Abstract
Fragment C (TetC) is a non-toxic 47 kDa polypeptide fragment of tetanus toxin that can be used as a subunit vaccine against tetanus. Expression of TetC in Escherichia coli and yeast was dependent on the availability of synthetic genes that were required to improve translation efficiency and stabilize the mRNA. To explore the feasibility of producing TetC in tobacco leaves, we attempted expression of both the bacterial high-AT (72.3% AT) and the synthetic higher-GC genes (52.5% AT) in tobacco chloroplasts. We report here that the bacterial high-AT mRNA is stable in tobacco chloroplasts. Significant TetC accumulation was obtained from both genes, 25 and 10% of total soluble cellular protein, respectively, proving the versatility of plastids for expression of unmodified high-AT and high-GC genes. Mucosal immunization of mice with the plastid- produced TetC induced protective levels of TetC antibodies. Thus, expression of TetC in chloroplasts provides a potential route towards the development of a safe, plant-based tetanus vaccine for nasal and oral applications.
INTRODUCTION
There is much interest in plant-based vaccines, which may be produced from genes stably incorporated in the nuclear genome, from plant viral vectors or by transient Agrobacterium-mediated gene delivery (1–4). Initial developments in the field of plant-based vaccines have been limited by low-level expression of immunogen from nuclear genes. An alternative method is to express vaccine antigens from the plastid genome, offering significant advantages over nuclear gene expression (5–7).
Our objective was to explore the feasibility of producing a mucosal tetanus vaccine by expressing the Fragment C domain of the tetanus toxin (TetC) in tobacco chloroplasts. TetC is a non-toxic 47 kDa polypeptide fragment shown to induce a protective immune response following parenteral immunization with preparations from Escherichia coli (8), yeast (9) and insect cells (10). Expression of TetC in E.coli was shown to be limited by the unfavorable codon bias of the highly AT-rich Clostridium tetani coding sequence; expression levels could be increased from a synthetic gene to ∼14% of cell protein (11). Since proteins expressed in E.coli may contain toxic cell wall pyrogens, TetC expression was also attempted in the non-toxic host Saccharomyces cerevisiae (9). The AT-rich C.tetani DNA could not be expressed in yeast due to the presence of several fortuitous polyadenylation sites which gave rise to truncated mRNAs. TetC accumulation was obtained when a codon-optimized high-GC gene, lacking the polyadenylation sites, was expressed in yeast. However, the yeast-produced TetC secreted in the culture medium was inactive as an immunogen due to glycosylation.
We report here that, in tobacco plastids, mRNAs expressed from both the high-AT bacterial and high-GC synthetic genes are stable. Significant TetC accumulation was obtained from both genes, 25 and 10% of TSP, respectively, proving the versatility of plastids for the expression of both high-AT and high-GC genes. Immunization of mice with the plastid-produced TetC induced protective levels of TetC antibodies, confirming the potential of chloroplasts for the production of a plant-based mucosal vaccine.
MATERIALS AND METHODS
Construction of transformation vectors
The TetC polypetide was expressed in chloroplasts from two different coding regions: the native AT-rich bacterial gene (tetC-AT) and a synthetic, relatively GC-rich sequence (tetC-GC). The tetC coding regions were PCR amplified to introduce an NdeI site including the translation initiation codon (ATG) and an XbaI site downstream of the stop codon. The tetC-AT coding region was PCR amplified with primers 5′-CGGGTACCCATATGAAAAATCTGGATTGTTGGGTCGACAATGAAG-3′and 5′-CGTCTAGAAATTAATCATTTGTCCATC-3′. The tetC-GC coding region was PCR amplified with primers 5′-CGGGTACCCATATGAAAAACCTTGATTGTTGG-3′ and 5′-GCTCTAGATTAGTCGTTGGTCCAACCT-3′. Templates for PCR amplification were plasmid pcDNA3/ntetC (tetC-AT) (12) and pcDNA3/tetC (tetC-GC) (13).
