Abstract
Gene conversion is a common outcome of double-strand break (DSB) repair in yeast. Prior studies revealed that DSB-induced gene conversion tracts are often short (<53 bp), unidirectional, and biased toward promoter-proximal (5′) markers. In those studies, broken ends had short, non-homologous termini. For the present study we created plasmid × chromosome, chromosomal direct repeat and allelic recombination substrates in which donor alleles carried mutant HO sites (HOinc—not cleaved) at the same position as cleavable HO sites in recipient alleles. In these substrates, broken ends are almost completely homologous to donor alleles, differing only at the three HOinc mutations. These mutations serve as markers very close to, or within, the four-base overhang produced by HO nuclease. We identified extremely short tracts (<12 bp) and many tracts were highly directional, extending <2 bp on one side of the DSB. We thought that terminal homology would promote bidirectional tracts, but found instead that unidirectional tracts were more frequent. Interestingly, substrates with terminal homology displayed enhanced 3′ conversion, and in several cases conversion bias was reversed toward 3′ markers. These results are discussed in relation to factors that may influence tract length and directionality, including heteroduplex DNA formation, transcription, replication and mismatch repair.
INTRODUCTION
In the yeast Saccharomyces cerevisiae DNA double-strand breaks (DSBs) are usually repaired by homologous recombination resulting in gene conversion with or without an associated crossover (1); this repair mechanism is also common in higher eukaryotes (2,3). Gene conversion involves non-reciprocal information transfer from a donor to a recipient (broken) allele. Most or all DSB-induced meiotic and mitotic conversion in yeast involves mismatch repair of heteroduplex DNA (hDNA) (4–7). DSBs are processed to 3′ single-stranded tails, which are converted to Rad51 nucleoprotein filaments that can invade a homologous duplex elsewhere in the genome. Rad51 activity is facilitated by Rad52, Rad54, Rad55 and Rad57 (reviewed in 1). hDNA may be formed during strand invasion (synapsis) and by branch migration of Holliday junctions. Resolution of recombination intermediates includes mismatch repair of hDNA and/or resolution of Holliday junctions. In cases where alleles differ at multiple sites, conversion tracts are usually continuous (4,5), and this can be explained by a mechanism in which mismatched bases in hDNA are repaired in single excision repair tract, as in Escherichia coli (8,9).
When alleles are marked at multiple sites and events are initiated by a DSB at a specific site, several conversion tract parameters can be defined. The minimum tract length is defined as the distance between the most distal converted markers. Tracts can be continuous or discontinuous, and they can be uni- or bidirectional. Prior studies in yeast have shown that for ectopic DSB-induced gene conversion in plasmid × chromosome (P × C) and chromosomal direct repeat substrates, tracts are usually unidirectional and ∼15% are short (<53 bp in length) (10,11). In contrast, only 20% of allelic conversion tracts are unidirectional and the shortest tracts are at least 227 bp in length (7). In other studies of mitotic and meiotic allelic conversion, bidirectional tracts were generally less frequent (12–15) but this probably reflects the greater distances between DSBs and the closest flanking markers in those systems. Tract directionality likely reflects several factors, including end invasion (one- or two-ended), the extent of hDNA, and mismatch repair. End invasion might be affected by the presence of non-homologous sequences at ends (‘terminal non-homology’), as when DSBs are induced in HO sites inserted into target genes. Rad1/Rad10 endonuclease processes terminal non-homologies and this function becomes more important as the length of non-homology increases (16–18).
In this study we analyzed conversion tracts in P × C, direct repeat, and allelic substrates with markers very near or within the overhang produced by HO nuclease. To place markers this close to the DSB required mutant (uncleaved) HO sites in donor alleles. Thus, in these substrates broken ends were almost completely homologous to donor sequences, differing only at three bases within the HO site. We show that broken ends with terminal homology give rise to tracts that can be extremely short (<12 bp) and highly directional (extending <2 bp into homology on one side of the DSB). Prior studies in which broken ends had terminal non-homology showed marked biases toward conversion of promoter-proximal (5′) markers (7,10,11). Here we demonstrate the opposite conversion bias in P × C and direct repeat substrates when broken ends have terminal homology. The allelic substrate also showed enhanced 3′ conversion, but this was insufficient to reverse the 5′ bias. These results are discussed in relation to factors that may regulate gene conversion tract lengths and directionality.
MATERIALS AND METHODS
Plasmid DNAs and yeast strain construction
Plasmids were manipulated and prepared as described (19,20). Yeast culture and chromosome modification were described earlier (7,10,11). Yeast strains are listed in Table 1, and structures of chromosomal recombination substrates are shown in Figure 1A. Strains carry an HO nuclease gene regulated by the GAL1 promoter (GALHO). ura3 alleles were manipulated as 1.2 kb HindIII fragments. The ura3R-HO432 allele has a 24 bp HO recognition site present in an EcoRI linker in the natural NcoI site at position 432, and nine silent mutations that create restriction fragment length polymorphisms (RFLPs). Plasmid pUraRHO432TAC is a pUC19 derivative with ura3RHO432, TRP1, ARS1 and CEN4 (10). Plasmids bearing ura3 and HIS3 were constructed with HO sites, also located at position 432; these HO sites were identical to HO432 except that they carried point mutations which prevent cleavage by HO nuclease (HOinc sites). Each HOinc site has three mutations: HOinc2L has two mutations left, and one right, of the DSB; HOinc2R has two mutations right, and one left, of the DSB; HOincLMR has mutations left, middle and right of the DSB; the middle mutation is within the four base overhang produced by HO nuclease. The pUraHOincHis plasmids were transformed into strain DY3436 (11) to create strains with ura3-HOinc alleles (JN3449, RL3640 and RL3642) by two-step replacement as described (10). Transformation of ura3-HOinc strains with pUraRHO432TAC creates Trp+ strains with P × C recombination substrates (JN3450, RL3641 and RL3643). Plasmid pUCUraR-HO432Δ5′HLeu is a pUC19 derivative with ura3RHO432 and LEU2 (11). Transformation of JN3449 with pUCUraR-HO432Δ5′HLeu creates strain JN3457 carrying a direct repeat recombination substrate. The haploid P × C and direct repeat strains have MATa-inc alleles to prevent MAT cleavage and mating-type switching/diploidization.
