Abstract
The transient expression of three novel plant amber suppressors derived from a cloned Nicotiana tRNASer(CGA), an Arabidopsis intron-containing tRNATyr(GTA) and an Arabidopsis intron-containing tRNAMet(CAT) gene, respectively, was studied in a homologous plant system that utilized the Agro bacterium-mediated gene transfer to Arabidopsis hypocotyl explants. This versatile system allows the detection of β-glucuronidase (GUS) activity by histochemical and enzymatic analyses. The activity of the suppressors was demonstrated by the ability to suppress a premature amber codon in a modified GUS gene. Co-transformation of Arabidopsis hypocotyls with the amber suppressor tRNASer gene and the GUS reporter gene resulted in ∼10% of the GUS activity found in the same tissue transformed solely with the functional control GUS gene. Amber suppressor tRNAs derived from intron-containing tRNATyr or tRNAMet genes were functional in vivo only after some additional gene manipulations. The G3:C70 base pair in the acceptor stem of tRNAMet(CUA) had to be converted to a G3:U70 base pair, which is the major determinant for alanine tRNA identity. The inability of amber suppressor tRNATyr to show any activity in vivo predominantly results from a distorted intron secondary structure of the corresponding pre-tRNA that could be cured by a single nucleotide exchange in the intervening sequence. The improved amber suppressors tRNATyr and tRNAMet were subsequently employed for studying various aspects of the plant-specific mechanism of pre-tRNA splicing as well as for demonstrating the influence of intron-dependent base modifications on suppressor activity.
INTRODUCTION
Suppressor tRNAs provide valuable tools for studying the expression of individual members of a tRNA gene family in vivo. This is especially helpful in higher plants where conventional analyses are hampered because all 45 tRNA isoacceptors are encoded by multigene families encompassing up to 20 members which frequently comprise identical coding regions and similar flanking sequences (1,2). Although indispensable for many investigations, there are a number of features that might restrict the generation of functional suppressor tRNAs such as impaired transcription of the gene, defective processing of precursors and most likely insufficient charging with the cognate amino acid of the mature tRNA due to the altered anticodon.
The expression of suppressors in plant systems has been developed essentially by two groups who studied the ability of tRNALeu, tRNATrp or tRNALys with mutated anticodons complementary to any of the three stop codons to mediate suppression of a premature stop codon in suitable reporter genes (3–9). None of the tRNA genes selected for these studies contained an intron, since a further complication arises if the corresponding pre-tRNAs carry intervening sequences whose efficient removal is an absolute prerequisite for the production of a functional tRNA.
During the last years we have intensively studied the mechanism of pre-tRNA splicing in two plant in vitro systems derived from wheat germ and tobacco cells, and have found that plant splicing endonuclease is highly specific for homologous substrates quite in contrast to the vertebrate and yeast enzymes (10–14). In plant nuclear genomes only two intron-containing tRNA gene families exist which code for tRNATyr and elongator tRNAMet (13,15,16). Possibly as a result of this low number, unique features of the mature tRNA domain and an extremely conserved intron secondary structure contribute to the specificity of plant splicing endonuclease as revealed by in vitro analyses of mutated pre-tRNAs (12,13,16,17). In plant intron-containing pre-tRNAsTyr and pre-tRNAsMet the 5′ and 3′ splice sites are located in single-stranded regions which are commonly separated by a helical structure of 4 bp formed by the interaction between bases of the anticodon loop and the intron. Disruption of the base-paired region severely impairs pre-tRNA splicing in both plant extracts (13,17). The generation of an amber suppressor derived from an Arabidopsis intron-containing tRNATyr gene results in the disturbance of the conserved intron structure, simply by the single nucleotide exchange in the anticodon (i.e., GTA→CTA) and the derived pre-tRNATyr was, therefore, a poor substrate for the wheat germ splicing endonuclease. In order to demonstrate that this feature caused the reduced splicing activity, we introduced a second point mutation in the intron which restored the helical intron structure. As expected, this substrate was efficiently spliced in vitro (17). Similarly, we have constructed an amber suppressor derived from an Arabidopsis intron-containing tRNAMet gene. The corresponding pre-tRNAMet(CUA) was spliced effectively in vitro, possibly because in this case the exchange of two nucleotides within the anticodon did not lead to a severe distortion of the intron secondary structure (13).
In this report we have studied the expression of amber suppressors derived from Arabidopsis intron-containing tRNATyr and tRNAMet genes in vivo. In addition we have investigated the activity of an amber suppressor derived from an intron-less tRNASer gene. As a novel system for transient expression, we have utilized the Agrobacterium-mediated gene transfer to Arabidopsis hypocotyls, a system commonly used for the generation of transgenic plants. We show here that all three novel suppressors can function in vivo, provided that single mutations have been introduced in the corresponding genes which either favour correct splicing of the pre-tRNAs or the aminoacylation of the mature tRNAs.
MATERIALS AND METHODS
Plant material
Seeds of Arabidopsis thaliana ecotype Wassilewskija were surface-sterilized with 10% (v/v) bleach (Haitar; Kao Co., Ltd, Tokyo)/0.02% (v/v) Triton X-100 for 10 min, rinsed five times with distilled water and placed on dishes with germination medium (18). Plants were grown at 23°C under 16 h of daily illumination for ∼2 weeks.
Construction of GUS reporter gene
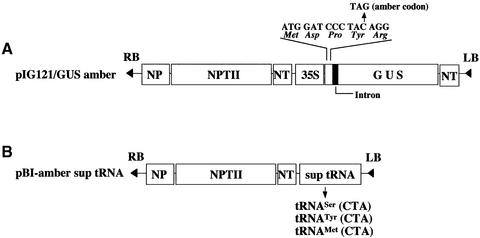
The β-glucuronidase (GUS) gene (gusA) in pIG121Hm contains a functional plant intron that is inserted at the N-terminal coding region (19). In order to introduce a premature amber codon, the intron-gusA ORF was first cleaved from pIG121Hm with XbaI/SacI. In parallel, pBI221 (Clontech) was digested with the same restriction enzymes to remove the region of a gusA ORF. The excised fragment from pIG121Hm was ligated into the corresponding sites of the gusA-deleted pBI221 vector DNA, resulting in pIG221. Next, the N-terminal and intron region of gusA (226 bp) was amplified from pIG221 by PCR using forward primer (5′-GCTCTAGAACATGGATCCCTAGAGGGT-3′) and reverse primer (5′-CGAGTCGACGGTTCTGTAAC-3′). Underlined regions indicate a BamHI and a SalI site, respectively. A substitution of C→G (indicated by a bold letter) changes the tyrosine codon (TAC) to an amber codon (TAG) (Fig. 1A). The PCR fragment was digested with BamHI/SalI and ligated into the corresponding sites of pBluescript II SK (+) DNA (Stratagene), in order to confirm mutagenesis by sequencing. The BamHI/SalI fragment of pIG221 was then replaced by the mutated fragment resulting in pIG221/amber. Finally, a XbaI/SnaBI fragment of the intron-gusA ORF was excised from pIG221/amber and cloned into the relevant sites of the gusA-deleted tumor-inducing (Ti) plasmid pBI121 (Clontech), yielding pIG121/GUS amber (Fig. 1A).
Figure 1.