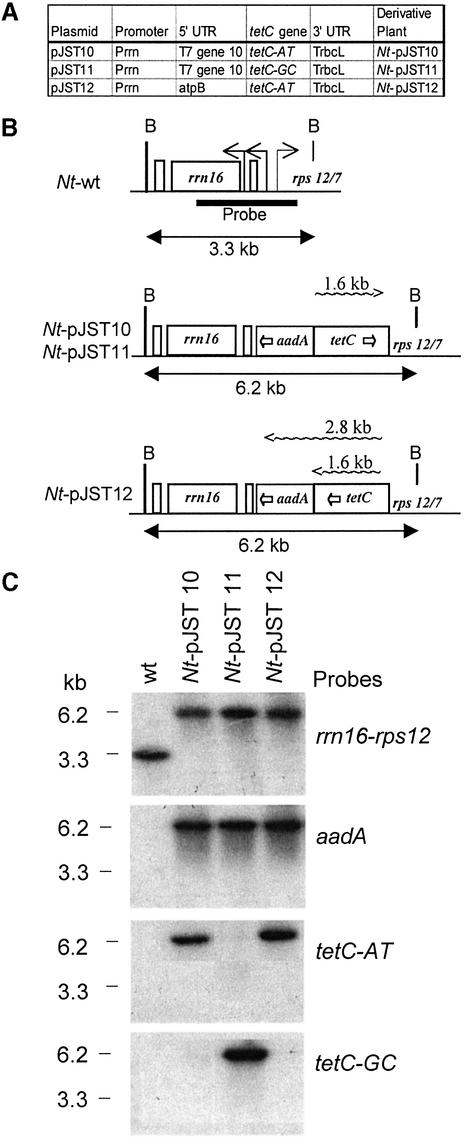
Plasmids pJST10 and pJST11 were obtained by replacing the neo coding region in plasmid pHK40 with the tetC-AT and tetC-GC coding regions as NdeI–XbaI fragments, respectively. Plasmid pHK40 is a plastid transformation vector derived from plasmid pPRV111A (14), with a spectinomycin resistance (aadA) gene as a selective marker and a neo gene expressed in a cassette consisting of a PrrnLT7g10 cassette and the rbcL 3′-UTR (TrbcL). The tetC genes are divergently oriented relative to the rrn operon (Fig. 1B).
Figure 1.
Transformed plastid genomes with tetC gene. (A) The plastid tetC genes. (B) Map of wild-type (Nt-wt) and transformed plastid genomes. Horizontal arrows mark promoters in wild-type plastid genomes. Horizontal wavy lines represent TetC transcripts. Abbreviations: rrn16 and rps12/7 are plastid genes; aadA, spectinomycin resistance gene; tetC, tetC gene; B, BamHI site. (C) DNA gel blot analysis confirms integration of tetC in plastid genome. Total cellular DNA was digested with the BamHI restriction enzyme and the blots were probed with targeting region (rrn16-rps12 probe) and the aadA, tetC-AT and tetC-GC probes. Note that tetC gene probes do not cross-hybridize due to differences in codon usage.
Plasmid pJST12 was obtained by replacing the neo coding region in plasmid pHK73 with the tetC-AT coding region as an NdeI–XbaI fragment. Plastid transformation vector pHK73 is a pPRV111B vector derivative in which the neo coding region is expressed in a cassette consisting of a PrrnLatpB cassette (plastid rrn operon promoter fused with atpB leader and an NdeI site including the ATG) and TrbcL. The PrrnLatpB cassette is identical to the cassette in plasmid pHK11 (15), except that an NdeI site was created by replacing AT with a CA at the –3/–2 position upstream of the ATG. The tetC gene in plasmid pJST12 is in tandem orientation with the rrn operon (Fig. 1B).
Plastid transformation
Plastid transformation was carried out as described previously (16). DNA for plastid transformation was prepared using the QIAGEN Plasmid Maxi Kit (Qiagen Inc., Valencia, CA). Transforming DNA was introduced into leaf chloroplasts on the surface of tungsten particles (1 µm) using the Du Pont PDS1000He Biolistic gun. Transplastomic plants were selected on RMOP medium containing 500 mg/l spectinomycin dihydrochloride. The transgenic plants were grown on Murashige-Skoog (MS) medium (17) containing 3% (w/v) sucrose and 0.6% (w/v) agar in sterile culture condition. A uniform population of transformed plastid genome copies was confirmed by DNA gel blot analysis. Double-stranded DNA probes were prepared by random-primed 32P-labeling using the Ready-To-Go DNA Labeling Beads (Amersham Pharmacia Biotech, Piscataway, NJ). The probes were: rrn16-rps12 plastid targeting region, ApaI–EcoRV ptDNA fragment; aadA, NcoI–XbaI coding region; tetC-AT and tetC-GC coding region, NdeI–XbaI fragments.
