Abstract
α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (AMPARs) are a major subtype of ionotropic glutamate receptors (iGluRs) that mediate rapid excitatory synaptic transmission in the vertebrate brain. Putative AMPARs are also expressed in the nervous system of invertebrates. In Caenorhabditis elegans, the GLR-1 receptor subunit is expressed in neural circuits that mediate avoidance behaviors and is required for glutamate-gated current in the AVA and AVD interneurons. Glutamate-gated currents can be recorded from heterologous cells that express vertebrate AMPARs; however, when C. elegans GLR-1 is expressed in heterologous cells, little or no glutamate-gated current is detected. This finding suggests that other receptor subunits or auxiliary proteins are required for function. Here, we identify Ce STG-1, a C. elegans stargazin-like protein, and show that expression of Ce STG-1 together with GLR-1 and the CUB-domain protein SOL-1 reconstitutes glutamate-gated currents in Xenopus oocytes. Ce STG-1 and homologues cloned from Drosophila (Dro STG1) and Apis mellifera (Apis STG1) have evolutionarily conserved functions and can partially substitute for one another to reconstitute glutamate-gated currents from rat, Drosophila, and C. elegans. Furthermore, we show that Ce STG-1 and Apis STG1 are primarily required for function independent of possible roles in promoting the surface expression of invertebrate AMPARs.
Keywords: C. elegans, SOL-1, transmembrane AMPA receptor regulatory protein (TARP), GLR-1
The majority of fast synaptic neurotransmission in the vertebrate central nervous system is mediated by ionotropic glutamate receptors (iGluRs) that are gated by the neurotransmitter glutamate. The non-NMDA class of iGluRs are selectively gated by the ligands α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) or kainate (1). Although much has been learned about iGluR function in recent years, our understanding is still incomplete. For example, functional vertebrate AMPA receptors (AMPARs) can be expressed in Xenopus oocytes; indeed, the first AMPAR subunit was cloned by functional expression in oocytes (2). In contrast, although invertebrate iGluRs have been cloned from nematodes and insects (3–5) and are known to mediate neurotransmission at invertebrate synapses (6–8), little or no glutamate-gated current can be recorded when genes encoding invertebrate iGluRs are expressed in heterologous cells, (3, 9). This absence of current indicates either that the correct iGluR subunit composition has not been identified or that iGluRs require one or more auxiliary proteins to be fully functional.
In Caenorhabditis elegans, mutations in the AMPAR subunits GLR-1 (4, 5) and GLR-2 (8) eliminate or reduce glutamate-gated currents in interneurons that mediate avoidance behaviors (10, 11). In addition, SOL-1, a CUB-domain transmembrane protein that interacts with GLR-1, is required for non-NMDA-type glutamate-gated currents. In sol-1 mutants, behaviors dependent on the GLR-1 AMPAR subunit are disrupted, and no fast glutamate-gated or kainate-gated currents can be detected after pressure application of ligand (12). However, coexpression of SOL-1 with C. elegans GLR-1 is not sufficient to reconstitute glutamate-gated currents in Xenopus oocytes (9).
In mice, the transmembrane protein stargazin associates with AMPARs and has been reported to regulate several aspects of iGluR expression and function (13). A mutation in stg, which encodes stargazin, eliminates all AMPAR-mediated synaptic transmission, greatly reduces the response to exogenously applied AMPA, and eliminates surface expression of AMPARs in cerebellar granule cells, but has no effect on NMDA-mediated currents (14, 15). Stargazin shares no amino acid sequence identity with SOL-1, but rather has sequence identity with γ calcium channel subunits. Stargazin is the founding member of a small family of transmembrane AMPA receptor regulatory proteins (TARPs) (16). Stargazin seems to have at least three functions: it serves as a chaperone protein for the exit of AMPARs from the endoplasmic reticulum, it targets AMPARs to the synapse by binding to PSD-95 (15, 17–19), and it modifies the electrophysiological characteristics of AMPARs (20–23). No stargazin-like proteins have been identified to date in invertebrates.
To address the requirements for invertebrate iGluR function, we have attempted reconstitution of C. elegans glutamate-gated currents in Xenopus oocytes. In C. elegans, we identified STG-1, a protein that is related to vertebrate stargazin, and show that we can record glutamate-gated currents in Xenopus oocytes that express Ce STG-1 together with SOL-1 and GLR-1. We have also identified Ce STG-1 homologues from Drosophila (Dro STG1) and Apis mellifera (honey bee, Apis STG1). We show that nematode and insect stargazin-like proteins have conserved function and when coexpressed with vertebrate GluR1 enhance the glutamate-gated current. In reconstitution experiments, we found that considerable surface expression of invertebrate iGluRs occurs in the absence of stargazin-like proteins yet no glutamate-gated current is detected, suggesting that an evolutionarily conserved role of stargazin-like proteins is to promote the function of AMPARs.