Schematic representation of constructs containing the GUS reporter or a suppressor tRNA gene. (A) The plasmid pIG121/GUS amber contains the gusA-coding sequence. The alteration made to introduce a premature TAG codon at the position of the first tyrosine codon (i.e., codon four of the fusion protein) is shown above. LB and RB indicate the left and right border of the T-DNA region. NP, nopaline synthase promoter; NPTII, neomycin phosphotransferase II; 35S, cauliflower mosaic virus 35S promoter; NT, nopaline synthase terminator. (B) The plasmid pBI-amber sup tRNA contains amber suppressor tRNA genes derived from either tRNASer (CGA), tRNATyr (GUA) or tRNAMet (CAU) genes, respectively.
Construction of suppressor tRNA genes
Three different kinds of plant tRNA genes and their derivatives were used in this study to generate amber suppressor tRNA genes. The first plasmid pNtS2 contains a nuclear tRNASer gene with CGA anticodon from Nicotiana rustica on a 256 bp RsaI fragment cloned into pUC19 vector DNA (20). Using this recombinant plasmid as a template, a CTA anticodon was introduced in the relevant site (Fig. 2A) by in vitro site-directed mutagenesis with an appropriate primer according to Carneiro et al. (4). The second plasmid pAtM1 carrying an Arabidopsis nuclear elongator tRNAMet gene on a 310 bp DNA fragment cloned between the XbaI/HindIII sites of pBluescript was used for a series of primary constructions of mutant derivates described elsewhere (13). In order to introduce the identity element of tRNAAla into tRNAMet by substituting C70→T70 of the mutant genes in these constructs, a primer complementary to the 3′-end region in the nuclear tRNAMet gene (5′-TGGAGTGAGAGAGGCTCG-3′, bold letter indicates the target site) was synthesized and used for the same mutagenesis procedure described above. Resulting fragments were subcloned in the pBluescript vector and their sequences confirmed by sequencing. The third plasmid used for the generation of an amber suppressor tRNATyr gene was pAtY3II which carries an A.thaliana tRNATyr gene on a 1398 bp RsaI-fragment cloned into pUC19 vector DNA (21). The two mutant tRNATyr genes, i.e., AtY3II-am and AtY3II-amiG7, were constructed as described previously (17). The intron-less derivative of AtY3II-am was called AtY3IIΔ-am. These mutant plant tRNA genes with CTA anticodon were all subcloned into the XbaI/SacI sites of gusA-deleted pBI121 (Fig. 1B). The Ti plasmids were immobilized in the agrobacterial strain GV3101 (22).
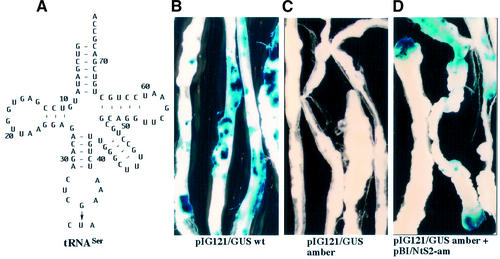
Figure 2.
Histochemical analyses of GUS activity in Arabidopsis hypocotyl-derived calli after Agrobacterium-mediated transformation. The agrobacteria contained the vector pIG121/GUS wt, carrying the wildtype gusA gene (B) or the vector pIG121/GUS amber, carrying a mutated gusA gene with a premature amber codon (C) as indicated in Figure 1A or the calli were co-cultivated with two different strains of Agrobacterium, harboring either the pIG121/GUS amber plasmid or a tobacco amber suppressor tRNASer gene (NtS2-am) in the vector pBI (D) as illustrated in Figure 1B. After co-cultivation for 3 days, the explants were washed in liquid MS medium several times in order to remove the bacteria, followed by incubation in X-Gluc-containing phosphate buffer overnight. The samples were decolored with 70% ethanol before the microscopic evaluation. The left panel (A) shows the cloverleaf structure of the amber suppressor tRNASer expressed in the transformed Arabidopsis explants (D). The nucleotide sequence is deduced from NtS2, which contains a Nicotiana tRNASer gene with CGA anticodon. The modified nucleosides of the tRNASer isoacceptor are not indicated and are presented elsewhere (20). A single mutation at position 35 in the anticodon (G→U) converts the tRNASer into an amber suppressor with CUA anticodon.
Plant tissue culture and transient expression of tRNA genes via agrobacteria
Hypocotyl explants (∼1 cm in length) excised from seedlings were transferred to petri dishes containing callus-inducing medium (CIM) supplemented with 20 µM acetosyringone and incubated for 7 or 8 days under the conditions described above. Infection of the explants with agrobacteria and co-cultivation were essentially performed as described by Akama et al. (18). Single colonies of agrobacterial strains carrying appropriate recombinant vectors were inoculated in 2 ml Luria-broth containing 100 µg/ml kanamicyn sulfate and incubated at 28°C overnight with shaking. Pre-culture (5– 10 ml) was transferred to 2–4 ml of the same media to start main culture under identical growth conditions. After determination of the absorbance of the overnight culture at 600 nm, bacteria were collected and suspended in 0.5× Murashige and Skoog (MS) salts (23) to adjust at A600 = 1. Bacterial suspensions carrying pIG121/GUS amber and a Ti plasmid with one of the mutant tRNA genes were mixed at a ratio of 0.3:0.7 in petri dishes, and then the callus-induced hypocotyl explants were immersed, followed by vacuum infiltration for 30 min and transfer to the same CIM plate. Co-cultivation was carried out at 23°C for 3 days. The tissues were transferred to dishes with liquid 0.5× MS salts and were gently shaken for 30 min three times to remove bacteria. Before GUS analyses, they were rinsed with 0.1 M sodium phosphate (pH 7.0).
GUS analyses
Histochemical and fluorometric GUS analyses were essentially done according to Jefferson (24). For GUS staining, the hypocotyls were transferred to 0.5 mg/ml of 5-bromo-4-chloro-β-d-glucuronide (X-Gluc) in 0.1 M sodium phosphate buffer (pH 7.0), incubated at 37°C overnight and decolored with 70% ethanol. For fluorometric analyses, five pieces of tissues were transferred to a microtube and ground in GUS extraction buffer (50 µM sodium phosphate buffer, pH 7.0, 10 mM β-mercaptoethanol, 10 mM EDTA and 0.1% N-laurosylsarcosine). Supernatants were recovered as crude protein extracts after centrifugation. The protein concentration was determined as described by Bradford (25). Proteins (5–10 µg) were incubated in GUS assay buffer (1 mM 4-methylumbelliferyl-β-d-glucuronide in GUS extraction buffer) for 30, 60 and 90 min. Aliquots were transferred to 0.2 M Na2CO3 to stop the reaction. GUS activity was measured with DynaQuant 200 (Amersham).
In vitro transcription with T7 RNA polymerase
For the in vitro transcription of plant tRNAMet genes, i.e., the intron-less wildtype AtM1Δ gene and the intron-less AtM1Δ-am gene, cloned into the pBluescript vector (13), the coding regions were amplified from these plasmids by PCR with primers (5′-CGGAATTCTAATACGACTCACTATAGGGGTGGTGGCGCAGTTG-3′ and 5′-GGGGTACCTGGTGGGGTGAGAGAGGCTCG-3′ or 5′-GGGGTACCTGGTGGA GTGAGAGAGGCTCG-3′). Underlined sequences indicate the promoter of T7 RNA polymerase and the BstNI site, respectively, and the bold A indicates the replacement of C70 with U70 in tRNAMet. This procedure results in the addition of the T7 RNA polymerase promoter at the 5′-end and a BstNI site at the 3′-end of the genes. All constructs were verified by DNA sequencing.