RNA gel blot analysis
RNA gel blot analysis was carried out as described previously by loading 3 µg total cellular RNA per lane (18). The templates for probing the tetC genes were NdeI–XbaI coding region fragments. The template for probing the tobacco cytoplasmic 25S rRNA was a PCR fragment amplified from total tobacco cellular DNA with primers 5′-TCACCTGCCGAATCAACTAGC-3′ and 5′-GACTTCCCTTGCCTACATTG-3′. Probes were prepared by random-primed 32P-labeling (see above). RNA hybridization signals were quantified using a Molecular Dynamics PhosphorImager and normalized to the 25S rRNA signal.
SDS–PAGE and immunoblotting
Leaves for protein extraction were taken from greenhouse plants. To obtain total soluble leaf protein, ∼200 mg of leaf was homogenized in 1 ml of buffer containing 50 mM HEPES–KOH (pH 7.5), 10 mM potassium acetate, 5 mM magnesium acetate, 1 mM EDTA, 1 mM or 10 mM DTT and 2 mM PMSF (10 mM DTT is 10 times more than we normally include in the extraction buffer). Insoluble material was removed by centrifugation. Protein concentrations were determined by the Bradford Protein Assay Reagent kit (Bio-Rad, Hercules, CA). Immunoblot analysis of TetC accumulation was carried out as described previously (19). In some experiments, in addition to mercaptoethanol (2.5%), 10 mM DTT was also included. The anti-TetC antibody was provided by Dr C. Turcotte, Imperial College, London. TetC was quantified on the immunoblots by densitometric analysis with the DensoSpot program of Alpha Imager 2000 (Alpha Innotech, San Leandro, CA) by comparison of the experimental samples with a recombinant TetC (rTetC) dilution series. Purified His6-tagged rTetC, expressed in a Novagen pET28a vector, was kindly provided by Mr O. Qazi, Imperial College, London.
Nasal immunization of mice
All mice were obtained from B & K suppliers (Scunthorpe, UK). For nasal immunization, a 15 µl volume of concentrated protein solution from plants expressing TetC or appropriate controls was applied to BALB-C female mice, 7.5 µl per nare. Protein extracts were prepared as for SDS–PAGE and concentrated using a Centriprep YM-10 (Millipore Ltd, Watford, UK). Cholera toxin (CT; 1 µg per dose) was added to nasal vaccines to act as mucosal adjuvant. Control TetC (rTetC) for mucosal immunization was prepared from E.coli. Mice were tail-bled at day 27 and samples stored at –20°C until assayed. On day 45 the mice were sacrificed by exsanguinations of the heart to collect serum samples. Lung and gut washes were performed, with 1 ml PBS containing protease inhibitors, Roche Complete protease inhibitor cocktail (Roche Diagnostics Ltd, Mannheim, Germany). All samples were stored at –20°C until required. For tetanus toxin challenge, mice were injected sub-cutaneously in the right flank with 0.5 ml PBS containing 50 PD50 of tetanus toxin (kindly provided by Dr Thea Sesardic, NIBSC) and sacrificed as soon as they showed symptoms of paralysis.
ELISAs for anti-TetC antibodies
Samples were assayed for anti-TetC IgG in serum and anti-TetC IgA in mucosal surface washes as described previously (20). For IgG measurements, samples were serially diluted in PBS containing 0.05% Tween-20 (PBST); for IgA readings, samples were serially diluted in PBST containing protease inhibitors (Roche Complete protease inhibitor cocktail). Microtiter plates were coated with 3 µg/µl TetC. Serum anti-TetC was determined with goat anti-mouse IgG (γ-chain specific) antisera conjugated with alkaline phophatase (Sigma-Aldrich, Dorset, UK). Mucosal anti-TetC IgA was determined with goat antiserum against mouse IgA (α-chain specific) conjugated to streptavidin (Sigma-Aldrich); this was then detected with biotinylated alkaline phosphatase (Dako, Ely, UK). Response was measured against the color change of OPD reagent (Sigma-Aldrich). Antibody titer was defined as the reciprocal of the dilution of antibody that produces an A490 of 0.3 for IgG readings and A490 of 0.2 for IgA readings.