Results
Ce stg-1 Encodes a Stargazin-Like protein That Is Expressed in the Nervous System.
In the C. elegans nervous system, SOL-1 and the iGluR subunits GLR-1 and GLR-2 are coexpressed in many neurons (4, 5, 12, 24), suggesting that these proteins may be sufficient to reconstitute functional receptors in heterologous cells. However, we were unable to record appreciable glutamate-gated currents from Xenopus oocytes that expressed GLR-1 alone or coexpressed with SOL-1 (9). Nor were we able to record currents when GLR-2 was also coexpressed (data not shown). Therefore, we reasoned that reconstitution might require a second auxiliary protein in addition to SOL-1. Based on weak sequence identity to vertebrate stargazin, we identified predicted ORFs in the C. elegans (C18D1.4), honey bee (Apis mellifera; XM 397021), and Drosophila (CG33670) genomes. The predicted Apis sequence was incomplete; therefore, we used PCR amplification and RACE to clone the complete ORF from A. mellifera first-strand cDNA.
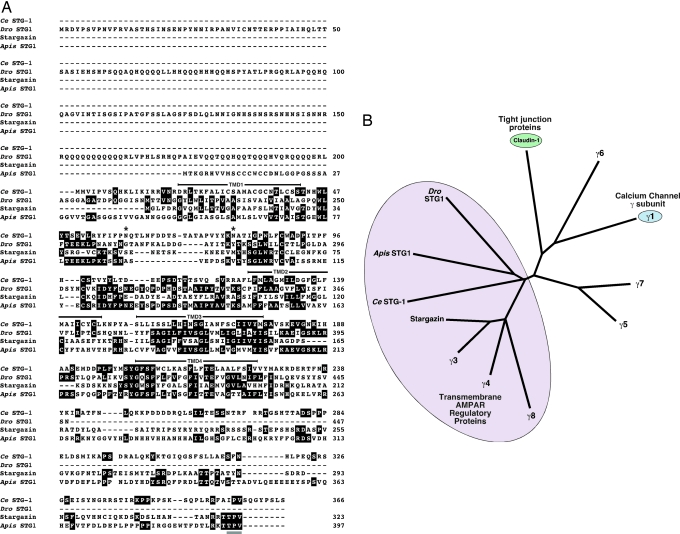
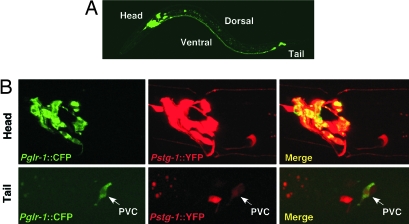
C. elegans, Apis, and Drosophila encode predicted 366 (Ce STG-1), 397 (Apis STG1), and 447 (Dro STG1) amino acid transmembrane proteins (Fig. 1A), and share ≈21% (Ce STG-1), 24% (Apis STG1), and 25% (Dro STG1) sequence identity with vertebrate stargazin. Hydropathy analysis identified four putative transmembrane domains. Because no N-terminal signal sequences are evident, we predict that the N and C termini of Ce STG-1, Apis STG1 and Dro STG1 are intracellular. Dro STG1 has a predicted 5′ region that is longer by ≈200 aa than the other stargazin-like proteins. In addition, its 3′ region is significantly shorter (Fig. 1A). These differences may indicate that the particular clone we have identified is alternatively spliced. The amino acid sequences of these predicted proteins were analyzed by generating a neighbor-joining tree of related vertebrate proteins, including TARPs, Ca2+ channel γ subunits, and claudin tight junction proteins (Fig. 1B). Ce STG-1, Apis STG1, and Dro STG1 seem only slightly more related to TARPs than to the Ca2+ channel γ subunits. To determine the cellular expression of Ce STG-1, we used the stg-1 promoter to drive the expression of GFP (Fig. 2A) or yellow fluorescent protein (YFP) (Fig. 2B) in transgenic worms. GFP and YFP were strongly expressed in the nervous system, with expression apparent in most of the neurons that normally express the GLR-1 subunit as indicated by coexpression of Pglr-1::CFP (cyan fluorescent protein) and Pstg-1::YFP (Fig. 2B). We did not observe expression in nonneuronal tissues such as muscle.
Fig. 1.
C. elegans STG-1, Drosophila STG1, and Apis STG1 are related to members of the TARP family. (A) The predicted amino acid sequences encoded by C. elegans stg-1, Drosophila stg1, vertebrate stg, and A. mellifera stg1. Amino acids are numbered beginning with the first predicted methionine. For Ce STG-1, bars and asterisks indicate predicted transmembrane domains and putative N-linked glycosylation sites, respectively. Underlined in gray are the potential type I PDZ-domain-binding sites for vertebrate stargazin and Apis STG1. (B) Phylogenetic tree of the amino acid sequences for stargazin-like proteins (figure modified from ref. 16).
Fig. 2.
