Abstract
The biogenesis of the photosynthetic thylakoid membranes inside plant chloroplasts requires enzymes at the plastid envelope and the endoplasmic reticulum (ER). Extensive lipid trafficking is required for thylakoid lipid biosynthesis. Here the trigalactosyldiacylglycerol2 (tgd2) mutant of Arabidopsis is described. To the extent tested, tgd2 showed a complex lipid phenotype identical to the previously described tgd1 mutant. The aberrant accumulation of oligogalactolipids and triacylglycerols and the reduction of molecular species of galactolipids derived from the ER are consistent with a disruption of the import of ER-derived lipids into the plastid. The TGD1 protein is a permease-like component of an ABC transporter located in the chloroplast inner envelope membrane. The TGD2 gene encodes a phosphatidic acid-binding protein with a predicted mycobacterial cell entry domain. It is tethered to the inner chloroplast envelope membrane facing the outer envelope membrane. Presumed bacterial orthologs of TGD1 and TGD2 in Gram-negative bacteria are typically organized in transcriptional units, suggesting their involvement in a common biological process. Expression of the tgd2-1 mutant cDNA caused a dominant-negative effect replicating the tgd2 mutant phenotype. This result is interpreted as the interference of the mutant protein with its native protein complex. It is proposed that TGD2 represents the substrate-binding or regulatory component of a phosphatidic acid/lipid transport complex in the chloroplast inner envelope membrane.
Keywords: Arabidopsis, membrane transporter, glycerolipids, ABC transporter, substrate, binding protein
As plant leaves expand the demand on the lipid biosynthetic machinery is high because leaf cells contain one of the most extensive membrane systems found in nature, the photosynthetic thylakoid membrane of chloroplasts. The most abundant thylakoid lipids include nonphosphorous galactolipids. Galactolipid biosynthesis involves the formation of phosphatidic acid (PA) in the plastid and at the endoplasmic reticulum (ER) in many plants (1, 2), including Arabidopsis. Fatty acids derived from de novo synthesis in the plastid are assembled into PA in the plastid or at the ER. In Arabidopsis, diacylglycerols derived from the plastid pathway or the ER pathway are present in galactolipids in approximately equal proportion (3). The Arabidopsis lipid galactosyltransferases MGD1 and DGD1, which successively galactosylate diacylglycerol, are associated with the inner and the outer chloroplast envelope membranes, respectively (4). The topology of the galactolipid biosynthetic machinery and the involvement of the ER pathway require extensive subcellular lipid trafficking, most of which is mechanistically not understood.
To date, two mutants of Arabidopsis have been described that affect lipid trafficking from the ER to the plastid. The act1(ats1) mutant is deficient in the plastidic glycerol 3-phopshate acyltransferase, and most of the galactolipids in this mutant are derived from the ER pathway (5). In contrast, galactolipids in the tgd1 mutant are primarily derived from the plastid pathway (6). This mutant shows a complex lipid phenotype, the accumulation of oligogalactolipids giving rise to its name, trigalactosyldiacylglycerol1, the accumulation of triacylglycerols in the leaves, a 5-fold increase in PA content, and an increase of 16-carbon fatty acids in the galactolipids, indicative of a change in molecular species toward those formed de novo in the plastid (6, 7). Pulse–chase labeling of leaves also was consistent with a disruption of the transfer of lipid molecular species from the ER to the plastid in the tgd1 mutant (6). Isolated tgd1 chloroplasts showed a decreased rate of conversion of labeled PA into galactolipids (7). The TGD1 protein resembles the permease component of bacterial ABC transporters and was shown to be an integral component of the inner chloroplast envelope membrane (7). Based on this wealth of data it was proposed that TGD1 is a component of a PA transporter in the inner chloroplast envelope and that it is crucial for the biosynthesis of ER-derived molecular species of galactolipids. Stronger alleles of tgd1 led to increased embryo arrest and seed abortion, suggesting that the affected biological process is essential. Here the trigalactosyldiacylglycerol2 (tgd2) mutant of Arabidopsis, the identification of the TGD2 gene, and the characterization of the TGD2 protein are described.
Results
Isolation of the tgd2-1 Mutant.
The tgd2-1 mutant was identified during a suppressor screen in the dgd1 mutant background using a chemically induced mutant population (6). The dgd1 mutant is deficient in DGD1 responsible for the bulk of digalactolipid biosynthesis (8). Presence of the tgd2-1 mutation in the dgd1 background partially alleviated the digalactolipid deficiency and caused the accumulation of a lipid cochromatographing with trigalactosyldiacylglycerol diagnostic for all tgd mutants. Crossing the double-homozygous dgd1/tgd1-1 and dgd1/tgd2 mutants gave rise to uniform plants in the F1 generation with a homozygous dgd1-like phenotype, suggesting that tgd1-1 and tgd2-1 are not allelic. The tgd2-1/dgd1 homozygous double mutant was crossed to Arabidopsis wild-type, ecotype Columbia-2 (Col-2). The F1 plants showed a wild-type lipid phenotype confirming that the tgd2-1 mutant allele is recessive. After selfing and lipid analysis, F2 plants homozygous at the tgd2-1 locus were genotyped at the DGD1 locus by using a derived cut amplified polymorphic sequence (dCAPS) marker (6) to test for loss of the dgd1 mutation. A homozygous tgd2-1 mutant line was backcrossed with wild type (Col-2) three times to reduce the chance of secondary mutations. Unless indicated otherwise, further analysis was done with this tgd2-1 mutant in the wild-type background.