Plasmids carrying a tRNAMet gene or its variants were digested with BstNI in order to generate transcripts encoding the CCA sequence. Linearized plasmid DNAs (∼1 µg) were added to the reaction mixture (40 mM Tris–HCl, pH 8.1, 6 mM MgCl2, 5 mM DTT, 1 mM spermidine, 1 mM each of GTP, CTP, UTP and ATP, and 400 U/mg T7 RNA polymerase) and incubated at 37°C for 1 h. Transcripts were fractionated on a 10% polyacrylamide/8 M urea gel. Elution of the tRNA transcripts from the gel pieces was performed as described by Stange and Beier (10).
In vitro aminoacylation assays
Aminoacylation of unlabeled tRNA transcripts (derived from transcription with T7 RNA polymerase) with alanine was carried out at 30°C in a 50 µl reaction mixture containing 100 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 55 mM KCl, 1 mM DTT, 5 mM ATP, 0.5 mM CTP, 5 µM alanine, 0.122 µM L-[3H]alanine (2 TBq/mmol) and 0.4–1 µM tRNA transcript. Aminoacylation with methionine was conducted in the same reaction buffer, containing 5 µM methionine and 1 nM L-[35S]methionine (37 TBq/mmol). The reaction was initiated by adding 4 µl dialysed wheat germ S100 extract (22 mg protein/ml). At appropriate time intervals, 5 µl aliquots were spotted on pieces of Whatman 3 MM paper which were incubated first in 10% trichloroacetic acid (TCA) for 10 min, washed twice in 5% TCA and twice in 96% ethanol. Radiolabeled aminoacyl-tRNA was then measured by liquid scintillation counting.
RESULTS
A novel in vivo system to measure activity of nonsense suppressor tRNAs
In order to study the efficiency of plant suppressor tRNAs in vivo, their expression has been mostly investigated in isolated tobacco protoplasts by direct gene transfer of the corresponding tRNA genes (4,6,9). Here we utilized a transient assay system based on Agrobacterium-mediated gene delivery. Briefly, hypocotyl explants from young seedlings of Arabidopsis are co-cultivated with agrobacteria containing the equivalent recombinant Ti plasmids (18). As a reporter gene, we employed a modified GUS gene which contains a 190 bp long functional plant intron within the N-terminal region (Fig. 1A). This intron harbours a termination codon in the same reading frame as the GUS coding sequence so that removal of the intron from the pre-mRNA is required for the generation of a functional GUS polypeptide. Unlike the original GUS gene, this Intron–GUS reporter gene did not express detectable GUS activity in Agrobacterium tumefaciens cells (19). Consequently, contamination of GUS activity by bacteria in transformed Arabidosis hypocotyls can be neglected.
The first tyrosine codon (i.e., codon number four of the recombinant GUS gene) was exchanged to an amber (TAG) stop codon that is flanked by an adenosine residue at the 3′ side (Fig. 1A). As will be discussed below, such a nucleotide context is supposed to impair UAG readthrough by natural suppressor tRNAs present in transformed cells (26,27). The Ti plasmid containing the mutated GUS gene was called pIG121/GUS amber as compared with the plasmid pIG121/GUS wt harbouring the wildtype GUS gene. We then examined the GUS activity in Arabidopsis hypocotyls transformed with either of the two Ti plasmids. The histochemical analyses revealed no blue staining at all in hypocotyls transformed with pIG121/GUS amber, in contrast to the intense GUS staining detected after transformation with pIG121/GUS wt (Fig. 2B and C). The quantitative evaluation of specific GUS activities indicated 30 000 pmol 4-methylumbelliferone (MU)/min/mg protein and 6 pmol 4-MU/min/mg protein in tissues transformed with GUS wt and GUS amber, respectively (Table 1). The low GUS activity (∼0.02%) measured in Arabidopsis hypocotyls co-transformed with pIG121/GUS amber confirms that the premature stop codon in GUS mRNA is effective and obviously not recognized by any natural endogenous suppressor tRNA.
Table 1. tRNASer-mediated suppression of an amber codon in gusA mRNA expressed in Arabidopsis calli transformed via Agrobacterium.
| GUS specific activitya | |
|---|---|
| (pmol 4-MU/min/mg protein) | |
| pIG121/GUS wt | 31 540 ± 10 480 (100%) |
| pIG121/GUS amber | 6.0 ± 0.6 (0.02%) |
| pIG121/GUS amber + pBI/NtS2-am | 3040 ± 640 (9.6%) |
aSpecific activities are the means of three experiments. ± Standard error.
We next constructed an amber suppressor tRNASer gene. All eukaryotic nuclear genomes encode multiple copies of four tRNASer isoacceptors with IGA, U*GA, CGA and GCU anticodon (20,28). We have chosen a tRNASer gene with CGA anticodon from N.rustica (i.e., NtS2) for the construction of a functional amber suppressor because (i) it has been previously shown that this gene is efficiently transcribed in vitro (20); (ii) the corresponding tobacco tRNASer isoacceptor has been isolated and sequenced (20); (iii) a single mutation at position 35 of the anticodon converts the tRNASer with CGA into an amber suppressor with CUA anticodon (Fig. 2A); and (iv) the major identity elements required for the specific aminoacylation by eukaryotic seryl-tRNA synthetases are the discriminator base and the long extra arm (29,30) implying that changes in the anticodon do not impair charging with the cognate amino acid. When Arabidopsis-derived hypocotyls were co-infected with agrobacteria carrying pIG121/GUS amber and the Ti plasmid pBI/NtS2-am, significant amounts of GUS activity were detected in histochemical (Fig. 2D) and enzymatic analyses (Table 1). Compared with Arabidopsis hypocotyls transformed with pIG121/GUS wt, 9.6% of GUS activity was measured after co-transformation with pIG121/GUS amber and pBI/NtS2-am (Table 1).
Transient GUS expression appears to be irregularly distributed in a number of transformed hypocotyls. (Fig. 2B and D). This observation is explained by a non-synchronized formation of calli preferentially at both ends of the hypocotyl explants which are most susceptible to infection with Agrobacterium upon co-cultivation. Furthermore, the subsequent proliferation of agrobacteria in the infected hypocotyls does not proceed homogenously. In order to minimize this kind of bias in individual transformed hypocotyls, we analysed routinely five explants altogether in a single assay and confirmed the expression pattern of a specific amber suppressor tRNA species in at least three independent experiments.