C. elegans STG-1 is expressed in the nervous system. Confocal images of transgenic worms that expressed Pstg-1::GFP (A) or both Pglr-1::CFP and Pstg-1::YFP (B) are shown. Expression is limited to neuronal cell bodies and processes.
GLR-1 Function, but Not Surface Expression, Depends on Ce STG-1 and SOL-1.
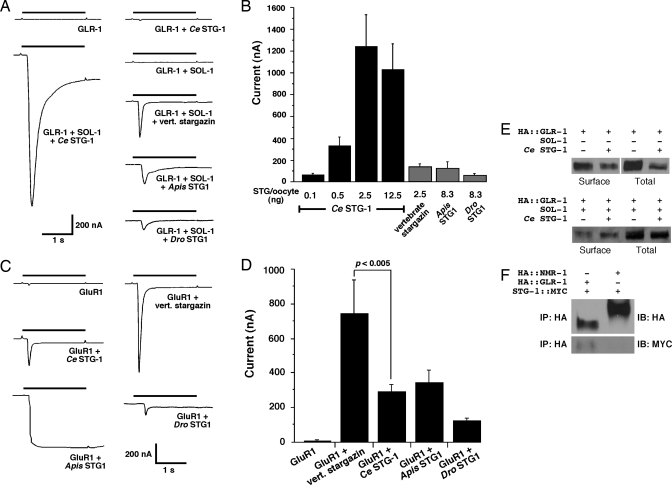
To attempt to reconstitute GLR-1 receptor activity, we injected cRNAs encoding C. elegans GLR-1 and auxiliary proteins into Xenopus oocytes and used standard voltage-clamp techniques to record the current in response to bath application of glutamate. We recorded no glutamate-gated current after expression of GLR-1 alone, and only negligible or very small currents after coexpression of GLR-1 with either SOL-1 (<3 nA) or Ce STG-1 (<8 nA) (Fig. 3A). In contrast, when GLR-1, Ce STG-1, and SOL-1 were coexpressed, the amplitude of the glutamate-gated current was increased by over two orders of magnitude (Fig. 3 A and B). We found that vertebrate stargazin, Apis STG1, and Dro STG1 could partially substitute for Ce STG-1 in reconstituting GLR-1-mediated currents in Xenopus oocytes (Fig. 3 A and B). The kinetics of current desensitization seemed dependent on which of the four stargazin-like proteins was coexpressed, indicating that these auxiliary proteins influence some aspect of receptor gating. The magnitude of GLR-1-mediated glutamate-gated current depended on the amount of injected Ce STG-1 cRNA, with maximal currents observed with ≈2.5 ng of cRNA per oocyte (Fig. 3B). A similar dose dependency was observed in a recent study of vertebrate stargazin (25).
Fig. 3.
GLR-1-mediated glutamate-gated currents in Xenopus oocytes are dependent on Ce STG-1 and SOL-1. (A) Currents recorded in response to 1 mM glutamate application in Xenopus oocytes injected with combinations of SOL-1, GLR-1, and C. elegans STG-1, vertebrate stargazin, Apis STG1, or Dro STG1 cRNA. Oocytes were voltage-clamped at a holding potential of −70 mV. (B) Average peak glutamate-gated current amplitude in oocytes coinjected with GLR-1, SOL-1, and Ce STG-1 cRNA as a function of Ce STG-1 (black). Values are also indicated for vertebrate stargazin, Apis STG1, and Dro STG1 (gray). GLR-1 plus SOL-1 plus 0.1 ng of Ce STG-1, n = 21; plus 0.5 ng of Ce STG-1, n = 19; plus 2.5 ng of Ce STG-1, n = 16; plus 12.5 ng of Ce STG-1, n = 18; plus vert. stargazin, n = 9; plus Apis STG1, n = 6; plus Dro STG1, n = 5. In A and B, oocytes were injected with 8.3 ng of GLR-1 and 8.3 ng of SOL-1 cRNA. (C) Glutamate-gated currents in Xenopus oocytes injected with rat GluR1 alone or coinjected with Ce STG-1, Apis STG1, Dro STG1, or vertebrate stargazin cRNAs. (D) Average peak glutamate-gated current amplitude as a function of stargazin cRNA. GluR1, n = 12; plus vert. stargazin, n = 5; plus Ce STG-1, n = 12; plus Apis STG1, n = 5; plus Dro STG1, n = 4. In C and D, oocytes were injected with 0.1 ng of rat GluR1 and 8.3 ng of the indicated stargazin cRNA. (E) Western blot showing the relative surface expression of C. elegans HA::GLR-1 in the presence or absence of Ce STG-1. (F) Coimmunoprecipitation of HA::GLR-1 and STG-1::MYC from HEK 293 cells. IB, immunoblot.