The Phenotypes of tgd2-1 and tgd1-1 Are Nearly Identical.
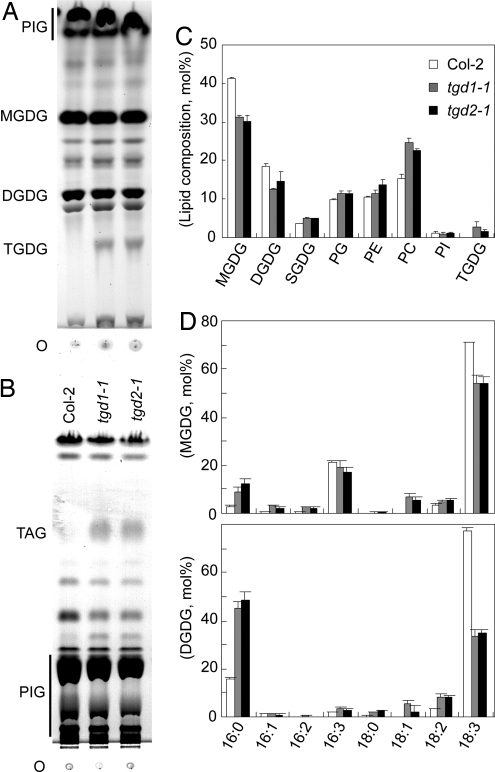
Compared to the wild type, tgd2-1 plants were consistently smaller and slightly pale, as was observed for the tgd1-1 mutants (7). Chlorophyll contents were reduced to a similar extent in the tgd1-1 and tgd2-1 mutants [chlorophyll (Chl) per gram of fresh weight (FW) ± SD, n = 4: wild type, 1,136 ± 138 μgChl·g−1FW; tgd1-1, 553 ± 115 μgChl·g−1FW; tgd2-1, 656 ± 145 μgChl·g−1FW]. Leaf lipid extracts of the wild type and the tgd1-1 and tgd2-1 mutants were compared by TLC. In the tgd2-1 sample a lipid staining positive for sugar and cochromatographing with authentic trigalactolipid of tgd1-1 (6) is present (Fig. 1A). A lipid cochromatographing with authentic triacylglycerol accumulating in tgd1-1 leaves was present in the tgd2-1 sample as well (Fig. 1B). Quantitative analysis of the polar lipids indicated similar changes in the two mutants with relative amounts of the monogalactolipid and digalactolipid reduced and relative amounts of phosphatidylcholine increased (Fig. 1C). In addition, trigalactolipid was present to a similar extent in both mutants (tgd1-1, 2.7 ± 1.4 mol%; tgd2-1, 1.6 ± 0.4 mol%; n = 4; data are ± SD) but was not detectable in the wild type. Analyzing the fatty acid composition of the two galactolipids indicated a reduction of 18-carbon fatty acids and an increase in 16-carbon fatty acids (Fig. 1D) to the same extent in both mutants. These overall fatty acid compositions for the tgd2-1 mutant imply a change in molecular species distribution in the two galactolipids consistent with a reduction of molecular species derived from the ER pathway as determined in detail for the tgd1-1 mutant (6). In addition, similar to the tgd1-1 mutant carrying a weak chemically induced mutant allele, the tgd2-1 mutant produced a fraction (≈43%, 281 of 651 in a representative sample) of aborted seeds.
Fig. 1.
Lipid phenotype of the tgd2-1 mutant compared with the tgd1-1 mutant and the Col-2 wild type. (A) Thin-layer chromatogram of polar lipids. Lipids were visualized by α-naphthol staining. (B) Thin-layer chromatogram of neutral lipids. Lipids were visualized by exposure to iodine vapor. (C) Polar lipid composition (relative mol%) determined by quantification of fatty acid methylesters derived from individual lipids. (D) Fatty acid composition of the two galactolipids MGDG and DGDG. The legend is the same as that for C. Fatty acids are indicated with number of carbons:number of double bonds. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; O, origin; PC, phosphatidylcholine; PE phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PIG, pigments; SQDG, sulfoquinovosyldiacylglycerol; TAG, triacylglycerol; TGDG, trigalactosyldiacylglycerol.
The TGD2 Gene Is Identical with Arabidopsis Gene At3g20320.