Expression of amber suppressor tRNATyr genes in Arabidopsis hypocotyls
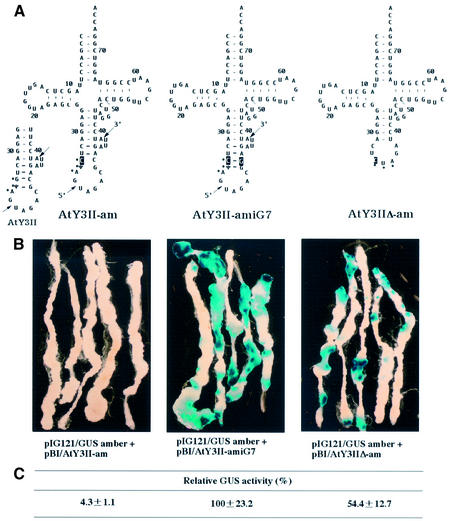
All eukaryotic nuclear-encoded tRNATyr genes contain intervening sequences (16,31–33). It has been shown by several groups that the intron is a prerequisite for the essential modification within the GΨA anticodon (i.e., U35→Ψ) of all cytoplasmic tRNAsTyr (17,32–34). Plant tRNA introns exhibit a highly conserved secondary structure that is formed by interactions between bases of the anticodon loop and the intron. Generally, the 5′ and 3′ splice sites are located in single stranded loops and are separated by an extended anticodon stem of 4 extra bp (Fig. 3A, AtY3II). We have previously investigated the features required by plant splicing endonuclease for the efficient and accurate intron excision in wheat germ (12,16) and in tobacco nuclear extract (13,14), and have found that distortion of the conserved intron structure drastically impairs splicing. By means of the established transformation of Arabidopsis hypocotyls, we now wanted to confirm these in vitro data also in vivo. We used a cloned Arabidopsis tRNATyr gene (i.e., AtY3II) for the generation of amber suppressors. This tRNATyr gene is efficiently transcribed in HeLa and tobacco nuclear extract. Moreover, the intron-containing pre-tRNATyr is efficiently processed and spliced in wheat germ and tobacco extract (35; Y.Yukawa, unpublished result). A single mutation at position 34 in the coding region of the tRNATyr gene converts the GTA to a CTA anticodon, which is complementary to an amber codon (Fig. 3A). It has previously been shown that this amber suppressor tRNATyr gene (AtY3II-am) was efficiently transcribed in vitro; however, intron excision was severely impaired (17). Co-infection of Arabidopsis hypocotyls with Ti plasmids pIG121/GUS amber and pBI/AtY3II-am did not result in any detectable GUS staining in histochemical analyses (Fig. 3B). The inability of tRNATyr derived from AtY3II-am to suppress the amber codon in GUS mRNA is very likely due to the observation made in vitro: that the intron of the corresponding pre-tRNATyr is not excised as a result of a distorted intron structure. The single nucleotide exchange in the anticodon to generate an amber suppressor tRNATyr destroys the third base pair in the extended anticodon stem of intron-containing pre-tRNATyr. A second mutation introduced at position number 7 of the intron (i.e., AtY3II-amiG7) restores this base pair (Fig. 3A). In vitro assays have shown that the corresponding pre-tRNATyr is in fact as efficiently spliced as pre-tRNATyr derived from the wildtype gene (17). Transformation of Arabidopsis-derived hypocotyls with Ti plasmids pIG121/GUS amber and pBI/AtY3II-amiG7 resulted in extensive GUS staining (Fig. 3B), suggesting that the pre-tRNATyr derived from the modified amber suppressor tRNATyr has been spliced as expected from the in vitro results. Interestingly, transformation with the intron-less amber suppressor tRNATyr gene (i.e., AtY3IIΔ-am) results in only half of the detectable GUS activity in histochemical and enzymatic analyses as compared with transformation with the intron-containing tRNATyr gene derived from AtY3II-amiG7 (Fig. 3B and C).
Figure 3.
Suppression of a premature stop codon in GUS mRNA mediated by amber suppressor tRNAsTyr. (A) Cloverleaf structures of intron-containing pre-tRNATyr with CΨA and of mature tRNATyr with CUA anticodon. The tRNAsTyr are derived from the recombinant plasmid AtY3II which contains a nuclear-encoded tRNATyr (GTA) gene with a 12 bp long intron (21,35). A single mutation at position 34 of the anticodon (G→C) converts the tRNATyr into a potential amber suppressor. The pre-tRNATyr derived from AtY3II-amiG7 contains an additional mutation in the intron (iC7→iG7) which restores a helical structure of 4 bp between the two splice sites. The recombinant plasmid AtY3IIΔ-am carries an amber suppressor tRNATyr gene, the intron of which has been removed. The introduced mutations are indicated by white letters on a black background. Arrows point to the 5′ and 3′ splice sites. Dots identifiy the anticodon. (B) Histochemical analyses of GUS activity in hypocotyl-derived Arabidopsis calli after transformation with Agrobacterium strains, each containing pIG121/GUS amber and, in addition, the corresponding Ti plasmids with amber suppressor tRNATyr genes as indicated below. (C) Relative GUS enzymatic activity in hypocotyl-derived Arabidopsis calli transformed accordingly. All values are displayed as the mean percentage of activity as compared to that of AtY3II-amiG7 after the removal of background activity (transformed with pIG121/GUS amber only).
Construction of an amber suppressor tRNAMet that is a substrate for alanyl-tRNA synthetase in vitro
In nuclear genomes a second intron-containing tRNA gene family exists that codes for elongator tRNAMet. In contrast to the intron-containing tRNATyr gene family, it is found exclusively in higher and lower plants (13,15) and not in other eukaryotic genomes. In addition to studying splicing of intron-containing pre-tRNAsTyr, we have also examined the features required for efficient intron excision from pre-tRNAsMet in wheat germ and tobacco nuclear extract (13,14) as a prerequisite for corresponding in vivo studies.
