Binding Assays.
Cells were detached from culture dishes in the presence of 0.2% trypsin, harvested by centrifugation, suspended in DMEM-F12 supplemented with ascorbic acid (50 μg/ml), and incubated in quadruplicate for 30 min at 30°C with [125I]iodosulpride (Amersham Pharmacia) at 0.1 nM and dopamine in increasing concentrations. Nonspecific binding was measured in the presence of enomapride (1 μM). Binding was stopped by filtration through GF/C filters coated with polyethyleneimine (0.3%), and radioactivity was counted on the filter by gamma scintigraphy. Dopamine inhibition curves were analyzed by nonlinear regression with a two-site model by using prism software (GraphPad, San Diego).
cAMP Accumulation Assays.
DRD3-expressing cells were transiently transfected with pCDNA3 containing cDNA encoding canine adenylyl cyclase type V, which specifically confers responsiveness to human DRD3 in HEK293 cells (23). Cells were preincubated in quintuplicate with 10 μM 3-isobutyl-1-methylxanthine in DMEM-F12 for 25 min and incubated with dopamine in increasing concentrations for 10 min in the presence of 0.1 μM forskolin. The reaction was stopped by addition of 50 μl of ice-cold 0.1 M HCl. The cells were mildly sonicated, and cAMP was assayed with the Rianen [125I]cAMP RIA kit (DuPont/NEN).
MAPK Activation Measurements.
After transfection, cells were cultured in 3-cm dishes and starved overnight in a serum-free culture medium containing 0.1% BSA. Cells were stimulated by dopamine and lysed in 350 μl RIPA buffer (10 mM Tris·HCl, pH 8/150 mM NaCl/1 mM EDTA/1% Nonidet P-40/10% glycerol/10 mM sodium fluoride/1 mM sodium orthovanadate/protein inhibitor mixture). Lysates were sonicated and cleared by centrifugation. Total protein concentrations were determined by the Bio-Rad protein assay. Equal amounts of proteins (80 μg) were resolved into 12% SDS/PAGE. Levels of phospho-MAPK were measured by the phospho-p44/42 MAPK (E10) antibody (1:2,000; Cell signaling Technology, Beverly, MA). Membranes were stripped and then reprobed with a p44/42 MAPK antibody (1:1,000; Cell signaling Technology) to control the total amount of MAPK.
Supplementary Material
Acknowledgments
We thank the following neurologists for providing us with access to patients for blood sampling and to their clinical records: Drs. A. Gordji, T. Champenois, and C. P. Jedynak. We thank Dr. C. W. Dessauer (Houston, TX) for the gift of pcDNA3 expressing adenylyl cyclase type V.
Abbreviations
- AAO
age-at-onset
- DRD3
dopamine D3 receptor
- ET
essential tremor
- GPCR
G protein-coupled receptor
- MAPK
mitogen-associated protein kinase
- TDT
transmission disequilibrium test
- YFP
yellow fluorescent protein
- HEK
human embryonic kidney.
Note Added in Proof.
Part of the genetic study included in this article (shown in Figs. 1 and 2) appeared in a report published in Clinical Genetics (40).