In a mapping population of 93 homozygous tgd2-1 F2 mutant plants (186 chromosomes) from a cross between the homozygous tgd2-1 mutant in the dgd1(Col-2) background and a plant from the ecotype Landsberg erecta the tgd2-1 mutant locus was mapped close to cut amplified polymorphic sequence (CAPS) marker ARLIM15.1 (www.arabidopsis.org) at ≈30 cM on chromosome 3 (Fig. 2). In an enlarged F2 mapping population from the same cross (3,506 chromosomes) the tgd2-1 mutant locus was mapped to an ≈45-kb fragment flanked by CAPS marker MQC12-3 and dCAPS marker MQC12-4 (Fig. 2A). This region falls onto the Arabidopsis bacterial artificial chromosome clone MQC12 (GenBank accession no. AB024036) and encompasses 14 predicted or confirmed genes (At3g20270–At3g20390). Notably, the translation product of At3g20320 was similar to the ttg2C protein (GenBank accession no. AAD17959; 25.0% identity over >100 aa) of Pseudomonas putida. This protein is predicted to be the substrate-binding protein of an ABC transporter, and its ORF is flanked by one encoding the ABC transporter permease ttg2B (GenBank accession no. AAD17958). Most notably, the Arabidopsis TGD1 protein is similar to ttg2B (29.6% identity over >100 aa) of P. putida. The predicted bacterial ABC transporter encoded by the ttg2 operon in P. putida has been genetically implicated in toluene resistance (9). The At3g20320 cDNA sequence obtained by RT-PCR from the Arabidopsis tgd2-1 mutant contained a G-to-A mutation (Fig. 2A) corresponding to position 7,088,870 of the assembled chromosome 3 sequence (GenBank accession no. NC_003074) and leading to a glycine-to-arginine change in the amino acid sequence. This mutation was confirmed by designing a tgd2-1 allele-specific dCAPS marker that was later used for genotyping (Fig. 2D).
Fig. 2.
Identification of the TGD2 locus. (A) Map position of the tgd2-1 mutation on chromosome 3 and structure of the TGD2 gene (At3g20320). Markers used for mapping and the respective number of recombinations are indicated. The TGD2 gene is indicated by a black box and expanded on the lowest line. The coding region of At3g20320 is shown as a shaded box. The darker shading indicates the predicted TMD. A region encoding an MCE domain is shown hashed. Introns are indicated by a line. Noncoding regions of the gene deduced from the cDNA are shown as open boxes. (B) Growth of different plants on soil (8 weeks old) with a genotype as indicated below the panel. Mutants were homozygous at all indicated loci. Three plants from independent transformation events expressing the TGD2 cDNA are indicated by “(c).” (C and D) Genotyping at the DGD1 locus (C) and at the TGD2 locus (D). Point mutation-specific dCAPS markers were used, and ethidium bromide-stained DNA diagnostic DNA fragments are shown with their respective lengths in base pairs. (E) Lipid phenotype of the six different plant lines. A section of thin-layer chromatogram stained for glycolipids is shown. DGDG, digalactosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
The TGD2 Protein Contains a Mycobacterial Cell Entry (MCE) Domain.
The TGD2 ORF of 1,146 bp (Fig. 2A, bottom) encodes a protein of 41.6 kDa. In addition to the similarity to bacterial substrate binding proteins, the TGD2 protein contains a MCE domain (amino acids 99–216). This domain is found in surface proteins of pathogenic mycobacteria. These proteins are essential virulence factors proposed to facilitate the bacterial entry into mammalian host cells (10). The mutation in tgd2-1 affects amino acid 234 just outside this MCE domain. A transmembrane-spanning domain (TMD) in TGD2 (amino acids 96–118) was predicted by using sosui software (11). A chloroplast targeting peptide of 45 N-terminal amino acids was predicted (score 0.545) by using chlorop with default settings (12).
Expression of the TGD2 cDNA Reverses the tgd2-1 Mutant Phenotype.
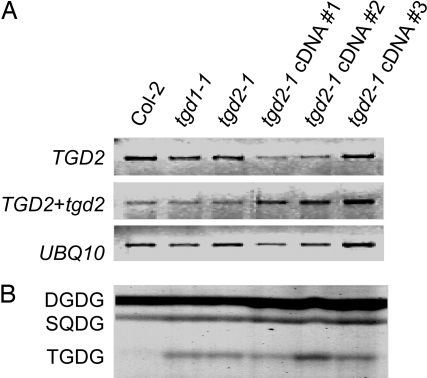
The tgd2-1 mutation in the dgd1 mutant background led to increased growth compared with the homozygous dgd1 mutant (Fig. 2B). This phenotype was reversed by expression of the wild-type TGD2 cDNA under the control of the 35S-CMV (cauliflower mosaic virus) promoter in the tgd2-1/dgd1 homozygous double mutant (Fig. 2B). The genotypes were confirmed by using mutant allele-specific dCAPS markers (Fig. 2 C and D). In the sample from the transgenic lines, two bands were present for the wild-type cDNA and the tgd2-1 genomic mutant locus, respectively (Fig. 2D). Reversion of the digalactolipid and the trigalactolipid phenotype of the tgd2-1/dgd1 double mutant to the homozygous dgd1 phenotype was observed as well (Fig. 1E). This complementation analysis confirmed the identity of the TGD2 gene as At3g20320.
Expression of the tgd2-1 cDNA in the Wild Type Recapitulates the tgd2-1 Phenotype.