RESULTS
Construction of transplastomic tobacco plants with tetC genes
The TetC polypeptide was expressed in tobacco chloroplasts from three different genes (Fig. 1A). Plastid vectors pJST10 and pJST12 encode the AT-rich tetC (tetC-AT; 72.3% AT) native coding region derived from C.tetani. Vector pJST11 encodes a more GC-rich synthetic tetC reading frame, successfully expressed in yeast (tetC-GC; 52.6% AT) (9). The tetC coding regions in plastids were expressed from the strong plastid rRNA operon (Prrn) promoter fused with a DNA segment encoding the T7 phage gene 10 (pJST10, pJST11) or the plastid atpB (pJST12) 5′-UTR. The engineered tetC genes were cloned in suitable plastid vectors (Fig. 1B) and introduced into the tobacco plastid genome by standard protocols. Incorporation of the tetC genes in the plastid genome was confirmed by DNA gel blot analysis (Fig. 1C). Several independently transformed lines were obtained with each of the constructs. The phenotype of plants transformed with vector pJST11 and pJST12 (Nt-pJST11, Nt-pJST12) was similar to the wild-type, non-transformed plants. The Nt-pJST10 plants grew slower in the greenhouse and had a chlorotic (pigment deficient) phenotype (Fig. 2). Since plants derived from independent transformation events were similar, data are shown only for one transformed clone per construct.
Figure 2.
High-level expression of TetC is detrimental to plants. Shown are an Nt-pJST10 shoot with chlorotic phenotype grafted onto wild-type plant (wt) and an Nt-pJST11 plant.
Expression of the tetC genes in chloroplasts
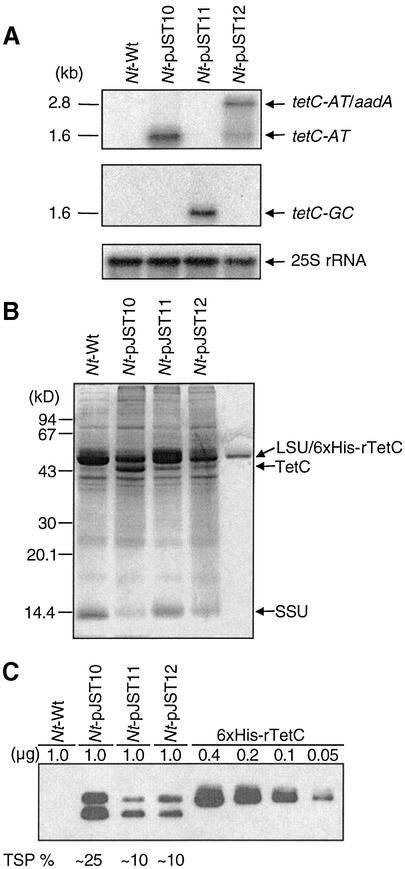
RNA gel blot analysis confirmed transcript accumulation for each of the three tetC genes (Fig. 3A). The Nt-pJST10 and Nt-pJST11 plants contained monocistronic messages, whereas the Nt-pJST12 plants accumulated both monocistronic and dicistronic transcripts due to read-through transcription through TrbcL (Fig. 1B). Steady-state tetC transcript levels in the Nt-pJST10 and Nt-pJST12 leaves were comparable (Fig. 3A). Relative amounts of tetC-GC transcripts in the Nt-pJST11 plants could not be determined, as the coding region probes did not cross-hybridize.
Figure 3.
Expression of tetC genes. (A) Accumulation of mRNA from the tetC genes. Monocistronic and dicistronic transcripts detected by the coding region probes are marked in Figure 1B. Relative amounts of mRNAs were quantified using cytoplasmic 25S rRNA as reference. (B) Coomassie Blue-stained protein gel with novel plant-produced 43 kDa TetC band. Control rTetC and the Rubisco large and small subunits (LSU, SSU) are also marked. (C) Immunoblots to quantify TetC in leaf protein extracts.