Table 1. Yeast strains.
| Name | Genotype | Source or Reference |
|---|---|---|
| DY3026 | MATa-inc, ade2-101, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-X764 | (10) |
| DY3028 | MATa-inc, ade2-101, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-X432 | (10) |
| DY3031 | MATα, ade2-101, his3-200, lys2-801, trp1-Δ1, leu2-Δ1 | (7) |
| DY3059 | MATα, ade2-101, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-X764 | This study |
| JD1002 | Diploid product of DY3026 × DY3031 | This study |
| JD1003 | Diploid product of DY3028 × DY3031 | (7) |
| DY3064 | MATα, ade2-101, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1 | This study |
| DY3066 | MATa-inc, ade2-101, his3-200, lys2-801, trp1-Δ1, leu2-Δ1 | (7) |
| DY3093 | MATα, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-X764 | This study |
| DY3436 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, URA3 | (11) |
| JN3449 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOinc2L | This study |
| JN3450 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOinc2L: pUraRHO432TAC (P × C substrate) | This study |
| JN3457 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOinc2L-LEU2-ura3R-HO432 (direct repeat substrate) | This study |
| SP3469 | MATα, ade2-101, his3-200, lys2-801::pUCGALHO::LYS2, trp1-Δ1, leu2-Δ1, RscRI-ura3R-HO432-LEU2 | This study |
| SP3470 | MATa-inc, ade2-101, his3-200, lys2-801, trp1-Δ1, leu2-Δ1, RscRI-ura3-HOinc2L-LEU2 | This study |
| SP3471 | Diploid product of DY3028 × DY3031 switched to MATa-inc/MATa-inc (allelic substrate) | This study |
| RL3640 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOinc2R | This study |
| RL3641 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOinc2R: pUraRHO432TAC (P × C substrate) | This study |
| RL3642 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOincLMR | This study |
| RL3643 | MATa-inc, ade2-101, his3-200, lys2-801::pHSSGALHO::LYS2, trp1-Δ1, leu2-Δ1, ura3-HOincLMR: pUraRHO432TAC (P × C substrate) | This study |
Figure 1.
Recombination substrates. (A) P × C, direct repeat, and allelic recombination substrates created for this study each have a recipient ura3 with a functional HO site and nine RFLP markers (indicated by shading; see panel C), and a donor ura3 with an HOinc site at the same position. Three prior studies (7,10,11) employed P × C, direct repeat, and allelic substrates that yield broken ends with terminal non-homology; these have the same ura3 recipient allele as above, and ura3-X764 as donor. In these diagrams, black bars indicate insertion mutations that inactivate ura3. (B) Locations of mutations within HOinc sites. The sequence of MATa near the Y/Z junction is shown. HO nuclease cleaves at the arrows, producing a four-base, 3′ overhang. Eight bases required for cleavage by HO nuclease are marked with asterisks; shading indicates HOinc mutations used in this study. The three HOinc sites are diagrammed, and the specific base changes are listed at the bottom. (C) Map of the 1.2 kb ura3 fragment; silent RFLPs are shown below, wild-type sites above.
DY3064 and DY3066 are meiotic products of JD1003 (7). Plasmid RscRI is a transplacement vector containing 2.0 kb and 0.9 kb regions up- and downstream of URA3, plus LEU2 and pUC19 (7). ura3R-HO432 or ura3-HOinc were inserted into RscRI, and the resulting vectors were used to replace URA3 in strains DY3064 and DY3066 with pUC19-ura3RHO432-LEU2 or pUC19-ura3-HOinc-LEU2, respectively, as described (7). The resulting strains, SP3469 and SP3470, were mated to produce a MATa-inc/MATα strain that was subsequently converted to MATa-inc/MATa-inc by brief induction of GALHO (21), creating strain SP3471. The ura3 alleles in SP3471 were amplified by PCR and mapped to confirm that none of the ura3 markers were altered during MAT switching. DY3059, a meiotic product of JD1002, carries ura3 inactivated by a +1 frameshift that creates an XbaI site (X764) (10). DY3093 is a spontaneous ADE2 revertant of DY3059.