The similarity of tgd1-1 and tgd2-1 mutant phenotypes and the organization of predicted bacterial orthologs of these two Arabidopsis genes in operons suggested that TGD1 and TGD2 act together in the same cellular process possibly as part of a larger lipid transfer complex. Expression of the tgd2-1 mutant cDNA under the control of the 35S-CMV promoter in the wild type led to the accumulation of a lipid cochromatographing with the trigalactolipid accumulating in the tgd1-1 and tgd2-1 mutants (Fig. 3B). Semiquantitative RT-PCR confirmed that this effect was not due to cosuppression of the genomic wild-type TGD2 gene and the tgd2-1 cDNA expression construct, because RNA derived from both genes was abundant in the transgenic lines (Fig. 3A). One interpretation of this dominant-negative effect is that the tgd2-1-encoded mutant protein is impaired in its activity but can still become part of its native protein complex, thereby disrupting overall function of the process involving the complex. In addition, this result provided independent corroboration for the identity of TGD2 with At3g20320.
Fig. 3.
Expression of the tgd2-1 mutant cDNA in the Col-2 wild type. The untransformed wild type (Col-2) and the untransformed tgd1-1 and tgd2-1 mutants are included for comparison. Three independent transformants are shown. (A) Semiquantitative RT-PCR of mRNA levels derived from the TGD2 wild-type gene (Top), the TGD2 wild-type gene and the tgd2-1 transgene (Middle), and the ubiquitin (UBQ10) control (Bottom). Negative images of ethidium bromide-stained gels are shown. (B) Polar lipid phenotype of the indicated plants. A section of the thin-layer chromatogram stained for glycolipids is shown. DGDG, digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
The TGD2 Protein Is Localized in the Inner Chloroplast Membrane.
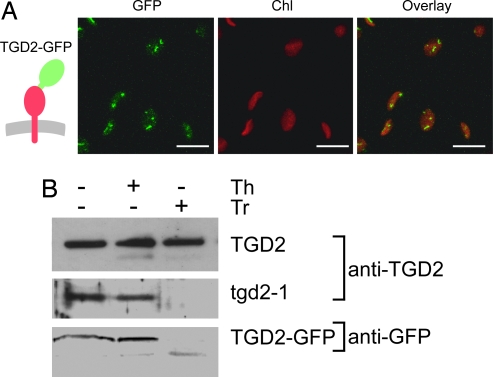
To determine the subcellular localization of the TGD2 protein, a construct encoding a full-length C-terminal fusion between the TGD2 protein and a GFP was transiently expressed in tobacco leaves. This TGD2–GFP fusion was visible in discrete spots at the periphery of chloroplasts (Fig. 4A). It should be noted that the equivalent experiment for the TGD1–GFP fusion construct showed a similar punctate fluorescence pattern at the chloroplast surface (7).
Fig. 4.
Subcellular localization and topology of TGD2 after transient expression in tobacco leaves. (A) Localization of full-length TGD2 protein fused to GFP (TGD2–GFP). The insertion of the respective protein into the membrane is schematically shown on the left. GFP, green fluorescence specific for GFP; Chl, red fluorescence of chloroplasts; the overlay of the two images is shown on the right. Confocal images are shown. (Scale bars: 10 μm.) (B) Topology of the TGD2 protein. The wild-type TGD2 protein, the tgd2-1 mutant protein, and the GFP fusion were transiently produced in tobacco leaves, and isolated chloroplasts were analyzed. The TGD2 and tgd2-1 proteins were detected by using a TGD2-specific antibody. The GFP fusion was detected by using a GFP-specific antibody. Samples were untreated with protease (−) or treated with thermolysin (+, Th) or with trypsin (+, Tr). Immunoblots are shown.
To further explore the association of the TGD2 protein with one of the two chloroplast envelope membranes and to determine its topology, chloroplasts were isolated from tobacco leaves expressing a wild-type TGD2 cDNA or the tgd2-1 mutant cDNA (Fig. 4B). The TGD2 wild type and the tgd2-1 mutant proteins were detected with a polyclonal antibody against TGD2. The chloroplasts were either untreated or treated with thermolysin, a protease unable to penetrate the outer envelope membrane, or trypsin, a protease able to penetrate the outer envelope but not the inner envelope membrane. Interestingly, the wild-type TGD2 protein was resistant to both proteases, whereas the mutant protein tgd2-1 was resistant to thermolysin but not trypsin (Fig. 4B Top and Middle). When the full-length wild-type TGD2 protein C-terminally fused to GFP was tested, the GFP tag detected by a GFP-specific antibody was resistant to thermolysin but not to trypsin (Fig. 4B Bottom). With the exception of the TGD2 wild-type protein, the result suggests that the TGD2 protein is associated with the inner envelope membrane with the C terminus facing the intermembrane space. The wild-type TGD2 is trypsin-resistant either because it is inside the plastid or, more likely, because it is in a complex or a membrane domain inaccessible to trypsin.
The TGD2 Protein Specifically Binds PA.