We also found that Ce STG-1, Apis STG1, and Dro STG1 can enhance vertebrate GluR1-dependent currents (Fig. 3 C and D). Thus, stargazin’s role in promoting glutamate-gated currents seems evolutionarily conserved. However, we noted different functional effects of stargazin-like molecules on GluR1-mediated currents. For example, the glutamate-gated current rapidly desensitized when GluR1 was coexpressed with vertebrate stargazin, but no desensitization was noted when coexpressed with Apis STG1 (Fig. 3C). This result suggests that a primary effect of stargazin-like molecules is to directly regulate iGluR function. Additional support for this interpretation comes from our analysis of the effects of Ce STG-1 on receptor surface expression. Coexpression of GLR-1 and Ce STG-1 increased GLR-1 surface expression ≈3-fold (see Materials and Methods) (Fig. 3E). This relatively small change in surface expression cannot explain the dramatic increase (at least two orders of magnitude) in glutamate-gated current observed when GLR-1 was coexpressed with Ce STG-1 (Fig. 3 A and B).
Vertebrate stargazin is known to associate with AMPARs (25, 26). To determine whether Ce STG-1 and GLR-1 form a complex when expressed in heterologous cells, we coexpressed HA-epitope-tagged GLR-1 (HA::GLR-1) with MYC-epitope-tagged Ce STG-1 (STG-1::MYC). Both constructs were fully functional when expressed in oocytes (data not shown). We also coexpressed STG-1::MYC with HA-tagged NMR-1 (HA::NMR-1), an NMDA receptor subunit that functions independently of SOL-1 (12). In vertebrates, stargazin is not required for NMDA-dependent currents (14, 15). Antibodies to HA precipitated HA::GLR-1 and HA::NMR-1, but STG-1::MYC coprecipitated only with HA::GLR-1 (Fig. 3F). These data indicate that Ce STG-1 associates with GLR-1.
Mutations in GLR-1 Partially Compensate for the Absence of Ce STG-1.
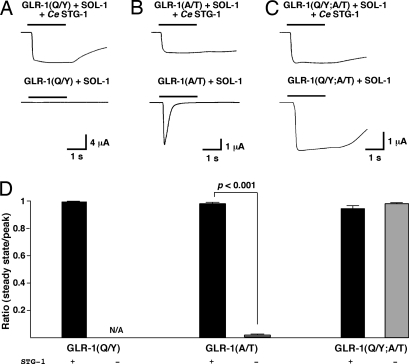
Because we observed only a small contribution of Ce STG-1 to the surface expression of GLR-1, we suspected that Ce STG-1 might be required for receptor function. For example, if the receptor rapidly desensitized, we would underestimate the true current magnitude because of the relatively slow application of glutamate. We tested this hypothesis by introducing a single amino acid change in the ligand-binding domain of the receptor that is known to slow desensitization (27). In the case of C. elegans, the corresponding mutation in GLR-1 is glutamine to tyrosine (Q552Y) (28). Glutamate-gated currents recorded from Xenopus oocytes that expressed GLR-1(Q552Y), Ce STG-1, and SOL-1 showed almost no desensitization (Fig. 4 A and D). In contrast, we did not observe significant glutamate-gated currents in oocytes that expressed GLR-1(Q552Y) and SOL-1 without Ce STG-1 (Fig. 4 A and D).
Fig. 4.
Glutamate-gated current in the absence of Ce STG-1 depends on modifying the gating properties of GLR-1. (A–C) Currents measured in response to 1 mM glutamate application in Xenopus oocytes coinjected with cRNAs for SOL-1 and GLR-1(Q552Y) (A), GLR-1(A687T) (B), or GLR-1(Q552Y; A687T) (C) both with (Upper) and without (Lower) Ce STG-1. Oocytes were voltage-clamped at −70 mV. (D) The ratio of the current amplitude 1 s after the start of glutamate application (steady state) to the peak current both with (black) and without (gray) Ce STG-1. GLR-1(Q/Y) plus SOL-1 plus Ce STG-1, n = 3; GLR-1(Q/Y) plus SOL-1, n = 3; GLR-1(A/T) plus SOL-1 plus Ce STG-1, n = 4; GLR-1(A/T) plus SOL-1, n = 4; GLR-1(Q/Y;A/T) plus SOL-1 plus Ce STG-1, n = 4; GLR-1(Q/Y;A/T) plus SOL-1, n = 6.
A single alanine to threonine (A/T) change was identified in the δ2 glutamate receptor subunit of the lurcher mouse, and expression of the mutated δ2 receptor in Xenopus oocytes was associated with constitutive activation of the receptor (29). Later studies showed that introduction of the lurcher (A/T) mutation into GluR1 slowed receptor kinetics, increased the apparent affinity for glutamate, and reduced the rate of desensitization (30–32). We have shown that the behavioral and electrophysiological defects of sol-1 mutants can be partially suppressed by transgenic expression of the GLR-1(A687T) lurcher variant in C. elegans (9, 12). To examine the effect of the lurcher mutation on GLR-1-dependent currents, we expressed GLR-1(A687T) in Xenopus oocytes together with SOL-1 and Ce STG-1. We observed two major changes in the glutamate-gated current: the oocytes had a substantial leak current (data not shown) and the kinetics of desensitization were markedly slowed (Fig. 4 B and D).