The fidelity of TetC protein expression in chloroplasts was confirmed by immunoblot analysis of protein extracts separated by SDS–PAGE (Fig. 3B and C). Two polypeptides were recognized by anti-TetC antisera: a 47-kDa protein that co-migrated with the Rubisco large subunit (LSU) and a 43-kDa protein, also visible in Coomassie Blue-stained SDS–PAGE gels (Fig. 3B). The 43-kDa band is the full-size TetC protein as it has both the intact TetC N- and C-termini based on micro-sequencing gel-purified protein (Edman degradation and ESI-MS/MS; data not shown) and has higher mobility than the His6-tagged rTetC control. The N-terminal amino acid sequence of the 43-kDa protein was MKNLD, indicating that the initiator methionine is not removed post-translationally. Thus, the 47 kDa protein is apparently a modified TetC. Other prominent bands on the gels are the Rubisco large and small subunits (LSU and SSU, respectively, in Fig. 3B) that together constitute ∼50% of TSP in wild-type plants.
Examination of western blots using densitometry and purified TetC standards showed TetC protein to be ∼25% TSP in the leaves of Nt-pJST10 plants and ∼10% TSP in Nt-pJST11 and Nt-pJST12 leaves (Fig. 3C). TetC levels in different experiments were in the 18–27% range in Nt-pJST10 leaves and in the 7–10% range in Nt-pJST11 and Nt-pJST12 leaves. The Nt-pJST10 plants, but not the Nt-pJST11 or Nt-pJST12 plants, have a mutant phenotype (Fig. 2). Since the Nt-pJST10 leaves contain the highest TetC level, ∼25% TSP, the mutant phenotype may be directly linked to TetC expression levels.
Immunization of mice with leaf protein extract harboring TetC
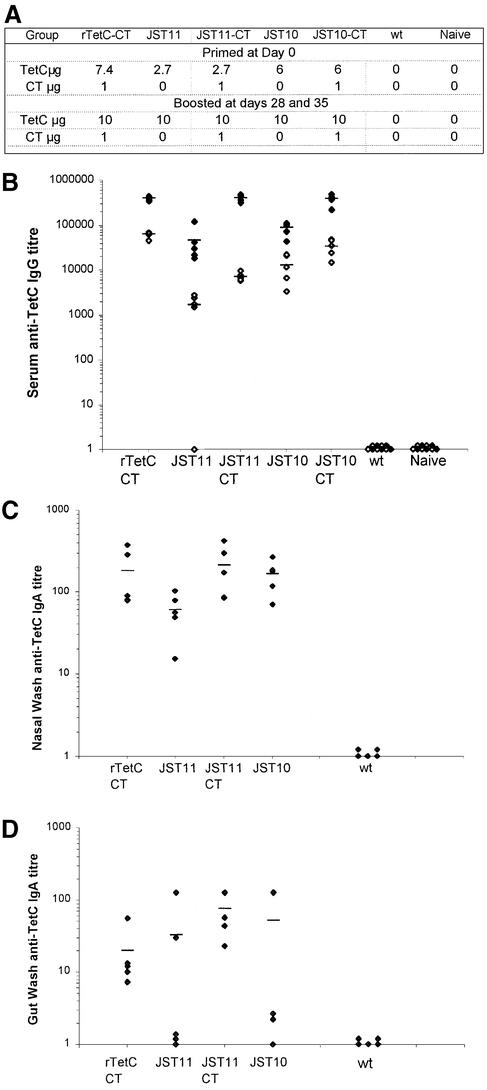
Groups of mice were immunized intranasally with vaccines harboring extracts from transgenic tobacco leaves mixed with small amounts of purified CT acting as adjuvant (Fig. 4A). The mice were primed on day 0. First blood samples were taken on day 27. The mice were boosted at days 28 and 35. Final mucosal surface wash samples and sera were collected on day 45. Control mice were immunized with recombinant TetC purified from E.coli (rTetC) and mixed with CT, non-transgenic leaf extract (wild-type) or PBS (naive mice sample).
Figure 4.
Response to nasal vaccination with plant-produced TetC. (A) Immunization schedule and dosing. (B) Serum anti-TetC IgG antibody titer indicates systemic immune response. (C) Nasal anti-TetC IgA titer. (D) Gut anti-TetC IgA titer. Open symbols are values after 27 days; filled symbols are values after 45 days. The bars are averages of five measurements.