Recombination assays
DSB-induced gene conversion frequencies were measured by using a non-selective assay (7,11) modified as follows. Cells from 2-day-old patches of parent strains were inoculated into 1.5 ml of rich medium with 2% glycerol as sole carbon source (YPGly) and incubated for 24 h. Cultures were divided, cells were harvested by centrifugation, suspended in 1.5 ml of YPD (glucose-grown, uninduced control) or YPGal (with 2% galactose; HO nuclease-induced), grown for 6 h, and counted with a Coulter Counter. With P × C substrates, cells were plated on synthetic complete medium lacking tryptophan (ensuring plasmid retention). Trp+ colonies were mated with lawns of DY3093 on YPD, and Trp+ Ade+ diploids were selected on medium lacking tryptophan and adenine. The resulting colonies were replica-plated to medium with 2% galactose and lacking tryptophan and adenine, then to YPGal, and finally to uracil omission medium. The sequential selection of Trp+ Ade+ diploids helps eliminate growth of haploids, and growth on galactose medium re-induces GALHO. In non-recombinant (parental) colonies, re-induction of GALHO introduces DSBs into ura3R-HO432, stimulating recombination with the ura3-X764 allele from DY3093, and producing Ura+ papillae within each colony. In recombinant colonies, however, the plasmid-borne ura3R-HO432 is converted to HOinc and they retain the donor ura3-HOinc allele (on chromosome V). These ‘HOinc/HOinc’ products are not cleaved upon GALHO induction and they remain Ura– (‘Ura– non-inducible’). Recombination frequencies were calculated as the number of Ura– non-inducible colonies per Trp+ colony. Converted plasmids were rescued from the original Trp+ colonies, RFLPs were mapped as described (10), and conversion of the HOinc markers was determined by DNA sequencing. All products that failed to convert one or both RFLPs flanking HO432 (Pml409 and Stu463) were sequenced to determine whether one, two or all three HOinc markers were transferred from the donor HOinc (chromosomal) allele to the recipient (plasmid-borne) HO432 allele. Because most conversion tracts are continuous, products that converted both Pml409 and Stu463 were presumed to have converted all three HOinc mutations; this was confirmed in three independent products of this type (data not shown). This assay scores gene conversion without associated crossing over since crossovers integrate CEN plasmids and produce lethal dicentric chromosomes. To determine the rate of plasmid loss, cells were also plated on YPD and the resulting colonies were replica-plated to tryptophan omission medium; under these conditions, ∼25% of glucose or galactose-grown cells lose ARS1/CEN4 plasmids (data not shown).
With the direct repeat substrate, gene conversion was assayed as above except that cells were initially plated on leucine omission medium (deletions resulting from associated crossovers or single-strand annealing are Leu– and were not scored in this study). Conversion tracts were determined by RFLP mapping and sequencing of recipient alleles amplified by PCR. With the allelic substrate, cells were initially plated on YPD. Because these cells were MATa-inc/MATa-inc, mating with DY3093 produced triploids that could be selected and screened for recombinants by using the same strategy as above. Converted recipient alleles (linked to pUC19) were excised from genomic DNA by digestion with ClaI, circularized with T4 DNA ligase, transformed into E.coli as described (7), and conversion tracts were determined by restriction mapping and sequencing the rescued plasmids as above.
In most experiments, product independence was assured because only one product was isolated per induced population. When multiple products were isolated per population, statistical analysis indicated that >95% of products were independent. Cell viability was not measured because there is little or no killing upon DSB induction in strains with related sub strates (21, and unpublished data). As seen previously (21), occasional Ura– non-inducible colonies were non-recombinant, i.e. HO432 was intact. These likely acquired defects in the HO nuclease gene or the galactose regulatory system. Statistical analyses were performed with Fisher exact tests.
RESULTS
Experimental design
Prior studies indicated that DSB-induced gene conversion tracts were often unidirectional and that a significant fraction were quite short (<53 bp), particularly for ectopic events (P × C and direct repeat substrates) (7,10,11). To increase the resolution of these measurements, we devised a strategy to examine conversion of markers within the HO nuclease recognition sequence (HO site). We examined conversion tracts in P × C, direct repeat, and allelic recombination substrates (Fig. 1A). A 24 bp sequence from MAT is sufficient for HO cleavage in vivo (22). Within this sequence are eight non-contiguous bases that are essential for cleavage (23) (Fig. 1B); mutant sites are called HOinc following the nomenclature for MAT-inc (inconvertible) mutations (24). Recombination substrates had wild-type HO sites in one copy of ura3, and HOinc sites at the same position in the second copy of ura3. DSBs were created in HO sites upon induction of GALHO, converting HO/HOinc parents to HOinc/HOinc recombinants. Since parents and recombinants are both Ura–, they were distinguished in a second round of HO-induced recombination following introduction (by mating) of a third, heteroallelic copy of ura3 (Fig. 2). Recipient alleles were isolated from recombinant products either by plasmid rescue or by PCR. This strategy for creating and identifying recombinants was used with only minor variations for P × C, direct repeat and allelic recombination substrates (see Materials and Methods).
Figure 2.

Recombination assay. Parent cells carry one ura3 allele with a wild-type HO site and another with an HOinc site. Cells that do not suffer a DSB, or that repair the DSB by precise non-homologous end-joining (NHEJ), retain the parental structure (left). DSB repair by gene conversion creates HOinc/HOinc recombinants (right). Cells are mated to DY3093 to introduce a third, heteroallelic copy of ura3 (ura3-X764). Reinduction of GALHO stimulates recombination in parental colonies, producing Ura+ papillae upon transfer to medium lacking uracil. Recombinant HOinc/HOinc colonies are not cleaved upon reinduction of GALHO and do not produce Ura+ papillae.