In contrast to GLR-1 or GLR-1(Q552Y), where we observed no current in the absence of Ce STG-1, we found that GLR-1(A687T) partially overcame the requirement for Ce STG-1. Thus, we observed large glutamate-gated currents in oocytes that expressed GLR-1(A687T) and SOL-1 (Fig. 4 B and D). In the presence of Ce STG-1, the current only slowly desensitized whereas, in the absence of Ce STG-1, the current fully desensitized during the time course of glutamate application. These results indicate that Ce STG-1 regulates receptor gating and provide additional evidence that GLR-1 is expressed on the cell surface in the absence of Ce STG-1.
To test whether Q552Y and A687T influenced different processes in GLR-1 receptor gating, we examined the effects of introducing both Q552Y and A687T mutations into GLR-1. The kinetics of desensitization of glutamate-gated current were markedly slowed in oocytes that coexpressed GLR-1(Q552Y; A687T), Ce STG-1, and SOL-1 (Fig. 4 C and D). In contrast to either single mutant alone, glutamate-gated currents recorded from oocytes that expressed GLR-1(Q552Y; A687T) and SOL-1 had slow kinetics of desensitization (Fig. 4 C and D). Thus, introducing two independent mutations into GLR-1, both of which affect receptor kinetics, had a nonadditive effect on receptor properties allowing function in the absence of Ce STG-1. Again, these data argue that GLR-1 must be expressed on the cell surface in the absence of Ce STG-1, because amino acid changes in GLR-1 that are known to affect receptor function rather than expression are sufficient to restore glutamate-gated current.
Vertebrate Stargazin, Apis STG1, Dro STG1, and C. elegans STG-1 Have Different Effects on Invertebrate Glutamate-Gated Currents.
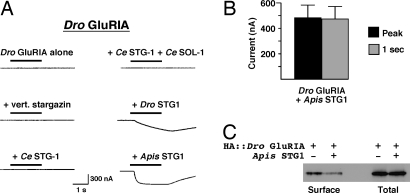
In Drosophila, a number of iGluRs have been cloned, including Dro GluRIA, which has significant identity with C. elegans GLR-1 and is expressed in the nervous system (3). However, only small kainate-gated and virtually no glutamate-gated currents can be recorded from oocytes that express Dro GluRIA (3). We also observed no glutamate-gated current when Dro GluRIA was expressed alone. We tested whether the lack of current was secondary to a requirement for a stargazin-like auxiliary protein. Unlike GLR-1-mediated currents (Fig. 3A), coexpression of Dro GluRIA with vertebrate stargazin, Ce STG-1, or SOL-1 and Ce STG-1 did not significantly increase glutamate-gated current (Fig. 5A). However, we were able to record small currents with coexpression of Dro GluRIA and Dro STG1, and significantly larger currents with coexpression of Dro GluRIA and Apis STG1 (Fig. 5 A and B). The onset of the current observed with coexpression of Dro STG1 was very slow, indicating that the majority of the current is likely a consequence of secondary activation of an endogenous Ca2+-activated chloride conductance (33). Consistent with this idea, substitution of barium for calcium in the extracellular solution dramatically reduced the amplitude of currents in response to glutamate application (data not shown). Our results indicate that there are important functional differences between vertebrate, C. elegans, and insect stargazin molecules in their ability to promote Dro GluRIA-mediated current. The large glutamate-gated current observed with coexpression of Apis STG1 could not be explained by surface delivery of receptors; in fact, we found that the fractional surface expression of Dro GluRIA alone was 2- to 3-fold higher than when coexpressed with Apis STG1 (Fig. 5C).
Fig. 5.
Apis and Drosophila STG1 differentially affect Dro GluRIA. (A) Currents measured in response to 1 mM glutamate application in Xenopus oocytes that expressed Dro GluRIA and various combinations of vertebrate stargazin, Ce STG-1, Dro STG1, Apis STG1, and Ce SOL-1. Oocytes were voltage-clamped at −70 mV. (B) Average peak current (black) and current amplitude 1 s after glutamate application (gray) for currents mediated by Dro GluRIA coexpressed with Apis STG1, n = 10. (C) Western blot showing the relative surface expression of HA::Dro GluRIA in the presence or absence of Apis STG1.