All groups of mice immunized with extract from transgenic plants expressing TetC had a significant systemic anti-TetC IgG response at day 27 (Fig. 4B). Levels of specific anti-TetC IgG were boosted after re-immunization at days 28 and 35. Anti-TetC IgA antibody responses were seen at the immunized site in nasal washes (Fig. 4C) and at a distal site in gut washes (Fig. 4D). Mice immunized with non-transgenic leaf extract or buffer alone (PBS) failed to mount a detectable TetC-specific antibody response. Levels of anti-TetC antibodies, comparable to those obtained here, have consistently been observed to confer solid protection against a toxin challenge (21).
In order to test if the anti-TetC antibody responses raised in the intranasal trial were protective, five mice of the JST10-CT group were challenged with tetanus toxin (50 PD50; 50× the dose required to cause paralysis in 50% of mice). IgG titers of the mice were: 364183, 489583, 413829, 218493 and 375292 (note that the values overlap on the plot). As a control group, five naive mice were used. The mice were monitored at 6 h time points (four times a day) after inoculation for 5 days. The first two positive mice in the naive group were seen at 24 h. The rest of the naive mice developed symptoms of paralysis by 42 h. All mice immunized with leaf extract harboring the plant-derived TetC survived the challenge and were free of symptoms.
DISCUSSION
We report here high-level TetC accumulation from both native bacterial high-AT (∼25% of TSP) and the synthetic high-GC (∼10% of TSP) tetC genes, confirming the utility of plastids for the expression of unmodified genes from diverse sources.
Expression of TetC from the native bacterial coding sequence was feasible, because the mRNA transcribed from the tetC-AT gene was stable. Although plants are eukaryotes, their plastids share some features of prokaryotic machinery for mRNA turnover (22,23), explaining why tetC mRNA is not degraded in plastids. Another example for a bacterial gene that is unstable in a eukaryotic (plant nucleus) environment but stable in plastids is the Bacillus thuringiensis (Bt) Cry1Ac insecticidal protein gene (24,25). Interestingly, mRNAs derived from the tetC-GC gene (this study) and the high-GC bacterial bar (68.3%) (26) were also stable, indicating the compatibility of plastid degradation machinery with mRNAs from diverse sources.
Studies in heterologous expression hosts have shown that codon usage can greatly influence the level of expression of foreign proteins (27,28). The importance of codon bias in plastids was investigated here by expressing the native bacterial AT-rich (72.3% AT) and codon-modified more GC-rich (52.6% AT) tetC genes. The AT-rich gene reproducibly yielded about twice as much TetC as the GC-rich gene in the same cassette: ∼25% of TSP compared with ∼10% of TSP. More efficient translation of the bacterial tetC-AT coding region reflects the plastid genome’s codon bias, which is also relatively AT-rich (29). A similar (1.5-fold) increase in protein level was observed when comparing expression from bacterial and codon optimized synthetic CP4 genes (30). Thus, the increase in protein levels that may be obtained by codon optimization is relatively modest, and it is in a few-fold range. A relatively small effect of codon modification on protein yields was predictable, as in tobacco plastids there is no example for low codon usage frequencies (1.4/2.1 per 1000 codons) that are correlated with problems of expressing eukaryotic cDNAs in E.coli (27,28). Based on 19 tobacco plastid gene sequences (4245 codons; NCBI Codon Usage Database), the least frequently used codons are UGC and CGC (3.5 per 1000 codons) followed by CCG, CGG and AGG (4.0, 4.2 and 4.9 per 1000 codons, respectively). The relatively modest (few-fold) gains obtainable by codon modification in Nicotiana tabacum, a higher plant, contrast with the 80-fold increase obtained by codon optimization in the chloroplasts of Chlamydomonas reinhardtii, a unicellular alga (31). In higher-plant plastids, the targets for engineering protein expression are the translation control signals, which may affect protein accumulation in the 10 000-fold range (reviewed in 6,7) (14,15,30).
The Nt-pJST10 plants are the first transplastomic plants in which high-level protein accumulation (∼25% of TSP) expressed from a transgene causes a mutant phenotype. A mutant phenotype in a different transplastomic line, with a low level of heterologous protein expression (0.26% of TSP), is caused by competition for a gene-specific mRNA maturation factor (32). We assume that abnormal chloroplast development in the Nt-pJST10 plants is due to high-level TetC accumulation, as expression of a different protein, neomycin phosphotransferase II, at ∼23.00 ± 5.4% TSP from the same cassette did not result in chlorophyll deficiency (14).