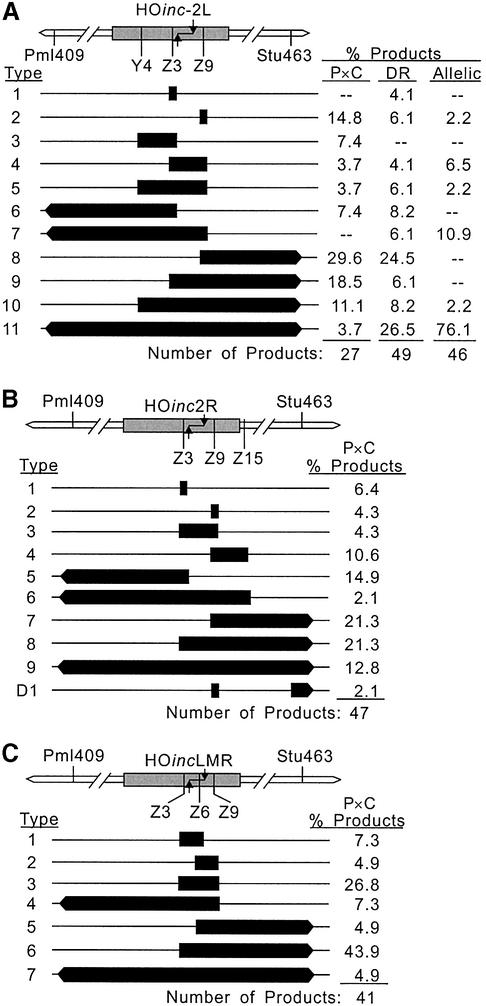
We created three HOinc sites, each carrying three mutations (Fig. 1B). All three mutations in HOinc2L and HOincLMR prevent cleavage. In HOinc2R, the Z3 and Z9 mutations prevent cleavage and the third mutation (Z15) does not, but this does not impose a serious limitation because most tracts are continuous, and Z15 is expected to co-convert with Z9 (see below). These arrangements of HOinc markers were designed to test specific hypotheses, as discussed below. The recombination substrates also carry nine phenotypically silent RFLP markers that flank the HO site at ∼100 bp intervals (Fig. 1C). Tract analysis thus involved sequencing of HOinc markers and restriction mapping of RFLPs, giving high-resolution and gross tract spectra, respectively. As in prior studies, DSBs in HO sites greatly enhanced gene conversion (Table 2). Although differences in substrate design and assay conditions preclude direct comparisons of conversion frequencies with terminal homology versus non-homology, we estimate that conversion is 3–80-fold more efficient with terminal homology (data not shown).
Table 2. Recombination frequencies.
| Recombination substrate | na | Conversion frequency (×104)b | Fold increasec | |
|---|---|---|---|---|
| Glucose | Galactose | |||
| P × C: HOinc-2L | 4/4 | 0 | 716 ± 320 | >700 |
| P × C: HOinc-2R | 3/3 | 0 | 101 ± 27 | >100 |
| P × C: HOinc-LMR | 3/3 | 0 | 87 ± 53 | >87 |
| Direct repeat: HOinc-2L | 10/10 | 0.3 ± 0.8 | 959 ± 389 | 3200 |
| Allelic: HOinc-2L | 5/25 | 0 | 305 ± 141 | >300 |
aNumber of determinations for glucose-/galactose-grown cells.
bAverage conversion frequencies (±SD) were calculated as the number of recombinant products per colony on medium lacking tryptophan (P × C) or leucine (direct repeat), or on YPD (allelic) for glucose- or galactose-grown cells (GALHO repressed or induced). For each determination 300 to 5000 colonies were scored.
cRatio of conversion frequencies for induced:repressed cultures.
Conversion tracts can be extremely short and highly directional
Our initial analysis of conversion within the HO site was performed with the HOinc2L substrates and the results are shown in Figure 3A. With the P × C substrate, 15% of tracts converted only one HOinc marker (Z9), and >80% of products failed to convert all three HOinc markers. The Z3 marker is between the DSB and the Y4 marker, and because all tracts were continuous, Y4 never converted without co-conversion of Z3. The direct repeat substrate gave similar results, with 10% of products converting only one HOinc marker, and 53% failing to convert all three markers. In two direct repeat products (type 1 in Fig. 3A), only the central (Z3) marker converted; these tracts are <12 bp in length. Another striking feature of these tract spectra is the large fraction of highly directional tracts. With the P × C and direct repeat substrate, ∼60% of products converted markers on only one side of the DSB. This is noteworthy because the markers flanking the DSB are immediately adjacent to, or only 1 bp from, the overhang produced by HO nuclease (Fig. 1B). In prior studies, short and unidirectional tracts were more prevalent in P × C and direct repeat substrates than in allelic substrates (7,10,11) reflecting the much more limited homology in ectopic substrates (1.2 kb versus the entire length of chromosome V). This trend was also apparent with the allelic HOinc2L substrate, as >90% of products converted all HOinc markers. However, we did identify one allelic product that converted only Z9, and three others that failed to convert Y4. These results indicate that conversion tracts can be extremely short and highly directional.
Figure 3.
High resolution tract spectra. (A) HOinc2L tract spectrum. The HOinc2L sequence within ura3 is shown by the shaded box, with mutations indicated below. The flanking RFLPs (Pml409 and Stu463) are also indicated. Black bars indicate converted markers; bars with arrowheads indicate tracts that reached Pml409 or Stu463 and may or may not extend to more distant RFLPs. The percentage of each tract type is listed for P × C, direct repeat (DR), and allelic recombination substrates. (B) HOinc2R tract spectrum (P × C only). (C) HOincLMR tract spectrum (P × C only).