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Findley L. J. Neurology. 2000;54:S8–S13. [PubMed] [Google Scholar]
- 2.Ondo W., Vuong K., Almaguer M., Jankovic J., Simpson R. K. Movement Disorders. 2001;16:1137–1142. doi: 10.1002/mds.1249. [DOI] [PubMed] [Google Scholar]
- 3.Ondo W. G., Jankovic J., Connor G. S., Pahwa R., Elble R., Stacy M. A., Koller W. C., Schwarzman L., Wu S. C., Hulihan J. F. Neurology. 2006;66:672–677. doi: 10.1212/01.wnl.0000200779.03748.0f. [DOI] [PubMed] [Google Scholar]
- 4.Deuschl G., Elble R. J. Neurology. 2000;54:S14–20. [PubMed] [Google Scholar]
- 5.Brin M. F., Koller W. Movement Disorders. 1998;13(Suppl. 3):55–63. doi: 10.1002/mds.870131310. [DOI] [PubMed] [Google Scholar]
- 6.Gulcher J. R, Jonsson P., Kong A., Kristjansson K., Frigge M. L., Karason A., Einarsdottir I. E., Stefansson H., Einarsdottir A. S., Sigurthoardottir S., et al. Nat. Genet. 1997;17:84–87. doi: 10.1038/ng0997-84. [DOI] [PubMed] [Google Scholar]
- 7.Higgins J. J., Pho L. T., Nee L. E. Movement Disorders. 1997;12:859–864. doi: 10.1002/mds.870120605. [DOI] [PubMed] [Google Scholar]
- 8.Higgins J. J., Lombardi R. Q., Pucilowska J., Jankovic J., Tan E. K., Rooney J. P. Neurology. 2005;64:417–421. doi: 10.1212/01.WNL.0000153481.30222.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng H., Le W. D., Guo Y., Huang M. S., Xie W. J., Jankovic J. Neurology. 2005;65:651–652. doi: 10.1212/01.wnl.0000173033.32535.23. [DOI] [PubMed] [Google Scholar]
- 10.Lannfelt L., Sokoloff P., Martres M. P., Pilon C., Giros B., Jonsson E., Sedvall G., Schwartz J.-C. Psychiatr. Genet. 1992;2:249–256. [Google Scholar]
- 11.Sokoloff P., Giros B., Martres M.-P., Bouthenet M.-L., Schwartz J.-C. Nature. 1990;347:146–151. doi: 10.1038/347146a0. [DOI] [PubMed] [Google Scholar]
- 12.Lerer B., Segman R. H., Fangerau H., Daly A. K., Basile V. S., Cavallaro R., Aschauer H. N., McCreadie R. G., Ohlraun S., Ferrier N., et al. Neuropsychopharmacology. 2002;27:105–119. doi: 10.1016/S0893-133X(02)00293-2. [DOI] [PubMed] [Google Scholar]
- 13.Ryoo H. L., Pierrotti D., Joyce J. N. Movement Disorders. 1998;13:788–797. doi: 10.1002/mds.870130506. [DOI] [PubMed] [Google Scholar]
- 14.Bezard E., Ferry S., Mach U., Stark H., Leriche L., Boraud T., Gross C., Sokoloff P. Nat. Med. 2003;6:762–767. doi: 10.1038/nm875. [DOI] [PubMed] [Google Scholar]
- 15.Hall H., Farde L., Halldin C., Hurd Y. L., Pauli S., Sedvall G. Psychopharmacology. 1996;128:240–247. doi: 10.1007/s002130050131. [DOI] [PubMed] [Google Scholar]
- 16.Diaz J., Levesque D., Lammers C. H., Griffon N., Martres M. P., Schwartz J.-C., Sokoloff P. Neuroscience. 1995;65:731–745. doi: 10.1016/0306-4522(94)00527-c. [DOI] [PubMed] [Google Scholar]
- 17.Louis E. D., Shungu D. C., Chan S., Mao X., Jurewicz E. C., Watner D. Neurosci. Lett. 2002;333:17–20. doi: 10.1016/s0304-3940(02)00966-7. [DOI] [PubMed] [Google Scholar]
- 18.Kruglyak L., Daly M. J., Reeve-Daly M. P., Lander E. S. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 19.Rajagopal R., Chen Z. Y., Lee F. S., Chao M. V. J. Neurosci. 2004;24:6650–6658. doi: 10.1523/JNEUROSCI.0010-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickery R. G., Von Zastrow M. J. Cell Biol. 1999;144:31–43. doi: 10.1083/jcb.144.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundstrom K., Turpin M. P. Biochem. Biophys. Res. Commun. 1996;225:1068–1072. doi: 10.1006/bbrc.1996.1296. [DOI] [PubMed] [Google Scholar]