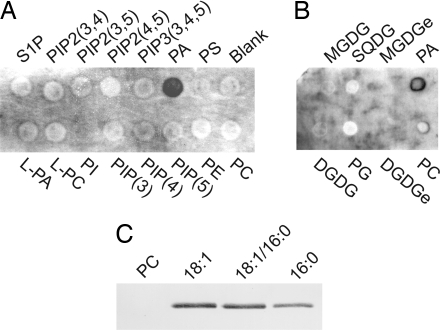
Because the TGD2 protein resembles substrate binding proteins of bacterial ABC transporters, and because the tgd2-1 phenotype was consistent with a defect in lipid transfer into the chloroplast, the TGD2 protein was tested for the specific binding of different lipids. To distinguish lipid binding to the TMD from lipid binding to a possible substrate site in the C-terminal domain, an N-terminally truncated version, TGD2-dTMD-His, was produced in Escherichia coli lacking the TMD. This protein was soluble. A C-terminal His tag was used for purification and detection of TGD2-dTMD-His by an anti-His tag antibody. A commercial membrane with different phospholipids and membranes with plant-specific lipids were used. Of all of the lipids tested, including diacylglycerol (data not shown), only PA bound to TGD2-dTMD-His (Fig. 5A). By employing an independent approach, the TGD2-dTMD-His protein bound phosphatidylcholine liposomes containing different molecular species of PA (Fig. 5B). Liposomes consisting of phosphatidylcholine alone did not bind. Binding was independent of the molecular species of PA at least at the semiquantitative immunoblot level. The results suggested that TGD2 contains a PA-specific binding domain in the C-terminal part of the protein.
Fig. 5.
Binding of recombinant TGD2 protein to lipids. The TGD2 protein is N-terminally truncated lacking the TMD to exclude lipid binding to this region of the protein. (A and B) Membrane binding assay with commercial phospholipid-containing membrane (A) or plant lipid-containing membrane (B). (C) Liposome binding assay. Liposomes consisted of phosphatidylcholine only (PC, first lane) or PC (60% wt/wt, second through fourth lanes) mixed with different molecular species of PA (40% wt/wt). PA molecular species tested were dioleoyl-PA (18:1), sn1-oleoyl, sn2-palmitoyl PA (18:1/16:0), and dipalmitoyl-PA (16:0). DGDG, prokaryotic digalactosyldiacylglycerol; DGDGe, eukaryotic digalactosyldiacylglycerol; L-PA, lysophosphatidic acid; L-PC, lysophosphatidylcholine; MGDG, prokaryotic monogalactosyldiacylglycerol; MGDGe, eukaryotic monogalactosyldiacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PIP(3), phosphatidylinositol 3-phosphate; PIP(4), phosphatidylinositol 4-phosphate; PIP(5), phosphatidylinositol 5-phosphate; PIP2(3,4), phosphatidylinositol 3,4-bisphosphate; PIP2(3,5), phosphatidylinositol 3,5-bisphosphate; PIP2(4,5), phosphatidylinositol 4,5-bisphosphate; PIP3(3,4,5), phosphatidylinositol 3,4,5-bisphosphate; PS, phosphatidylserine; S1P, sphingosine 1-phosphate; SQDG, sulfoquinovosyldiacylglycerol; TGDG, trigalactosyldiacylglycerol.
Discussion
Protein import into chloroplasts requires the interaction of protein complexes spanning the inner and outer chloroplast envelope membranes (13, 14). Currently, our knowledge about the protein import machinery dwarfs our mechanistic insights into lipid import into the plastid. Like protein import, the import of ER-derived lipids during chloroplast biogenesis is extensive and presumably requires transporters mediating the transfer of lipids between and through the involved membranes. The recently described TGD1 protein (6, 7) and the TGD2 protein discussed here have the hallmarks of components of a lipid transporter of the inner chloroplast envelope membrane. Although the analysis of the tgd1-1 mutant to date is far more extensive, it is apparent that the tgd2-1 mutation causes identical biochemical and physiological phenotypes: the accumulation of oligogalactolipids and triacylglycerols, the increase of 16-carbon fatty acids in plastid lipids indicative of reduced presence of ER-derived molecular species, and the increase in growth in the dgd1 background. We have not yet found a difference in phenotypes in the two tgd mutants, suggesting that the products of the two genes are involved in the same biological process, thylakoid lipid biosynthesis from ER-derived precursors.
The molecular analysis supports this interpretation. The TGD1 and TGD2 proteins are localized in the inner chloroplast envelope membrane, and expression of GFP fusions for both cause punctate fluorescence patterns in the periphery of plastids (7) (Fig. 4A). Moreover, the Arabidopsis TGD1 and TGD2 proteins are similar to the permease and substrate-binding protein of bacterial ABC transporters, respectively. The corresponding bacterial orthologs are found in clusters, which is a strong indicator for the function of the gene products in the same pathway or process and a key parameter used in metabolic reconstruction from bacterial genome sequences (15). Although unambiguous evidence for the direct interaction of TGD1 and TGD2 is not yet available, two findings suggest that TGD2 is active in a complex in Arabidopsis: (i) ectopic expression of the tgd2-1 mutant cDNA gives rise to the mutant phenotype, i.e., a dominant-negative mutation (Fig. 3); and (ii) the wild-type TGD2 protein is protected in isolated chloroplasts against trypsin whereas the TGD2 fusion protein is not (Fig. 4B). Both results can be interpreted as the association of the TGD2 protein with other proteins and/or specific lipid domains inaccessible to the protease.