Enhanced 3′ conversion with terminal homology
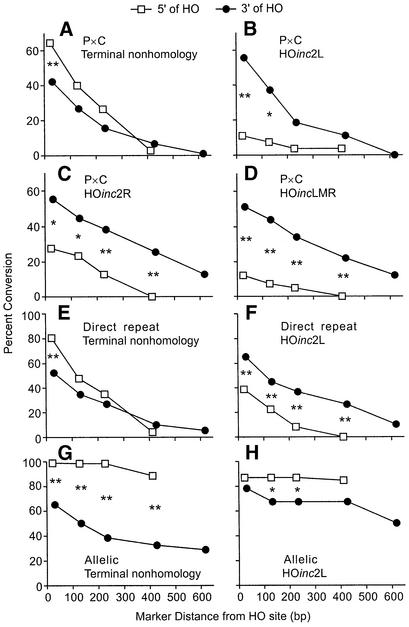
The present HOinc substrates carried a set of silent RFLP markers flanking the HO site identical to those used in prior studies of terminal non-homology substrates (7,10,11). By analyzing just the RFLPs, we can directly compare tract spectra from substrates with terminal homology versus terminal non-homology. Note that the terminal non-homology substrates have an additional marker (X764; Fig. 1A), so these tract spectra were reconstructed as if X764 were absent. From the complete gross tract spectra for HOinc2L P × C, direct repeat, and allelic substrates (not shown), we determined the percentages of uni- and bidirectional tracts, and conversion rates for each RFLP. Tract spectra from substrates with terminal non-homology show consistent 5′ conversion biases, seen both as an excess of 5′ unidirectional versus 3′ unidirectional tracts (Table 3), and as higher conversion rates of 5′ markers versus equidistant 3′ markers (Fig. 4A, E and G). (These biases are distinct because the fractions of unidirectional tracts exclude bidirectional tracts, whereas conversion rates for individual markers are determined from complete tract spectra.) Compared to substrates with terminal non-homology, the P × C and direct repeat HOinc2L substrates yielded more 3′ unidirectional tracts (each P < 0.0007), and fewer 5′ unidirectional tracts (each P < 0.02) than terminal non-homology substrates (Table 3). Similar results were obtained with the allelic substrates, with fewer 5′ unidirectional tracts with HOinc2L than with terminal non-homology (P = 0.007).
Table 3. Tract lengths and gross tract directionality.
| Substratea | nb | Average tract length (bp)c | Tract directionality (%)d | |||
|---|---|---|---|---|---|---|
| Unidirectional | HO Only | 5′ | 3′ | |||
| P × C: HOinc2L | 27 | 126 | 96.3 | 29.6 | 7.4 | 59.3 |
| P × C: HOinc2R | 47 | 231 | 87.2 | 25.5 | 17.0 | 44.7 |
| P × C: HOincLMR | 41 | 179 | 95.1 | 39.0 | 7.3 | 48.7 |
| P × C: TNHe | 74 | 160 | 76.8 | 12.5 | 41.6 | 22.9 |
| Direct repeat: HOinc2L | 49 | 219 | 75.5 | 20.4 | 14.3 | 40.8 |
| Direct repeat: TNHf | 86 | 219 | 54.7 | 11.8 | 35.3 | 7.6 |
| Allelic: HOinc2L | 46 | 743 | 23.9 | 10.9 | 10.9 | 2.1 |
| Allelic: TNHg | 75 | 617 | 34.5 | 0 | 33.3 | 1.2 |
aSubstrates with HOinc sites yield broken ends with terminal homology, others yield ends with terminal non-homology (TNH).
bNumber of products analyzed.
cAverage minimum tract length: calculated as the sum of the minimum tract lengths for all products divided by the number of products.
dPercentage of unidirectional tracts defined as those that failed to convert Pml409 and/or Stu463.
fData from Cho et al. (11).
gData from Nickoloff et al. (7).
Figure 4.
Conversion biases for substrates with terminal non-homology or terminal homology. In each graph, percent conversion of each RFLP is plotted as a function of distance from the DSB. 5′ markers are shown by open squares, 3′ markers by closed circles. Significant differences are shown by * (P < 0.05) or ** (P < 0.01). P × C substrates are shown in panels (A)–(D), direct repeats in panels (E)–(F), and allelic substrates in panels (G)–(H). Data for terminal non-homology substrates are from Sweetser et al. (10), Cho et al. (11) and Nickoloff et al. (7).
Enhanced 3′ conversion is also apparent from the analysis of individual marker conversion rates. In P × C and direct repeat substrates with terminal non-homology, markers 5′ of the DSB converted as often, or more often, than equidistant 3′ markers (Fig. 4A and E), but this was reversed in substrates with HOinc2L (Fig. 4B and F). The allelic substrate with terminal non-homology displayed a strong 5′ conversion bias (Fig. 4G), and although HOinc2L did not reverse this bias, 3′ conversion was clearly enhanced (compare filled circles in Figs. 4G and H).
Of the three markers within HOinc2L, two are 5′ and one is 3′ of the DSB (Fig. 1B). We reasoned that markers very close to a DSB might influence tract directionality, with the higher density of 5′ markers in HOinc2L shifting conversion toward the 3′ side of the DSB. For example, the enhanced 3′ conversion with HOinc2L might reflect reduced strand invasion or branch migration on the more densely marked 5′ side of the DSB. We tested this hypothesis with P × C substrates carrying HOinc2R (one 5′ marker, two 3′ markers), and with HOincLMR (one 5′ marker, one 3′ marker, and one marker within the overhang) (Fig. 1B). We chose P × C substrates for this study because they are easier to build and analyze, and because similar results were obtained with HOinc2L in P × C, direct repeat, and allelic substrates.
High-resolution tract spectra for HOinc2R and HOincLMR are shown in Figure 3B and C, and gross tract parameters are summarized in Figure 4C and D, and in Table 3. As with HOinc2L, HOinc2R and HOincLMR tracts were frequently short and significant fractions were highly directional. With HOinc2R, 10% of products converted only Z3 or Z9, and more than half did not convert all three markers. In HOincLMR, the central marker (Z6) is within the four-base overhang. Because invasion with either end would include Z6, we predicted that all HOincLMR products would convert Z6 and this was indeed the case. None of the HOincLMR products converted only one marker, and only 17% failed to convert all three markers (compared to 64% with HOinc2R and 82% with HOinc2L); these differences probably reflect the closer spacing (and hence more frequent co-conversion) of the HOincLMR markers. The HOinc2R and HOincLMR spectra do not support the hypothesis that conversion bias is affected by marker density near a DSB. Even though HOinc2R has two markers 3′ of the DSB and HOincLMR has markers evenly distributed on each side of the DSB, both spectra displayed strong 3′ conversion biases. Compared to the P × C substrate with terminal non-homology, the HOinc2R and HOincLMR substrates yielded more 3′ unidirectional tracts (each P < 0.02), fewer 5′ unidirectional tracts (each P < 0.005) (Table 3) and more frequent conversion of 3′ versus equidistant 5′ markers (Fig. 4C and D). We conclude that conversion bias is influenced by the presence or absence of non-homology at broken ends.