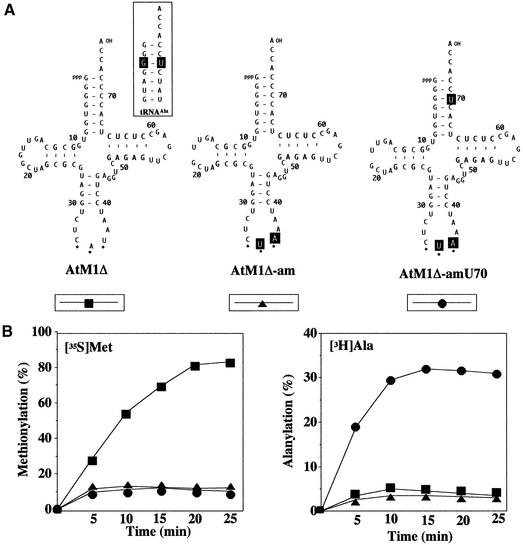
We cloned intron-containing and intron-less amber suppressor tRNAMet genes (AtM1-am and AtM1Δ-am) in the Ti vector plasmids (Fig. 1B) and examined their capability to suppress the premature amber codon of the reporter GUS gene (Fig. 1A) in transformed Arabidopsis hypocotyls. However, neither histochemical nor enzymatic analyses of GUS activity of tissue transformed with either of the two suppressor tRNAMet genes revealed any significant suppressor activity (not shown). Although the identity determinants of plant cytosolic tRNAsMet are not known, the outcome of this first series of experiments was not quite unexpected, since it has been demonstrated that all three anticodon bases of tRNAMet are important for the recognition by mammalian methionyl-tRNA synthetase (36–38) and the conversion of wildtype tRNAMet to an amber suppressor necessitates two base exchanges in the anticodon (Fig. 4A). In order to generate a functional tRNAMet suppressor we focused our attention on the G3:U70 base pair in the acceptor stem of tRNAAla which is the major recognition element of alanyl-tRNA synthetase in pro- and eukaryotes. Moreover, it has been demonstrated in vivo and in vitro that transfer of this base pair to a variety of other tRNA isoacceptors (mostly amber suppressors) conferred a complete or partial tRNAAla identity on these molecules (39,40). A comparison of the nucleotide sequences of the acceptor stem regions from Arabidopsis tRNAAla and elongator tRNAMet indicates that they are very similar. In fact, the first 4 bp (with the exception of the nucleoside at position 70) and the discriminator base are identical (Fig. 4A). This is of particular significance, because minor identity elements such as the three base pairs G1:C72; G2:C71; G4:C69 and the discriminator base A73 have been discussed to be also important for the recognition by alanyl-tRNA synthetase (41), suggesting that an identity switch from plant amber suppressor tRNAMet to tRNAAla should be feasible. In order to study first the effect of the C70→U70 substitution on tRNAMet aminoacylation in vitro, a T7 RNA polymerase promoter sequence and a BstNI recognition site were inserted upstream and downstream, respectively, of the coding regions of AtM1Δ, AtM1Δ-am and AtM1Δ-amU70. Run off transcription of the BstNI-cleaved plasmid DNAs with T7 RNA polymerase yielded the corresponding transcripts with mature CCA end (Fig. 4A). They were subsequently used as templates for in vitro aminoacylation assays in the presence of either [35S]methionine or 3H-labeled alanine and a crude preparation of aminoacyl-tRNA synthetase from wheat germ. As expected, the wildtype tRNAMet transcript was efficiently aminoacylated with methionine, whereas the two amber suppressor tRNAsMet (with and without the G3:U70 base pair) were very poorly charged with methionine (Fig. 4B), indicating that the two base exchanges in the anticodon of the suppressors prevent aminoacylation with the cognate amino acid. In contrast, assays carried out in the presence of [3H]alanine revealed that the suppressor tRNAMet transcript with the G3:U70 base pair (AtM1Δ-amU70) was charged with alanine with an efficiency of up to 30% whereas the other two transcripts derived from AtM1Δ and AtM1Δ-am were not significantly charged with alanine (Fig. 4B). The aminoacylation assays were routinely performed in the presence of saturating amounts of wheat germ synthetase, i.e., higher synthetase concentrations did not increase alanylation. Furthermore, all transcripts were denatured before aminoacylation by heating for 4 min at 68°C in 5 mM MgCl2 followed by slow cooling to room temperature. This procedure had considerably improved the charging of tRNASer and tRNALeu T7 transcripts (42) but was without effect on tRNAMet transcripts and its derivatives.
Figure 4.
Kinetics of methionine and alanine acceptance of tRNAMet-derived transcripts in the presence of wheat germ S100 extract. (A) Cloverleaf structures of mature and amber suppressor tRNAsMet generated by transcription with T7 RNA polymerase from the corresponding intron-less tRNA genes. Mutation of C70→U creates a putative identity element, i.e., the base pair G3:U70, recognized by alanyl-tRNA synthetase (39). The introduced mutations are indicated by reversed letters. Dots identify the anticodon. For comparison, the acceptor stem of tRNAAla from A.thaliana is shown in the inset as deduced from the known gene sequence (55). (B) Aminoacylation was measured with 1 µM RNA transcript and saturating amounts of crude wheat germ synthetase in the presence of [35S]methionine or [3H]alanine. Values are given as percent tRNA transcript charged with the indicated amino acid.
Expression of modified amber suppressor tRNAMet genes (G3:U70) in Arabidopsis hypocotyls
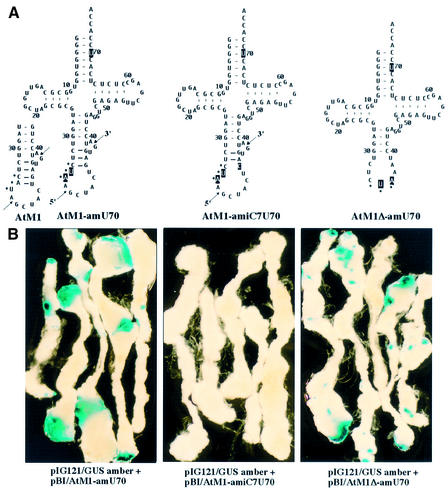
The modified intron-containing and intron-less amber suppressor tRNAMet genes containing the G3:U70 recognition element specific for alanyl-tRNA synthetase were cloned into the corresponding Ti plasmids (Fig. 5A). Agrobacterium-mediated transformation of Arabidopsis hypocotyls with pIG121/GUS amber and pBI/AtM1-amU70 or pBI/AtM1Δ-amU70 disclosed comparable GUS staining patterns in histochemical analyses, indicating that both types of tRNA genes (with and without intron) generated a functional amber suppressor in vivo (Fig. 5B). No GUS staining or GUS specific activity was detected in tissues that had been co-transformed with pIG121/GUS amber and pBI/AtM1-amiC7U70 (Fig. 5B). The latter intron-containing tRNAMet gene contains a mutation within the intron at position 7 (iG7→iC7). As a consequence, the third base pair of the extended anticodon stem formed by interactions between bases of the anticodon loop and the intron of pre-tRNAMet is disrupted (Fig. 5A). The absence of any suppressor activity in vivo of the tRNA derived from this substrate confirms our previous observations made in wheat germ and tobacco extract that intron excision is impaired due to the disturbed intron secondary structure (13).
Figure 5.
Suppression of a premature stop codon in GUS mRNA mediated by amber suppressor tRNAsMet. (A) Cloverleaf structures of intron-containing pre-tRNAsMet and of mature tRNAMet with CUA anticodon. The tRNAsMet are derived from the recombinant plasmid AtM1, which contains a nuclear-encoded elongator tRNAMet (CAT) gene with an 11 bp long intron (15). Two mutations at positions 35 and 36 of the anticodon convert the tRNAMet into a potential amber suppressor. The pre-tRNAMet derived from AtM1-amiC7 contains an additional mutation in the intron (iG7→iC7) which impairs the helical structure between the two splice sites. The recombinant plasmid AtM1Δ-am carries an amber suppressor tRNAMet gene, the intron of which has been removed. All tRNAsMet contain a uridine at position 70 (U70), which results in isoacceptors that are charged with alanine (Fig. 4). The introduced mutations are indicated by reverse letters. Arrows point to the 5′ and 3′ splice sites, respectively. Dots identify the anticodon. (B) Histochemical analyses of GUS activity in hypocotyl-derived Arabidopsis calli after transformation with Agrobacterium strains, each containing pIG121/GUS amber and, in addition, either of the Ti plasmids with amber suppressor tRNAMet genes as indicated below.
DISCUSSION
We have generated amber suppressor tRNA genes by site-directed mutagenesis of plant nuclear-encoded tRNASer (CGA), tRNATyr(GTA) and elongator tRNAMet(CAT) genes, and have studied their ability to suppress a premature amber stop codon within a modified GUS reporter gene in transformed Arabidopsis hypocotyls. All three types of amber suppressors have not been investigated in any plant system so far. However, the expression of tRNASer- and tRNATyr-derived suppressors has been studied in animal cells. For instance, amber and ochre suppressor tRNAs originating from a human tRNASer gene were shown to efficiently suppress nonsense mutations in viral mRNAs and in a chloramphenicol acetyltransferase (CAT) reporter gene in several mammalian cell lines (43,44). Furthermore, amber and ochre suppressors derived from a Xenopus and Drosophila, respectively, tRNATyr gene were active in monkey kidney cells (45,46) and in transformed Drosophila flies (47).