The extensive investigation of the tgd1-1 mutant indicating the accumulation of PA and the reduced incorporation of PA into glycolipids of isolated plastids led to the suggestion that the TGD1 protein is a component of a PA transporter (7). Consistent with the proposed interaction of TGD1 and TGD2 in a PA-transporting complex, the recombinant TGD2 protein lacking the membrane-spanning domain was found to specifically bind PA (Fig. 5). An alternative interpretation would be that TGD2 binds PA as an effector molecule modulating the activity of TGD1. More intriguing, TGD2 could remove a PA molecule from the outer envelope membrane and make it available to TGD1 for import into the plastid and conversion by the plastidic PA phosphatase. Because TGD2 appears to be tethered with its membrane-spanning domain to the inner envelope membrane, the PA binding domain might reach out to the inside of the outer envelope membrane either locally fusing the two membranes or extracting an ER-derived PA. Although to date there is no direct evidence for this hypothesis, one intriguing observation in support is derived from mycobacterial orthologs of TGD2 required for cell entry of the bacterium (10). Recombinant bacterial orthologs can mediate the uptake of latex beads into mammalian cells, a process requiring an interaction of the protein on the bacterial surface with the mammalian cell membrane. The MCE domains present in the MCE proteins or bacterial substrate binding proteins associated with ABC transporters have been delineated based on sequence. The finding that TGD2 specifically binds PA, possibly through its MCE domain, might also shed light on the function of these important bacterial proteins and their interaction with membrane lipids.
Materials and Methods
Plant Material.
Arabidopsis thaliana plants were of the ecotypes Columbia-2 (Col-2) or Landsberg erecta (Ler). The tgd1-1 and dgd1 mutants were previously isolated by us (6, 16). Standard growth conditions were used for surface-sterilized seeds on agar-solidified MS (17) medium supplemented with 1% (wt/vol) sucrose or for plants grown on soil as previously described (18).
Lipid Analysis.
Lipids were extracted, and fatty acid methylesters were prepared and quantified by gas chromatography as previously described (6). Polar lipids were analyzed on activated ammonium sulfate-impregnated silica gel TLC plates (Si250PA; Mallinckrodt, Baker, NJ) by using a solvent system of acetone/toluene/water (90/30/7, vol/vol). Neutral lipids were separated on untreated TLC plates and developed with petroleum ether/ether/acetic acid (70/30/1, vol/vol). Lipids were visualized by brief exposure to iodine vapor or staining with α-naphthol to detect glycolipids (19).
Markers for Genetic Mapping and Genotyping.
For fine mapping, 10 CAPS markers (20) and 1 dCAPS (MQC12-4) marker (21) were generated, taking advantage of the Monsanto Polymorphism and Ler Sequence Collection (www.arabidopsis.org/Cereon/index.jsp). Primers and restriction enzymes were as follows: MYF24, 5′-GACAGCCCACAAATTGATGG-3′ and 5′-CACCAACGCTCAATGCCTAC-3′, HinfI; MLD14, 5′-GGGGTCCTTAAAATAGAGAC-3′ and 5′-GGCCTTTTGAGTTGGGAAAAG-3′, HindIII; MIL23, 5′-GGGGGTGATATCTATCGTAG-3′ and 5′-GCACCCTGGATATTCTTTCG-3′, HinfI; MPN9, 5′-CGGTCATATGCTGGCTGAAG-3′ and 5′-GACAGCACACAAGTTCCAGG-3′, AluI; MPN9-2, 5′-GTGCTATGGTTCAGGAGTTC-3′ and 5′-CTTACCAGCCATGACGATTC-3′, AccI; MAL21, 5′-GAGAAGAAACACCGATTCCG-3′ and 5′-GTTGTGATACGAATGGTGGC-3′, RsaI; K10D20, 5′-GGACCTGCCTTTCCCATATC-3′ and 5′-GCCCAAGCCTCAAGATGTTG-3′, HindIII; MSA6, 5′-GGAAGAGGGAGGTTTTGTTC-3′ and 5′-CCAATTCGTCTCCTTTTCACC-3′, SpeI; MQC12-2, 5′-GTGAGACCAACAGTGTCAAC-3′ and 5′-CCAC AATACACCACCACTTG-3′, HinfI; MQC12-3, 5′-CCTCCGTCTCATACATCTAC-3′ and 5′-CCAATTCGGTTTCATCCAATCCTCT-3′, BfaI; MQC12-4, 5′-CATATGCATTGATGATAACTGAAATCGA-3′ and 5′-CTTCTAGATCTCCTCCTTTC-3′, EcoRI.
For genotyping of the tgd2-1 mutant, a dCAPS marker was generated: 5′-TGATCGTTTGTGATAGGCAGCCTATAAAA-3′ and 5′-CCTTGCTTCCTCAATAACCG-3′, cut with EcoNI. The dgd1 dCAPS marker was described in ref. 6.
Complementation and Dominant-Negative Mutation Analysis.