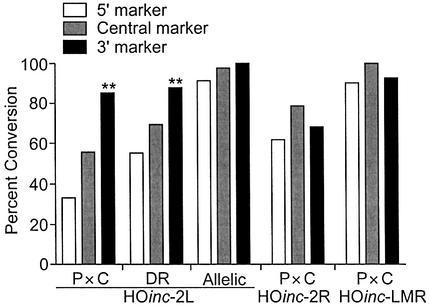
The conversion biases described above are defined by the RFLPs flanking the HO site. With the HOinc substrates, conversion bias also can be defined by the HOinc markers (Fig. 5). At this higher level of resolution, the P × C and direct repeat substrates with HOinc2L showed significantly higher conversion rates for the 3′ (Z9) marker than the 5′ (Y4) marker (both P < 0.0006). With the P × C HOinc2R substrate, the HOinc markers showed no conversion bias, suggesting that the additional 3′ marker in HOinc2R partially suppresses the 3′ bias. There were no biases with the allelic HOinc2L, and P × C HOincLMR substrates; in the majority of these products, all three HOinc markers were converted.
Figure 5.
Percent conversion of markers within the HO site. Data for P × C, direct repeat (DR), and allelic substrates from Figure 3. ** indicates P < 0.001.
Short and unidirectional tracts are more frequent with terminal homology than with terminal non-homology
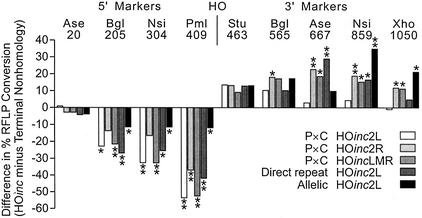
One model to account for frequent unidirectional tracts when broken ends have non-homologous termini holds that non-homologous ends inhibit strand invasion, and that this restricts two-ended invasions and limits bidirectional tracts. In this view, terminal homology at broken ends should promote two-ended invasions and decrease unidirectional tracts. However, we found that unidirectional tracts were more frequent in the HOinc substrates than in substrates with terminal non-homology (Table 3), and in three of the five cases, the increases were statistically significant (P × C HOinc2L, P = 0.02; direct repeat HOinc2L, P = 0.037; P × C HOincLMR, P = 0.01). In addition, substrates with terminal homology gave 2–3-fold higher frequencies of short tracts in which no RFLPs were converted (‘HO only’; Table 3), although with these sample sizes only the P × C HOincLMR and allelic HOinc2L substrates showed significant differences (both P < 0.009). Average tract lengths varied among the different substrates (Table 3), but none of these differences were statistically significant. The enhanced 3′ conversion could reflect altered conversion rates on only one side of the DSB. However, since tracts lengths were not altered, enhanced 3′ conversion reflects both decreased 5′ conversion and increased 3′ conversion. These changes are evident in plots of differences in percent conversion of each RFLP between HOinc substrates and terminal non-homology substrates (Fig. 6).
Figure 6.
Shift in conversion bias reflects decreased 5′ conversion and increased 3′ conversion. Percent conversion of each RFLP (listed at top) in each HOinc substrate was subtracted from the percent conversion in the analogous substrate with terminal non-homology, and the difference (positive or negative) is plotted. Significant differences in percent conversion are shown by * (P < 0.05) or ** (P < 0.01).
DISCUSSION
Short conversion tracts
Measurements of DSB-induced gene conversion tracts requires markers in recombination substrates. The resolution of these measurements is dependent on the number of markers, and their positions relative to each other and to the initiating DSB. It has long been known that conversion tracts can be quite long, extending 12 kb in one study (25), and >36 kb in another (26). We showed previously that ectopic conversion tracts are often <53 bp in length (10,11). Repeated regions in ectopic recombination substrates are limited in length (1.2 kb in our ura3 systems) and conversion is suppressed near homology borders (10,27). Homology is effectively unlimited in allelic recombination substrates, conversion tracts are longer, and allelic tracts <53 bp in length were not observed (7). In all of these prior studies, the HO site was present in only one copy of ura3, and broken ends terminated with 21 and 14 bp of non-homology on the 5′ and 3′ sides of the DSB, respectively. In the present study, broken ends were almost fully homologous to donor sequences, and mutations within HO sites served as markers, allowing the highest resolution analysis of yeast conversion tracts to date. A striking result from this analysis is that tracts can be extremely short (<12 bp).
For markers distant from a DSB, conversion tracts are the observable outcome of hDNA formation (strand invasion, repair synthesis/annealing, and branch migration) and mismatch repair in hDNA (including restoration-type and conversion-type repair) (28). Recent evidence indicates that 3′ ends at DSBs are very stable in yeast (29), so even when markers are very close to a broken end, they are unlikely to convert by gap repair. However, such markers may be lost soon after strand invasion if the invading end is processed by the mismatch repair system or by the proofreading function of the repair polymerase prior to 3′ end extension. The fact that gene conversion tracts can be <12 bp in length raises a question about the minimum length of hDNA required for an intermediate to be processed to a mature gene conversion product. Although no studies have directly addressed this issue, we believe it is unlikely that hDNA as short as 12 bp would provide sufficient stability. Instead, very short conversion tracts may arise from intermediates with longer hDNA regions through restoration repair of mismatches, and/or through reverse branch migration of Holliday junctions prior to mismatch repair.