Discussion
Two explanations are commonly put forth when receptors at the surface do not respond to a candidate ligand: either the ligand is inappropriate or the receptor requires one or more auxiliary subunits. We have shown that the latter is true for C. elegans GLR-1 and Drosophila GluRIA, both of which require a stargazin-like molecule for function. We have identified stargazin homologues in C. elegans (Ce STG-1), the honey bee (A. mellifera, Apis STG1), and Drosophila (Dro STG1), and demonstrated a conserved function in promoting glutamate-gated currents. We noted considerable expression of GLR-1 at the cell surface even in the absence of Ce STG-1, yet cell surface GLR-1 receptors produced significant responses to glutamate only in the presence of Ce STG-1. This result provides strong evidence that Ce STG-1 is critically required for some aspect of GLR-1 receptor function. Furthermore, the kinetics of GLR-1 and vertebrate GluR1 glutamate-gated currents were dependent on the coexpressed stargazin homolog, suggesting that regulation of iGluR gating is a conserved feature of STG function.
We found that two mutations in GLR-1, Q552Y and A687T, slowed the desensitization of the receptor, but only one of these mutations, GLR-1(A687T), partially bypassed the requirement for Ce STG-1. Interestingly, this suppression, or genetic redundancy, may explain why stg-1 has not yet been identified in our screens for suppressors of the lurcher phenotype. Our results indicate that the molecular rearrangements that lead to receptor desensitization may be more complicated than commonly believed. The crystal structure of the extracellular domain of an iGluR subunit revealed that two extracellular domains, S1 and S2, are arranged in a clamshell-like arrangement that undergoes a conformational change upon binding of ligand that leads to opening of the channel pore (34–36). A secondary rearrangement of the receptor dimer interface leads to the desensitization of the receptor (37). These rearrangements may in turn be modified by interactions with stargazin-like molecules. Interestingly, the currents measured with the double GLR-1(Q552Y; A687T) mutant seem essentially independent of Ce STG-1, indicating that these two mutations together bypass the functional effects of Ce STG-1.
Recently, several reports have suggested that vertebrate stargazin has a role in modulating AMPAR function in addition to its primary role in promoting surface expression (20–23). In contrast, our results indicate that invertebrate stargazin homologues have primarily a functional role, suggesting perhaps that the most evolutionarily conserved function of stargazin is to regulate receptor gating. In vertebrates, four TARPs have been identified, and these proteins seem to have redundant function but differential distribution in the central nervous system (16). Because the neuronal distribution of Ce STG-1 does not completely overlap that of GLR-1, we predict that other stargazin-like proteins might also be expressed in C. elegans.
The dependence of AMPAR function on the auxiliary proteins SOL-1 and Ce STG-1 provides for stringent control of glutamate-gated neurotransmission. Based on our findings in oocytes, we would predict that the synaptic currents mediated by GLR-1 receptors in the absence of these regulatory molecules would be dramatically reduced. In support of this hypothesis, synaptic communication and behavior are disrupted in sol-1 mutants (9, 12). The requirement for auxiliary subunits may provide a mechanism for protecting neurons from possible excitotoxicity or inappropriate depolarization by limiting functional receptors to the synapse.
AMPARs have a critical role in long-term potentiation (LTP), a cellular model of learning and memory. During LTP, the number of functional AMPARs is increased at synapses, thus increasing synaptic strength. A recent study of AMPAR trafficking predicted that “placeholder proteins” would regulate the number of functional receptors at the synapse (38). Our findings suggest that SOL-1 and Ce STG-1 may comprise or contribute to this placeholder function and conversely, that drugs that interfere with the function of these auxiliary proteins may have relatively selective modulatory effects on synaptic function.
Materials and Methods
General Methods and Strains.
All strains were raised at 20°C under standard conditions. Germ-line transformation was achieved as described by using the lin-15 clone pJM23 (40 ng·μl−1) as a transformation marker (5). lin-15(n765ts) mutants were used in all transgenic experiments and expressed one of the following extrachromosomal arrays: akEx478, pDM649 (Pstg-1::GFP); or akEx485, pDM666 (Pstg-1::YFP) plus pYZ262 (Pglr-1::CFP). We isolated full-length stg-1 and Apis stg1 by PCR amplification from C. elegans, and A. mellifera first-strand cDNAs (GenBank accession nos. DQ015968 and DQ015969). Drosophila stg1 cDNA (GenBank accession no. CG33670) was amplified from the di-cistronic clone GH12419 (AY122110). Analysis of predicted proteins was facilitated by the expasy suite of programs and clustalw (39).
Plasmid Constructs.
The oocyte expression plasmids were as follows: pSP64T, Drosophila GluRIA; p59/2-rat GluR1; pDM657, C. elegans glr-1; pDM862, C. elegans glr-1(A687T); pDM858, C. elegans glr-1(Q552Y); pDM863, C. elegans glr-1(Q552Y;A687T); pGEMHE-mouse stargazin; pDM921, A. mellifera stg1; pDM1035, Drosophila melanogaster stg1; pDM654, C. elegans stg-1; pDM350, C. elegans sol-1. Additional plasmids used for coimmunoprecipitation and surface labeling experiments were: pYZ253, HA::GLR-1; pDM473, HA::NMR-1; pDM1043, STG-1::MYC; pCSW141, HA::GLR-1; and pDM1009, HA::Dro GluRIA.