The ORFs for TGD2 and tgd2-1 were isolated by RT-PCR from mRNA preparations by using RNeasy and Omniscript kits (Qiagen, Valencia, CA) and standard PCR conditions. The following primers were used: 5′-GTCGACATGATTGGGAATCCAGTAATTCAAG-3′ and 5′-GTCGACTCATAGTAGCCTGCTTAGGG-3′. The fragments were ligated into pGEM-T Easy (Promega) and sequenced at the Michigan State University Genomics and Technology Facility. The resulting plasmids were digested with SalI and inserted into pCAMBIAmcs1300 (7) followed by transformation into Agrobacterium. Plants were transformed by the floral-dip method (22) and screened by resistance to hygromycin (25 μg/ml) on agar-solidified MS medium.
For semiquantitative PCR of TGD2 and tgd2 transcripts the following primers were used: TGD2-specific primer set, 5′-CGGCTTGCTCAAGGAAGTTG-3′ and 5′-CCAGTCTAAAATCTACAGGCTG-3′; TGD2 and tgd2-1, 5′-TGATCGTTTGTGATAGGCAGCCTATAAAA-3′ and 5′-CCTTGCTTCCTCAATAACCG-3′; UBQ10 5′-TCAATTCTCTCTACCGTGATCAAGATGCA-3′ and 5′-GTGTCAGAACTCTCCACCTCAAGAGTA-3′. Isolation of RNA and reverse transcription were done as described above. Amplification conditions were as follows: 94°C for 3 min followed by 25 cycles at 94°C for 0.5 min, 55°C for 0.5 min, and 72°C for 0.5 min followed by 3 min at 72°C.
TGD2–GFP Fusion and in Vivo Chloroplast Import Assay.
The sequence encoding the full-length TGD2 protein was amplified from the pCAMBIAmcs1300 plasmid derivative mentioned above by PCR using the following primers: forward, 5′-GTCGACATGATTGGGAATCCAGTAATTCAAG-3′; reverse, 5′-GTCGACTAGTAGCCTGCTTAGGGATTTG-3′. The fragment was inserted into the pGEM-T Easy vector, sequenced and digested with SalI, and inserted into pCAMBIAmcsGFP (7). In vivo analysis of the GFP-tagged protein was done by confocal fluorescence microscopy (7).
In vivo chloroplast import analysis using transient expression of the constructs in tobacco leaves was carried out exactly as described in ref. 7. For immunodetection of the TGD2 or tgd2-1 proteins, a polyclonal antibody was raised in rabbits (Cocalico Biologicals, Reamstown, PA) against the truncated TGD2 protein used also for the lipid binding assay. The anti-serum was purified with a Melon Gel IgG Purification Kit (Pierce). For TGD2 immunodetection, the purified anti-TGD2 antibody was used at a 1:2,000 dilution. For GFP immunodetection, a rabbit anti-GFP antibody (Molecular Probes) was used at a 1:3,000 dilution. The antibodies were detected with an anti-rabbit horseradish peroxidase-coupled antibody (Bio-Rad) at a dilution of 1:60,000 followed by development with Chemiluminescent Peroxidase Substrate (Sigma).
Recombinant TGD2 Protein Production and Purification.
The sequence encoding N-terminally truncated TGD2-dTMD protein (from Gly-119 to stop codon) lacking the targeting peptide and the TMD was PCR-amplified by using primers 5′-GTCGACGGTTTTCAAATGCGGTCGAAG-3′ and 5′-GTCGACTCATAGTAGCCTGCTTAGGG-3′. This fragment was inserted into pPICT2 plasmid (23) and sequenced. After digestion with SalI, the insert was ligated into pQE31 (Qiagen). An overnight preculture of LB medium (1 ml) was used to start a 500-ml culture in M9 medium as described (24). The protein was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside at an OD600 of 0.4 at 22°C, and growth was continued overnight. Cultures were cooled to 4°C, washed twice, and resuspended in lysis buffer (50 mM Tris·HCl, pH 7.5/600 mM NaCl/20 mM imidazole). The suspensions were lysed by sonication followed by brief centrifugation at 1,500 × g to eliminate cell debris. The supernatants were centrifuged at 20,000 × g and applied to a Ni-NTA agarose column (Qiagen). The His-tagged protein was eluted with lysis buffer containing 250 mM imidazole. Samples were dialyzed in the lysis buffer lacking imidazole. Protein concentration was determined according to Bradford (25) by using BSA as a standard.
Lipid Binding Assays.
Membrane strips prespotted with lipids were purchased from Echelon Biosciences (Salt Lake City, UT). Prokaryotic phosphatidylcholine and PA were purchased from Avanti Polar Lipids. Prokaryotic monogalactolipid, digalactolipid, sulfolipid, and phosphatidylglycerol were purified from Synechocystis PCC6803 by TLC of lipid extracts (see Lipid Analysis). Eukaryotic monogalactolipid and digalactolipid was isolated from pea leaves. Approximately 5 μg of lipids were spotted onto a Hybond-C membrane (Amersham Pharmacia Biosciences). The membranes were first blocked with 3% BSA in TBST (10 mM Tris·HCl, pH 8.0/150 mM NaCl/0.1% Tween 20) for 1 h and incubated in 0.5 μg/ml TGD2 protein solution in the blocking buffer at 4°C overnight. The blots were washed five times with TBST and soaked in 3% BSA in TBST with a Penta-His mouse monoclonal antibody (Qiagen) at a 1:1,000 dilution at room temperature overnight. The membranes were washed twice with TBST and soaked in 3% BSA in TBST with alkaline phosphatase-conjugated anti-mouse antibody (Jackson ImmunoResearch) at a 1:5,000 dilution for 1 h at room temperature. After washing with TBST twice, the protein was detected by using the Immun-Star AP detection system (Bio-Rad).