Highly directional conversion tracts
With substrates yielding broken ends with terminal non-homology, unidirectional tracts comprised ∼80% of P × C, ∼60% of direct repeat, and ∼20% of allelic tracts; resolution in these studies was 23–31 bp (7,10,11). In another study, we increased the resolution of the P × C measurements by placing markers closer to the HO site, and identified unidirectional tracts that extended <6 bp into homology on one or both sides of the HO site (18). In the present study we show that tracts are often highly directional, in some cases extending <2 bp from a broken end. We thought that the frequent unidirectional tracts seen with terminal non-homology might reflect the inhibition of strand invasion. In this view, terminal homology would be expected to promote two-ended invasion and decrease the fraction of unidirectional tracts. Instead we found that unidirectional tracts increased significantly with terminal homology. Unidirectional tracts also predominate when conversion is initiated by homeologous termini (71% homologous) (18). The results with terminal homeology (18), and the present results with terminal homology, are inconsistent with uni- and bidirectional tracts arising from one- and two-ended invasions, respectively. As discussed previously (18), both uni- and bidirectional tracts can result from one- or two-ended invasions. Although the ratio of uni- and bidirectional tracts appears to be influenced by the presence or absence of non-homology at broken ends, this ratio, and other tract parameters, may be more strongly affected by other factors such as replication or transcription (as discussed below).
Conversion bias with terminal homology or non-homology
In past studies with substrates with terminal non-homology, we observed a consistent 5′ conversion bias: 5′ unidirectional tracts arose far more often than 3′ unidirectional tracts, and 5′ markers converted more often than equidistant 3′ markers (7,10,11). Surprisingly, all three HOinc2L substrates (with terminal homology) displayed enhanced 3′ conversion, and in the ectopic substrates this reversed the conversion bias. In HOinc2L, two of three markers within the HO site are 5′ of the DSB. Heterologies inhibit spontaneous recombination (30–34), and although heterologies have little or no effect on the efficiency of DSB-induced recombination (7), it was possible that enhanced 3′ conversion with HOinc2L reflected enhanced conversion toward the less densely marked 3′ side of the DSB. However, this is not the case because strong 3′ biases were also observed when two of three markers were 3′ of the DSB (HOinc2R), and when markers were symmetric around the DSB (HOincLMR) (Table 3). Thus, conversion bias is not affected by a few markers near broken ends, but rather by the presence of homologous or non-homologous termini.
It is unclear how conversion bias is affected by the presence or absence of non-homology at termini. Although the HO site is asymmetric, a previous study indicated that HO site orientation does not affect tract spectra (10). Furthermore, unlike the meiotic Spo11 nuclease, which is covalently bound to one end after creating a DSB (35), there is no evidence that HO binds to broken ends, nor is HO nuclease a member of the Spo11/topoisomerase enzyme family. Instead, conversion bias may involve polar processes like transcription and replication, or functions of mismatch repair or other proteins. We previously tested the effects of transcription on DSB-induced gene conversion in ura3 direct repeats and found that transcription of donor alleles increases the 5′ conversion bias, and that increased transcription in recipient alleles reduces the frequency of unidirectional tracts (36). There is increasing evidence of interactions between replication and recombination (37,38) and we are currently testing whether the direction of replication regulates conversion bias. It is possible that the differential conversion bias with terminal homology versus non-homology involves Rad1/10 endonuclease, perhaps in conjunction with mismatch repair proteins. Rad1/10 is a structure-specific endonuclease with roles in nucleotide excision repair, mismatch repair, and recombination (17,39–42). Rad1/10 directly interacts with the Msh2 mismatch recognition protein (43), and both Rad1/10 and Msh2 are involved in removal of terminal non-homology in recombination intermediates (17,40), and in the repair of loop mismatches (42). Although conversion tract spectra were similar in substrates with terminal homeology in wild-type and rad1 mutants (18), additional studies will be required to determine if Rad1/10 differentially influences conversion bias in substrates with homologous and non-homologous termini.
The regulation of conversion tract lengths and directionality is important because conversion causes local loss of heterozygosity, and because long conversion tracts are preferentially associated with crossovers (44,45). Crossovers pose special risks to genome stability because they can cause expansions and contractions of linked repeats and translocations. Further more, although crossovers between homologous chromosomes have no immediate genetic effect, these events lead to loss of heterozygosity for all markers centromere-distal to the crossover point in 50% of subsequent mitotic divisions (46). Such large-scale loss of heterozygosity may be important for inactivation of tumor suppressor genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Guru Jot Khalsa, Heather Hough and Kimberly Paffet for technical assistance, and Jennifer Clikeman for many helpful comments. This work was supported by grant CA55302 to J.A.N. from the National Cancer Institute of the NIH. R.L. was supported by NIH/MARC Undergraduate Research Program grant GM08751.
REFERENCES
- 1.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taghian D.G. and Nickoloff,J.A. (1997) Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol., 17, 6386–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang F., Han,M.G., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petes T.D., Malone,R.E. and Symington,L.S. (1991) Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. I, pp. 407–521.
- 5.Nickoloff J.A. and Hoekstra,M.F. (1998) Double-strand break and recombinational repair in Saccharomyces cerevisiae. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair, Vol. 1: DNA Repair in Prokaryotes and Lower Eukaryotes. Humana Press, Totowa, NJ, pp. 335–362.
- 6.Weng Y.-S. and Nickoloff,J.A. (1998) Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics, 148, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickoloff J.A., Sweetser,D.B., Clikeman,J.A., Khalsa,G.J. and Wheeler,S.L. (1999) Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics, 153, 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modrich P. (1991) Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet., 25, 229–253. [DOI] [PubMed] [Google Scholar]
- 9.Rasmussen L.J., Samson,L. and Marinus,M.G. (1998) Dam-directed DNA mismatch repair. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair. Humana Press, Totowa, NJ, pp. 205–228.