Immunoprecipitation.
HEK 293 cells (TSA201 cells, a gift from M. F. Sheets, Cardiovascular Research and Training Institute, University of Utah, Salt Lake City) were cultured in DMEM nutrient medium with 10% bovine fetal serum. The cells were transiently transfected by using Lipofectamine 2000 (Invitrogen). After 48 h, transfected cells were collected and lysed in ice-cold immunoprecipitation (IP) buffer [25 mM Tris, pH 7.4/150 mM NaCl/10% glycerol/1% Triton X-100/protease inhibitor mixture (Roche Diagnostics)]. The lysates were incubated with agarose-conjugated antibodies (Santa Cruz Biotechnology) in IP wash buffer (25 mM Tris, pH 7.4/150 mM NaCl/7% glycerol/1% Triton X-100/1% sodium deoxycholate/0.1% SDS/protease inhibitor mixture (Roche). Samples were washed six times in ice-cold IP wash buffer and then boiled in SDS/PAGE sample buffer for 5 min. The precipitated proteins were resolved on a 7% SDS/PAGE gel, transferred to nitrocellulose membranes, probed with either mouse anti-HA (12CA5) or anti-MYC (9E10) primary antibodies (University of Utah Antibody Core Facility), followed by peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch), and analyzed with the SuperSignal West Femto Chemiluminescence kit (Pierce).
Microscopy.
GFP, CFP, and YFP images from transgenic worms were acquired by using confocal microscopy with a Zeiss LSM 510 microscope.
Electrophysiological Studies.
Xenopus oocyte recordings were carried out by using standard two-electrode voltage clamp as described (40). Statistical significance was determined by using the standard Student’s t test. Error bars represent the SEM.
Surface Protein Detection.
HA::GLR-1 and HA::Dro GluRIA surface expression in Xenopus oocytes followed previously published protocols (40, 41). Proteins were resolved on a 7% SDS/PAGE gel, probed with mouse anti-HA antibody at 1:1,000 (University of Utah Antibody Core Facility), followed by peroxidase-conjugated goat anti-mouse secondary antibody at 1:10,000 (Jackson ImmunoResearch), and analyzed as indicated above. Signal densities were quantified by scanning the film followed by densitometry of the bands using imagej. The total (loading control) is 3% of the amount that was loaded in the surface lane.
Acknowledgments
We thank M. Vetter and members of the Maricq laboratory for comments on the manuscript, L. Jack for help generating transgenic strains, Heinrich Betz and Bertram Schmitt (Max Planck Institute, Frankfurt) for the Dro GluRIA clone, Michael Hollmann (Ruhr-Universität Bochum, Bochum, Germany) for the vertebrate GluR1 clone and pSGEM, David Bredt and Roger Nicoll (University of California, San Francisco) for the vertebrate stargazin clone, Gene Robinson and Thomas Newman (Universtiy of Illinois at Urbana-Champaign, Urbana) and Ryszard Maleszka (Australian National University, Canberra) for Apis first-strand cDNA, and Andrew Fire (Standford Universtiy, Stanford, CA) for C. elegans expression vectors. We thank the Caenorhabditis Genetics Center [funded by the National Institutes of Health (NIH)] for providing worm strains and the Drosophila Genomics Resource Center (Indiana University) for Drosophila cDNA. This research was made possible by support from the Burroughs Wellcome Foundation and NIH Grants NS35812 (to A.V.M.) and DA016754 (to M.M.F.) and by the Deutsche Forschungsgemeinschaft (N.S.-S.).
Abbreviations
- iGluR
ionotropic glutamate receptor
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- TARP
transmembrane AMPA receptor regulatory protein
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein.