The amber suppressor tRNASer deduced from the cloned Nicotiana tRNASer gene NtS2 revealed a high readthrough activity. Arabidopsis hypocotyls co-transformed with the GUS reporter gene and the suppressor tRNASer gene exhibited reproducibly ∼10% of the GUS activity found in tissue transformed solely with the GUS wildtype gene (Fig. 2, Tables 1 and 2) and hence proved to be a powerful plant amber suppressor.
Table 2. Suppression of an amber codon in gusA mRNA mediated by three novel amber suppressor tRNAs in transformed Arabidopsis calli.
| Relative GUS activity (%)a | |
|---|---|
| pIG121/GUS amber | 0.01 ± 0.00 |
| pIG121/GUS amber + pBI/AtM1-amU70 (tRNAMet suppressor) | 0.09 ± 0.02 |
| pIG121/GUS amber + pBI/AtY3II-amiG7 (tRNATyr suppressor) | 0.61 ± 0.03 |
| pIG121/GUS amber + pBI/NtS2-am (tRNASer suppressor) | 10.45 ± 1.47 |
| pIG121/GUS wt | 100.00 ± 6.92 |
aActivities are the means of three experiments. ± Standard error.
One of the most efficient plant amber suppressors has been generated from a bean tRNALeu(CAA) gene. This tRNALeu gene is readily transcribed in heterologous and homologous systems, and in vitro synthesized transcripts derived from the suppressor tRNALeu(CTA) gene are excellent substrates for plant leucyl-tRNA synthetase (48). Tobacco protoplasts co-electroporated with the amber suppressor tRNALeu gene and a GUS reporter gene showed up to 20–25% of the activity found in protoplasts transfected with the functional control GUS gene (4).
Since the anticodon is very often a major determinant needed for the interaction of the tRNA molecule with its cognate synthetase (37,41), mischarging or inefficient charging of the suppressor tRNAs is a severe handicap that reduces their activity drastically. In eukaryotes, cytosolic tRNAsSer and tRNAsLeu are exceptional in that their unique long extra arm displays the major identity element (29,30,42). Consequently, changes in the anticodon of these two isoacceptors do not impair their charging with the cognate amino acid and therefore active suppressors are easily established without any further manipulations. In many cases, however, tRNA isoacceptors rely on their complete anticodon sequences for efficient aminoacylation (37). One elegant device to overcome this obstacle is to introduce the simple identity element of alanyl-tRNA synthetase, i.e., a G3:U70 base pair, into the corresponding suppressor tRNA. Thus, a successful identity switch has been achieved by the conversion of a G3:C70 to a G3:U70 base pair within Arabidopsis amber suppressor tRNAPhe. The mutated tRNAPhe was shown to be a good substrate for alanyl-tRNA synthetase in vitro and in vivo (40). Accordingly, we have exchanged the G3:C70 to a G3:U70 base pair in Arabidopsis amber suppressor tRNAMet (Fig. 4A). The wildtype tRNAMet transcript (AtM1Δ) was efficiently aminoacylated in vitro with [35S]methionine, whereas the mutant transcripts derived from AtM1Δ-am and AtM1Δ-amU70, respectively, were only poorly charged with the cognate amino acid. Conversely, aminoacylation assays in the presence of [3H]alanine revealed that only the transcript synthesized from AtM1Δ-amU70 was a substrate for alanyl-tRNA synthetase; however, the efficiency of charging of the chimeric tRNAMet was impaired (Fig. 4B). The presence of inactive T7 transcripts due to misfolded structures cannot be ruled out completely, although the transcripts were denatured and renatured before aminoacylation. Alternatively, unknown antideterminants within the chimeric RNA molecule might not allow optimal alanylation. Nevertheless, examination in vivo of the modified amber suppressor tRNAMet, containing the alanine-specific determinant, confirmed that it is able to significantly suppress the premature UAG codon in the GUS reporter gene (Fig. 5, Table 2).
Our main purpose to generate a new class of functional suppressors was to study subsequently the mechanism of pre-tRNA splicing in vivo. In plant nuclear genomes, only two tRNA families coding for tRNATyr and elongator tRNAMet contain intervening sequences (13,15,16). We have selected the cloned Arabidopsis intron-containing tRNA genes AtY3II and AtM1 for the generation of amber suppressors because their efficient transcription in vitro had been previously verified (13,35). An absolute prerequisite for effective and accurate splicing of pre-tRNAs by plant splicing endonuclease is a defined intron secondary structure that is formed by interactions between bases in the intron and in the anticodon. As a result, the 3′ and 5′ splice sites are located in single stranded regions and the anticodon helix is elongated by 3–5 bp (Fig. 3A). This conserved intron structure is found in all known tRNATyr and tRNAMet genes from lower and higher plants (13,15,16,49,50). The generation of an amber suppressor tRNATyr with CΨA instead of a GΨA anticodon results in the disruption of the third base pair in the extended anticodon stem of pre-tRNATyr (Fig. 3A), whereas the construction of an amber suppressor tRNAMet with CUA instead of a CAU anticodon creates a pre-tRNAMet in which an elongated anticodon helix of 3 bp is still maintained (Fig. 5A). Consequently, tRNAMet derived from AtM1-amU70 was an active amber suppressor in vivo (Fig. 5B), whereas tRNATyr derived from AtY3II-am showed no activity in transformed Arabidopsis hypocotyls (Fig. 3B). For the generation of a functional amber suppressor tRNATyr we had to modify the intron sequence in a way that restored the elongated anticodon helix of 4 bp (Fig. 3A). This modified tRNATyr gene (i.e., AtY3II-amiG7) is in fact a potent amber suppressor in vivo (Fig. 3B, Table 2). Conversely, we have demonstrated that disruption of the third base pair in pre-tRNAMet derived from AtM1-amiC7U70 abolished suppressor activity in vivo (Fig. 5). The observation that the two pre-tRNAs synthesized from the genes AtY3II-am and AtM1-amiC7U70 are not functional in vivo, whereas the same tRNAs derived from intron-less genes are active suppressors (Figs 3 and 5), confirms unambiguously that the inability of the former to suppress amber codons is solely due to a defect in pre-tRNA splicing caused by a distorted intron structure.
The minimal structural requirement for efficient tRNA intron excision is apparently less stringent in non-plant organisms. A variety of intron structures is accepted by the yeast, mammalian and Drosophila splicing endonuclease. In accordance with this observation, generation of functional amber or ochre suppressor tRNAsTyr has not necessitated additional manipulations within the intron of the corresponding tRNATyr genes (34,45–47).