- 10.Sweetser D.B., Hough,H., Whelden,J.F., Arbuckle,M. and Nickoloff,J.A. (1994) Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol., 14, 3863–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho J.W., Khalsa,G.J. and Nickoloff,J.A. (1998) Gene conversion tract directionality is influenced by the chromosome environment. Curr. Genet., 34, 269–279. [DOI] [PubMed] [Google Scholar]
- 12.McGill C.B., Shafer,B.K., Derr,L.K. and Strathern,J.N. (1993) Recombination initiated by double-strand breaks. Curr. Genet., 23, 305–314. [DOI] [PubMed] [Google Scholar]
- 13.Schultes N.P. and Szostak,J.W. (1990) Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics, 126, 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbertson L.A. and Stahl,F.W. (1996) A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics, 144, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter S.E., White,M.A. and Petes,T.D. (1993) Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics, 134, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prado F. and Aguilera,A. (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10 and RAD52 genes. Genetics, 139, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fishman-Lobell J. and Haber,J.E. (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science, 258, 480–484. [DOI] [PubMed] [Google Scholar]
- 18.Nelson H.H., Sweetser,D.B. and Nickoloff,J.A. (1996) Effects of terminal nonhomology and homeology on double-strand break-induced gene conversion tract directionality. Mol. Cell. Biol., 16, 2951–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghian D.G. and Nickoloff,J.A. (1996) Subcloning strategies and protocols. In Harwood,A. (ed.), Basic DNA and RNA Protocols. Humana Press, Totowa, NJ, Vol. 58, pp. 221–235. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Clikeman J.A., Khalsa,G.J., Barton,S.L. and Nickoloff,J.A. (2001) Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKu-dependent and -independent mechanisms. Genetics, 157, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickoloff J.A., Chen,E.Y.C. and Heffron,F. (1986) A 24-base-pair sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl Acad. Sci. USA, 83, 7831–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickoloff J.A., Singer,J.D. and Heffron,F. (1990) In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol. Cell. Biol., 10, 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiffenbach B., Rogers,D.T., Haber,J.E. and Zoller,M. (1983) Deletions and single base pair changes in the yeast mating type locus that prevent homothallic mating type conversions. Proc. Natl Acad. Sci. USA, 80, 3401–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Symington L.S. and Petes,T. (1988) Expansions and contractions of the genetic map relative to the physical map of yeast chromosome III. Mol. Cell. Biol., 8, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golin J.E. and Esposito,M.S. (1984) Coincident gene conversion during mitosis in Saccharomyces cerevisiae. Genetics, 107, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn B.-Y., Dornfeld,K.J., Fagrelius,T.J. and Livingston,D.M. (1988) Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol. Cell. Biol., 8, 2442–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirkpatrick D.T., Dominska,M. and Petes,T.D. (1998) Conversion-type and restoration-type repair of DNA mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Genetics, 149, 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank-Vaillant M. and Marcand,S. (2002) Transient stablity of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell, 10, 1189–1199. [DOI] [PubMed] [Google Scholar]
- 30.Chambers S.R., Hunter,N., Louis,E.J. and Borts,R.H. (1996) The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol., 16, 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datta A., Adjiri,A., New,L., Crouse,G.F. and Jinks-Robertson,S. (1996) Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta A., Hendrix,M., Lipsitch,M. and Jinks-Robertson,S. (1997) Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl Acad. Sci. USA, 94, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negritto M.T., Wu,X., Kuo,T., Chu,S. and Bailis,A.M. (1997) Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol. Cell. Biol., 17, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selva E.M., New,L., Crouse,G.F. and Lahue,R.S. (1995) Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics, 139, 1175–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keeney S., Giroux,C.N. and Kleckner,N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell, 88, 375–384. [DOI] [PubMed] [Google Scholar]
- 36.Weng Y.-S., Xing,D., Clikeman,J.A. and Nickoloff,J.A. (2000) Transcriptional effects on double-strand break-induced gene conversion tracts. Mutat. Res., 461, 119–132. [DOI] [PubMed] [Google Scholar]
- 37.Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or “gimme a break”. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- 38.Haber J.E. (1999) DNA recombination: the replication connection. Trends. Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- 39.Prakash S. and Prakash,L. (2000) Nucleotide excision repair in yeast. Mutat. Res., 451, 13–24. [DOI] [PubMed] [Google Scholar]
- 40.Bardwell A.J., Bardwell,L., Tomkinson,A.E. and Friedberg,E.C. (1994) Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science, 265, 2082–2085. [DOI] [PubMed] [Google Scholar]
- 41.Saparbaev M., Prakash,L. and Prakash,S. (1996) Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics, 142, 727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirkpatrick D.T. and Petes,T.D. (1997) Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature, 387, 929–931. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand P., Tishkoff,D.X., Filosi,N., Dasgupta,R. and Kolodner,R.D. (1998) Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl Acad. Sci. USA, 95, 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jinks-Robertson S., Michelitch,M. and Ramcharan,S. (1993) Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 3937–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symington L.S., Kang,L.E. and Moreau,S. (2000) Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in nuclease-deficient strains of Saccharomyces cerevisiae. Nucleic Acids Res., 28, 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickoloff J.A. (2002) Bertino,J.R. (ed.), Encyclopedia of Cancer, 2nd Edn. Elsevier Science, San Diego, USA. Vol. 4, pp. 49–59.
- 47.Weng Y.-S., Whelden,J., Gunn,L. and Nickoloff,J.A. (1996) Double-strand break-induced gene conversion: examination of tract polarity and products of multiple recombinational repair events. Curr. Genet., 29, 335–343. [DOI] [PubMed] [Google Scholar]