Footnotes
References
- 1.Dingledine R., Borges K., Bowie D., Traynelis S. F. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Hollmann M., O’Shea-Greenfield A., Rogers S. W., Heinemann S. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- 3.Ultsch A., Schuster C. M., Laube B., Schloss P., Schmitt B., Betz H. Proc. Natl. Acad. Sci. USA. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart A. C., Sims S., Kaplan J. M. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 5.Maricq A. V., Peckol E., Driscoll M., Bargmann C. I. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 6.Usherwood P. N., Machili P., Leaf G. Nature. 1968;219:1169–1172. doi: 10.1038/2191169a0. [DOI] [PubMed] [Google Scholar]
- 7.Patlak J. B., Gration K. A., Usherwood P. N. Nature. 1979;278:643–645. doi: 10.1038/278643a0. [DOI] [PubMed] [Google Scholar]
- 8.Mellem J. E., Brockie P. J., Zheng Y., Madsen D. M., Maricq A. V. Neuron. 2002;36:933–944. doi: 10.1016/s0896-6273(02)01088-7. [DOI] [PubMed] [Google Scholar]
- 9.Zheng Y., Brockie P. J., Mellem J. E., Madsen D. M., Walker C. S., Francis M. M., Maricq A. V. Proc. Natl. Acad. Sci. USA. 2006;103:1100–1105. doi: 10.1073/pnas.0504612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockie P. J., Maricq A. V. Neurosignals. 2003;12:108–125. doi: 10.1159/000072159. [DOI] [PubMed] [Google Scholar]
- 11.de Bono M., Maricq A. V. Annu. Rev. Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y., Mellem J. E., Brockie P. J., Madsen D. M., Maricq A. V. Nature. 2004;427:451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
- 13.Nicoll R. A., Tomita S., Bredt D. S. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto K., Fukaya M., Qiao X., Sakimura K., Watanabe M., Kano M. J. Neurosci. 1999;19:6027–6036. doi: 10.1523/JNEUROSCI.19-14-06027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Chetkovich D. M., Petralia R. S., Sweeney N. T., Kawasaki Y., Wenthold R. J., Bredt D. S., Nicoll R. A. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 16.Tomita S., Chen L., Kawasaki Y., Petralia R. S., Wenthold R. J., Nicoll R. A., Bredt D. S. J. Cell Biol. 2003;161:805–816. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell E., Sizemore M., Karimzadegan S., Chen L., Bredt D. S., Nicoll R. A. Proc. Natl. Acad. Sci. USA. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chetkovich D. M., Chen L., Stocker T. J., Nicoll R. A., Bredt D. S. J. Neurosci. 2002;22:5791–5796. doi: 10.1523/JNEUROSCI.22-14-05791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredt D. S., Nicoll R. A. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 20.Yamazaki M., Ohno-Shosaku T., Fukaya M., Kano M., Watanabe M., Sakimura K. Neurosci. Res. 2004;50:369–374. doi: 10.1016/j.neures.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Priel A., Kolleker A., Ayalon G., Gillor M., Osten P., Stern-Bach Y. J. Neurosci. 2005;25:2682–2686. doi: 10.1523/JNEUROSCI.4834-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita S., Adesnik H., Sekiguchi M., Zhang W., Wada K., Howe J. R., Nicoll R. A., Bredt D. S. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- 23.Turetsky D., Garringer E., Patneau D. K. J. Neurosci. 2005;25:7438–7448. doi: 10.1523/JNEUROSCI.1108-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brockie P. J., Madsen D. M., Zheng Y., Mellem J., Maricq A. V. J. Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomita S., Fukata M., Nicoll R. A., Bredt D. S. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa T., Cheng Y., Ramm E., Sheng M., Walz T. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 27.Stern-Bach Y., Russo S., Neuman M., Rosenmund C. Neuron. 1998;21:907–918. doi: 10.1016/s0896-6273(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 28.Brockie P. J., Mellem J. E., Hills T., Madsen D. M., Maricq A. V. Neuron. 2001;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 29.Zuo J., De Jager P. L., Takahashi K. A., Jiang W., Linden D. J., Heintz N. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 30.Kohda K., Wang Y., Yuzaki M. Nat. Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 31.Klein R. M., Howe J. R. J. Neurosci. 2004;24:4941–4951. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taverna F., Xiong Z. G., Brandes L., Roder J. C., Salter M. W., MacDonald J. F. J. Biol. Chem. 2000;275:8475–8479. doi: 10.1074/jbc.275.12.8475. [DOI] [PubMed] [Google Scholar]
- 33.Galzi J. L., Devillers-Thiery A., Hussy N., Bertrand S., Changeux J. P., Bertrand D. Nature. 1992;359:500–505. doi: 10.1038/359500a0. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong N., Sun Y., Chen G. Q., Gouaux E. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 35.Armstrong N., Mayer M., Gouaux E. Proc. Natl. Acad. Sci. USA. 2003;100:5736–5741. doi: 10.1073/pnas.1037393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin R., Banke T. G., Mayer M. L., Traynelis S. F., Gouaux E. Nat. Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y., Olson R., Horning M., Armstrong N., Mayer M., Gouaux E. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 38.Shi S., Hayashi Y., Esteban J. A., Malinow R. Cell. 2001;105:331–343. doi: 10.1016/s0092-8674(01)00321-x. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strutz-Seebohm N., Werner M., Madsen D. M., Seebohm G., Zheng Y., Walker C. S., Maricq A. V., Hollmann M. J. Biol. Chem. 2003;278:44691–44701. doi: 10.1074/jbc.M305497200. [DOI] [PubMed] [Google Scholar]
- 41.Chen L., El-Husseini A., Tomita S., Bredt D. S., Nicoll R. A. Mol. Pharmacol. 2003;64:703–706. doi: 10.1124/mol.64.3.703. [DOI] [PubMed] [Google Scholar]