The amber suppressor tRNATyr derived from the modified intron-containing gene AtY3II-amiG7 was about twice as efficient in vivo as the same suppressor synthesized from the intron-less gene AtY3IIΔ-am (Fig. 3C). This observation can be explained by the presumed presence and absence, respectively, of pseudouridine (Ψ35) in the anticodon of the corresponding mature tRNAsTyr which is thought to strengthen conventional and unconventional anticodon–codon interactions (27). It has been shown that the conversion of U35 to Ψ by Ψ35 synthase requires intron-containing pre-tRNAs as substrates in yeast (34,51), Xenopus oocytes (33), HeLa cells (32) and wheat germ (52). Since the nucleotide at the first position of the anticodon (N34) is unimportant for the activity of Ψ35 synthase, amber or ochre suppressor tRNAsTyr derived from intron-containing genes carry a CΨA or UΨA anticodon, respectively, whereas suppressor tRNAsTyr synthesized from the intron-less counterparts have a CUA or UUA anticodon. The transformation of an appropriate yeast strain with an ochre suppressor tRNATyr gene resulted in significant lower suppressor activity relative to the unaltered gene, if the intervening sequence had been deleted (34) in accordance with our own observations.
The amber suppressor tRNAMet (AtM1-amU70) was not significantly more active than tRNAMet synthesized from the intron-less gene AtM1Δ-amU70 (Fig. 5). We conclude from these results that Ψ35 synthesis has not occurred in the former, although the corresponding pre-tRNAMet carries almost all necessary features required by plant Ψ35 synthase such as the presence of an intron and the consensus sequence surrounding U35 (52).
The three new amber suppressor tRNAs that are active in a plant in vivo system after some modifications differ in their competence by the order of two magnitudes. The amber suppressor tRNASer reads the premature UAG stop codon in the GUS mRNA with an efficiency of 10%, and therefore can be regarded as a potent plant amber suppressor comparable to the known suppressors tRNALeu (4), tRNATrp (3) and tRNALys (9), whereas the tRNATyr (AtY3II-amiG7) and tRNAMet (AtM1-amU70) suppressors assert readthrough values of only 0.6 and 0.1%, respectively (Table 2). Above all, the relatively low activities of the latter two can be explained by impaired aminoacylation as already discussed above for amber suppressor tRNAMet (Fig. 4). The major tyrosine identity determinants are the base pair C1:G72 and the discriminator residue A73; however, the three anticodon bases G34, U35 and A36 also contribute to the tyrosine identity with G34 having the most pronounced effect (53), suggesting that suppressor tRNATyr with CΨA anticodon might be charged with a reduced rate by tyrosyl-tRNA synthetase.
Although the modified amber suppressors tRNATyr and tRNAMet are relatively weak suppressors in vivo, we could study various aspects of pre-tRNA splicing, aminoacylation and modification by means of tRNA-dependent suppression, because of two reasons. First, the mutant GUS gene used in this study expressed GUS activities not significantly above background (Tables 1 and 2). This implies that natural endogenous suppressor tRNAs have not interacted with the premature UAG stop codon in GUS mRNA. A powerful natural UAG suppressor present in all plant tissues (albeit to a different extent) is normal cytosolic tRNATyr with GΨA anticodon (54). However, the activity of this isoacceptor depends strongly on a favorable codon context, that is, only the sequence CARYYA at the 3′ side of the suppressed stop codon confers leakiness in vivo and in vitro (26,27). Second, the versatile system, i.e., the Agrobacterium-mediated gene delivery to Arabidopsis hypocotyl explants (18), used here for the first time to study the transient expression of suppressor tRNA genes has proven very valuable because it allows both histochemical and enzymatic GUS analyses.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Kenzo Nakamura (Nagoya University, Japan) for kindly providing the Ti plasmid pIG121/Hm. We are grateful to H. J. Gross for many stimulating discussions and suggestions and for critical reading of the manuscript. This work was supported by Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology (no. 13680766) and from the Kato Memorial Bioscience Foundation to K.A.
REFERENCES
- 1.Small I., Akama,K., Akashi,K., Chapron,A., Choisne,N., Dietrich,A., Drouard,L., Duchene,A.M., Giritch,A., Lancelin,D., Peeters,N., Souciet,G. and Wintz,H. (1999) taaRSAt, a database of tRNAs and aminoacyl-tRNA synthetases from Arabidopsis thaliana. Biochimie, 81, s341. [Google Scholar]
- 2.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 3.Franklin S., Lin,T.Y. and Folk,W.R. (1992) Construction and expression of nonsense suppressor tRNAs which function in plant cells. Plant J., 2, 583–588. [DOI] [PubMed] [Google Scholar]
- 4.Carneiro V.T.C., Pelletier,G. and Small,I. (1993) Transfer RNA-mediated suppression of stop codons in protoplasts and transgenic plants. Plant Mol. Biol., 22, 681–690. [DOI] [PubMed] [Google Scholar]
- 5.Ulmasov B. and Folk,W. (1995) Analysis of the role of 5′ and 3′ flanking sequence elements upon in vivo expression of the plant tRNATrp genes. Plant Cell, 7, 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choisne N., Martin-Canadell,A. and Small,I. (1997) Transactivation of a target gene using a suppressor tRNA in transgenic tobacco plants. Plant J., 11, 597–604. [DOI] [PubMed] [Google Scholar]
- 7.Choisne N., Carneiro,V.T.C., Pelletier,G. and Small,I. (1997) Implication of 5′-flanking sequence elements in expression of a plant tRNALeu gene. Plant Mol. Biol., 36, 113–123. [DOI] [PubMed] [Google Scholar]
- 8.Betzner A.S., Oakes,M.P. and Huttner,E. (1997) Transfer RNA-mediated suppression of amber stop codons in transgenic Arabidopsis thaliana. Plant J., 11, 587–595. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Ulmasov,B. and Folk,W.R. (1998) Nonsense and missense translational suppression in plant cells mediated by tRNALys. Plant Mol. Biol., 36, 163–170. [DOI] [PubMed] [Google Scholar]
- 10.Stange N. and Beier,H. (1987) A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J., 6, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stange N., Gross,H.J. and Beier,H. (1988) Wheat germ splicing endonuclease is highly specific for plant pre-tRNAs. EMBO J., 7, 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stange N., Beier,D. and Beier,H. (1992) Intron excision from tRNA precursors by plant splicing endonuclease requires features of the mature tRNA domain. Eur. J. Biochem., 210, 193–203. [DOI] [PubMed] [Google Scholar]
- 13.Akama K., Junker,V., Yukawa,Y., Sugiura,M. and Beier,H. (2000) Splicing of Arabidopsis tRNAMet precursors in tobacco cell and wheat germ extracts. Plant Mol. Biol., 44, 155–165. [DOI] [PubMed] [Google Scholar]
- 14.Yukawa Y., Fan,H., Akama,K., Beier,H., Gross,H.J. and Sugiura,M. (2001) A tobacco nuclear extract supporting transcription, processing, splicing and modification of plant intron-containing tRNA precursors. Plant J., 28, 583–594. [DOI] [PubMed] [Google Scholar]
- 15.Akama K. and Kashihara,M. (1996) Plant nuclear tRNAMet genes are ubiquitously interrupted by introns. Plant Mol. Biol., 32, 427–434. [DOI] [PubMed] [Google Scholar]
- 16.Akama K., Naß,A., Junker,V. and Beier,H. (1997) Characterization of nuclear tRNATyr introns: their evolution from red algae to higher plants. FEBS Lett., 417, 213–218. [DOI] [PubMed] [Google Scholar]