The liposome binding assay was performed according to ref. 26. Lipids (phosphatidylcholine or a mixture of phosphatidylcholine and PA at 6:4 wt/wt) were incubated in TBS (50 mM Tris·HCl, pH 7/0.1 M NaCl) at 37°C for 1 h followed by vigorous vortexing for 5 min. The liposomes were precipitated at 20,000 × g and washed twice with ice-cold TBS. Liposomes (200 μg) were mixed with purified TGD2 protein lacking the TMD (10 μg/ml) and TBS to make 100 μl of solution. The mixture was incubated at 30°C for 30 min and washed twice with ice-cold TBS by centrifugation at 20,000 × g at 4°C. The liposome pellet mixed with sample buffer was analyzed by SDS/PAGE (27). Immunodetection of the His-tagged protein was accomplished by using the above-mentioned Penta-His antibody at 1:6,000 and the anti-mouse antibody at 1:10,000 dilution. The BCIP/NBT Kit from Bio-Rad was used for color detection.
Acknowledgments
This work was funded in part by U.S. Department of Energy Grant DE-FG02-98ER20305 and U.S. National Science Foundation Grant MCB-0453858 (to C.B.). K.A. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad from the Ministry of Education, Sports, Science, and Culture of Japan.
Abbreviations
- PA
phosphatidic acid
- ER
endoplasmic reticulum
- MCE
mycobacterial cell entry
- CAPS
cut amplified polymorphic sequence
- dCAPS
derived CAPS
- TMD
transmembrane-spanning domain.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. NM_202616).
References
- 1.Browse J., Somerville C. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991;42:467–506. [Google Scholar]
- 2.Roughan P. G., Slack C. R. Annu. Rev. Plant Physiol. 1982;33:97–132. [Google Scholar]
- 3.Browse J., Warwick N., Somerville C. R., Slack C. R. Biochem. J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benning C., Ohta H. J. Biol. Chem. 2005;280:2397–2400. doi: 10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- 5.Kunst L., Browse J., Somerville C. Proc. Natl. Acad. Sci. USA. 1988;85:4143–4147. doi: 10.1073/pnas.85.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C., Fan J., Riekhof W., Froehlich J. E., Benning C. EMBO J. 2003;22:2370–2379. doi: 10.1093/emboj/cdg234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu C., Fan J., Froehlich J., Awai K., Benning C. Plant Cell. 2005;17:3094–3110. doi: 10.1105/tpc.105.035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dörmann P., Balbo I., Benning C. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 9.Kim K., Lee S., Lee K., Lim D. J. Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chitale S., Ehrt S., Kawamura I., Fujimura T., Shimono N., Anand N., Lu S., Cohen-Gould L., Riley L. W. Cell. Microbiol. 2001;3:247–254. doi: 10.1046/j.1462-5822.2001.00110.x. [DOI] [PubMed] [Google Scholar]
- 11.Hirokawa T., Boon-Chieng S., Mitaku S. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 12.Emanuelsson O., Nielsen H., von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutensohn M., Fan E., Frielingsdorf S., Hanner P., Hou B., Hust B., Klosgen R. B. J. Plant Physiol. 2006;163:333–347. doi: 10.1016/j.jplph.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis P., Robinson C. Curr. Biol. 2004;14:R1064–R1077. doi: 10.1016/j.cub.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 15.Overbeek R., Begley T., Butler R. M., Choudhuri J. V., Chuang H. Y., Cohoon M., Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murashige T., Skoog F. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 18.Xu C., Härtel H., Wada H., Hagio M., Yu B., Eakin C., Benning C. Plant Physiol. 2002;129:594–604. doi: 10.1104/pp.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benning C., Huang Z. H., Gage D. A. Arch. Biochem. Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 20.Konieczny A., Ausubel F. M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 21.Neff M. M., Neff J. D., Chory J., Pepper A. E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 22.Clough S. J., Bent A. F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi J., Kii I., Sugiyama Y., Takeshita S., Kudo A. J. Bone Miner. Res. 2001;16:260–269. doi: 10.1359/jbmr.2001.16.2.260. [DOI] [PubMed] [Google Scholar]
- 24.Duffieux F., Annereau J. P., Boucher J., Miclet E., Pamlard O., Schneider M., Stoven V., Lallemand J. Y. Eur. J. Biochem. 2000;267:5306–5312. doi: 10.1046/j.1432-1327.2000.01614.x. [DOI] [PubMed] [Google Scholar]
- 25.Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Sano H., Kuroki Y., Honma T., Ogasawara Y., Sohma H., Voelker D. R., Akino T. J. Biol. Chem. 1998;273:4783–4789. doi: 10.1074/jbc.273.8.4783. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]