- 22.Griffon N., Pilon C., Sautel F., Schwartz J.-C., Sokoloff P. J. Neurochem. 1997;68:1–9. doi: 10.1046/j.1471-4159.1997.68010001.x. [DOI] [PubMed] [Google Scholar]
- 23.Robinson S. W., Caron M. G. Mol. Pharmacol. 1997;52:508–514. doi: 10.1124/mol.52.3.508. [DOI] [PubMed] [Google Scholar]
- 24.Beom S., Cheong D., Torres G., Caron M. G., Kim K. M. J. Biol. Chem. 2004;279:28304–28314. doi: 10.1074/jbc.M403899200. [DOI] [PubMed] [Google Scholar]
- 25.Deuschl G., Bain P., Brin M. Movement Disorders. 1998;13(Suppl. 3):2–23. doi: 10.1002/mds.870131303. [DOI] [PubMed] [Google Scholar]
- 26.Christopoulos A., Kenakin T. Pharmacol. Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 27.Colley N. J., Cassill J. A., Baker E. K., Zuker C. S. Proc. Natl. Acad. Sci. USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fishburn C. S., Elazar Z., Fuchs S. J. Biol. Chem. 1995;270:29819–29824. doi: 10.1074/jbc.270.50.29819. [DOI] [PubMed] [Google Scholar]
- 29.Kim K. M., Valenzano K. J., Robinson S. R., Yao W. D., Barak L. S., Caron M. G. J. Biol. Chem. 2001;276:37409–37414. doi: 10.1074/jbc.M106728200. [DOI] [PubMed] [Google Scholar]
- 30.Bouvier M. Nat. Rev. Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 31.Morfis M., Christopoulos A., Sexton P. M. Trends Pharmacol. Sci. 2003;24:596–601. doi: 10.1016/j.tips.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Ceravolo R., Salvetti S., Piccini P., Lucetti C., Gambaccini G., Bonuccelli U. Movement Disorders. 1999;14:468–472. doi: 10.1002/1531-8257(199905)14:3<468::aid-mds1013>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Yetimalar Y., Irtman G., Gurgor N., Basoglu M. Eur. J. Neurosci. 2003;10:79–82. doi: 10.1046/j.1468-1331.2003.00534.x. [DOI] [PubMed] [Google Scholar]
- 34.Pilla M., Perachon S., Sautel F., Garrido F., Mann A., Wermuth C. G., Schwartz J.-C., Everitt B. J., Sokoloff P. Nature. 1999;400:371–375. doi: 10.1038/22560. [DOI] [PubMed] [Google Scholar]
- 35.Watner D., Jurewicz E. C., Louis E. D. Movement Disorders. 2002;17:378–381. doi: 10.1002/mds.10085. [DOI] [PubMed] [Google Scholar]
- 36.Louis E. D., Ottman R. Neurology. 1996;46:1200–1205. doi: 10.1212/wnl.46.5.1200. [DOI] [PubMed] [Google Scholar]
- 37.Fahn S., Elton R. In: Recent Developments in Parkinson's Disease, Fahn S., Marsden C. D., Calne D. B., Goldstein M., editors. Vol. 2. Florham Park, NJ: Macmillan Health Care Information; 1987. pp. 153–163. [Google Scholar]
- 38.Spielman R. S., Ewens W. J. Am. J. Hum. Genet. 1998;62:450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilon C., Levesque D., Dimitriadou V., Griffon N., Martres M. P., Schwartz J.-C., Sokoloff P. Eur. J. Pharmacol. 1994;268:129–139. doi: 10.1016/0922-4106(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 40.Lucotte G., Lagarde J. P., Funalot B., Sokoloff P. Clin. Genet. 2006;69:437–440. doi: 10.1111/j.1399-0004.2006.00600.x. (lett.) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.