- 17.Szweykowska-Kulinska Z. and Beier,H. (1991) Plant nonsense suppressor tRNATyr genes are expressed at very low levels in vitro due to inefficient splicing of the intron containing pre-tRNAs. Nucleic Acids Res., 19, 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akama K., Shiraishi,H., Ohta,S., Nakamura,K., Okada,K. and Shimura,Y. (1992) Efficient transformation using hypocotyl explants of Arabidopsis thaliana: comparison of the efficiencies with various organs, plant ecotypes and Agrobacterium strains. Plant Cell Rep., 12, 7–11. [DOI] [PubMed] [Google Scholar]
- 19.Ohta S., Mita,S., Hattori,T. and Nakamura,K. (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol., 31, 805–813. [Google Scholar]
- 20.Teichmann T., Urban,C. and Beier,H. (1994) The tRNASer-isoacceptors and their genes in Nicotiana rustica: genome organization, expression in vitro and sequence analyses. Plant Mol. Biol., 24, 889–901. [DOI] [PubMed] [Google Scholar]
- 21.Beier D., Stange,N., Gross,H.J. and Beier,H. (1991) Nuclear tRNATyr genes are highly amplified at a single chromosomal site in the genome of Arabidopsis thaliana. Mol. Gen. Genet., 225, 72–80. [DOI] [PubMed] [Google Scholar]
- 22.Deblaere R., Bytebier,B., De Greve,H., Deboeck,F., Schell,J., Van Montagu,M. and Leemans,J. (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res., 13, 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant, 15, 473–497. [Google Scholar]
- 24.Jefferson R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep., 5, 387–405. [Google Scholar]
- 25.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 26.Skuzeski J.M., Nichols,L.M., Gesteland,R.F. and Atkins,J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- 27.Zerfaß K. and Beier,H. (1992) Pseudouridine in the anticodon GΨA of plant cytoplasmic tRNATyr is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res., 20, 5911–5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X.-Q. and Gross,H.J. (1993) The long extra arm of human tRNA(Ser)Sec and tRNASer function as major identity elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res., 21, 5589–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Achsel T. and Gross,H.J. (1993) Identity determinants of human tRNASer: sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J., 12, 3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman H.M., Olson,M.V. and Hall,B.D. (1977) Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc. Natl Acad. Sci. USA, 74, 5453–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Tol H. and Beier,H. (1988) All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res., 16, 1951–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choffat Y., Suter,B., Behra,R. and Kubli,E. (1988) Pseudouridine modification in the tRNATyr anticodon is dependent on the presence, but independent of the size and sequence, of the intron in eucaryotic tRNATyr genes. Mol. Cell. Biol., 8, 3332–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson P.F. and Abelson,J. (1983) The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature, 302, 681–687. [DOI] [PubMed] [Google Scholar]
- 35.Stange N., Beier,D. and Beier,H. (1991) Expression of nuclear tRNATyr genes from Arabidopsis thaliana in HeLa cell and wheat germ extracts. Plant Mol. Biol., 16, 865–875. [DOI] [PubMed] [Google Scholar]
- 36.Meinnel T., Mechulam,Y., Fayat,G. and Blanquet,S. (1992) Involvement of the size and sequence of the anticodon loop in tRNA recognition by mammalian and E.coli methionyl-tRNA synthetases. Nucleic Acids Res., 20, 4741–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallanck L., Pak,M. and Schulman,L.H. (1995) tRNA discrimination in aminoacylation. In Söll,D. and RajBhandary,U.L. (eds), tRNA, Structure, Biosynthesis, and Function. ASM Press, Washington, DC, pp. 371–394.
- 38.Drabkin H.J., Estrella,M. and RajBhandary,U.L. (1998) Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell. Biol., 18, 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou Y.-M. and Schimmel,P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature, 333, 140–145. [DOI] [PubMed] [Google Scholar]
- 40.Carneiro V.T.C., Dietrich,A., Maréchal-Drouard,L., Cosset,A., Pettetier,G. and Small,I. (1994) Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol. Biol., 26, 1843–1853. [DOI] [PubMed] [Google Scholar]
- 41.Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitschopf K., Achsel,T., Busch,K. and Gross,H.J. (1995) Identity elements of human tRNALeu: structural requirements for converting human tRNASer into a leucine acceptor in vitro. Nucleic Acids Res., 23, 3633–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capone J.P., Sharp,P.A. and RajBhandary,U.L. (1985) Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J., 4, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capone J.P., Sedivy,J.M., Sharp,P.A. and RajBhandary,U.L. (1986) Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol. Cell. Biol., 6, 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laski F.A., Belagaje,R., RajBhandary,U.L. and Sharp,P.A. (1982) An amber suppressor tRNA gene derived by site-specific mutagenesis: cloning and function in mammalian cells. Proc. Natl Acad. Sci. USA, 79, 5813–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laski F.A., Belagaje,R., Hudziak,R.M., Capecchi,M.R., Norton,G.P., Palese,P., RajBhandary,U.L. and Sharp,P.A. (1984) Synthesis of an ochre suppressor tRNA gene and expression in mammalian cells. EMBO J., 3, 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laski F.A., Ganguly,S., Sharp,P.A., RajBhandary,U.L. and Rubin,G.M. (1989) Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 86, 6696–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Small I., Maréchal-Drouard,L., Masson,J., Pettetier,G., Cosset,A., Weil,J.H. and Dietrich,A. (1992) In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants—towards novel ways of influencing mitochondrial gene expression. EMBO J., 11, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs T., Beier,D. and Beier,H. (1992) The tRNATyr multigene family of Nicotiana rustica: genome organization, sequence analyses and expression in vitro. Plant Mol. Biol., 20, 869–878. [DOI] [PubMed] [Google Scholar]
- 50.Arends S., Kraus,J. and Beier,H. (1996) The tRNATyr multigene family of Triticum aestivum: genome organization, sequence analyses and maturation of intron-containing pre-tRNAs in wheat germ extract. FEBS Lett., 384, 222–226. [DOI] [PubMed] [Google Scholar]
- 51.Grosjean H., Szweykowska-Kulinska,Z., Motorin,Y., Fasiolo,F. and Simos,G. (1997) Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie, 79, 293–302. [DOI] [PubMed] [Google Scholar]
- 52.Szweykowska-Kulinska Z. and Beier,H. (1992) Sequence and structure requirements for the biosynthesis of pseudouridine (Ψ35) in plant pre-tRNATyr. EMBO J., 11, 1907–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fechter P., Rudinger-Thirion,J., Théobald-Dietrich,A. and Giegé,R. (2000) Identity of tRNA for yeast tyrosyl-tRNA synthetase: tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry, 39, 1725–1733. [DOI] [PubMed] [Google Scholar]
- 54.Beier H., Barciszewska,M. and Sickinger,H.-D. (1984) The molecular basis for the differential translation of TMV RNA in tobacco protoplasts and wheat germ extracts. EMBO J., 3, 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akama K. and Tanifuji,S. (1990) Sequence analysis of three tRNAPhe nuclear genes and a mutated gene, and one gene for tRNAAla from Arabidopsis thaliana. Plant Mol. Biol., 15, 337–346. [DOI] [PubMed] [Google Scholar]