Anti TetC-antisera detected two TetC forms. We believe the 43 kDa form to be the intact TetC, based on intact N- and C-termini, and mobility that is faster than that of the His-tagged control rTetC. The reason for the altered mobility of the slower (47 kDa) TetC form is unknown. Increasing the concentration of DTT in the isolation buffer and in SDS–PAGE did not prevent formation of the two bands (data not shown), a treatment that prevented formation of an artefact in E.coli-produced TetC (8). Glycosylation was also considered as the reason for the mobility shift since TetC has seven potential sites for Asn-linked glycosylation. Modification of these sites in yeast shifted TetC mobility to 65 kDa or higher (9). We consider glycosylation of TetC in plastids unlikely as the observed shift was minor and the enzymatic machinery for glycosylation is localized to the endoplasmic reticulum and Golgi apparatus (33).
We report here that TetC produced in tobacco leaf is effective for mucosal immunization of mice when applied nasally and orally (data not shown), and protects mice from a tetanus toxin challenge. Thus, TetC is a new addition to the list of vaccines that may be produced in plants.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr O. Qazi for the purified rTetC protein, Dr C. Turcotte for the anti-TetC antibody and Dr Thea Sesardic for the gift of tetanus toxin. Research at IC was supported by a BBSRC studentship to J.S.T., a BBSRC grant to P.N. and an EU grant MUCIMM and a Wellcome Trust grant awarded to G.D. Research at Rutgers was supported by Rutgers F&A Special Project Grant to P.M.
REFERENCES
- 1.Ma J.K. (2000) Genes, greens, and vaccines. Nat. Biotechnol., 18, 1141–1142. [DOI] [PubMed] [Google Scholar]
- 2.Giddings G., Allison,G., Brooks,D. and Carter,A. (2000) Transgenic plants as factories for biopharmaceuticals. Nat. Biotechnol., 18, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 3.Walmsley A.M. and Arntzen,C.J. (2000) Plants for delivery of edible vaccines. Curr. Opin. Biotechnol., 11, 126–129. [DOI] [PubMed] [Google Scholar]
- 4.Stoger E., Sack,M., Fischer,R. and Christou,P. (2002) Plantibodies: applications, advantages and bottlenecks. Curr. Opin. Biotechnol., 13, 161–166. [DOI] [PubMed] [Google Scholar]
- 5.Bock R. (2001) Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol., 312, 425–438. [DOI] [PubMed] [Google Scholar]
- 6.Maliga P. (2002) Engineering the plastid genome of higher plants. Curr. Opin. Plant Biol., 5, 164–172. [DOI] [PubMed] [Google Scholar]
- 7.Staub J.M. (2002) Expression of recombinant proteins via the plastid genome. In Parekh,S.R. and Vinci,V.A. (eds.), Handbook of Industrial Cell Culture: Mammalian, Microbial and Plant Cells. Humana Press Inc., Totowa, NJ, pp. 261–280.
- 8.Makoff A.J., Ballantine,S.P., Smallwood,A.E. and Fairweather,N.F. (1989) Expression of tetanus toxin fragment C in E. coli: its purification and potential use as a vaccine. Biotechnology, 7, 1043–1046. [Google Scholar]
- 9.Romanos M.A., Makoff,A.J., Fairweather,N.F., Beesley,K.M., Slater,D.E., Rayment,F.B., Payne,M.M. and Clare,J.J. (1991) Expression of tetanus toxin fragment C in yeast: gene synthesis is required to eliminate fortuitous polyadenylation sites in AT-rich DNA. Nucleic Acids Res., 19, 1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles I.G., Rodgers,B.C., Makoff,A.J., Chatfield,S.N., Slater,D.E. and Fairweather,N.F. (1991) Synthesis of tetanus toxin fragment C in insect cells by use of a baculovirus expression system. Infect. Immun., 59, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makoff A.J., Oxer,M.D., Romanos,M.A., Fairweather,N.F. and Ballantine,S. (1989) Expression of tetanus toxin fragment C in E. coli: high level expression by removing rare codons. Nucleic Acids Res., 17, 10191–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stratford R., Douce,G., Zhang-Barber,L., Fairweather,N., Eskola,J. and Dougan,G. (2001) Influence of codon usage on the immunogenicity of a DNA vaccine against tetanus. Vaccine, 19, 810–815. [DOI] [PubMed] [Google Scholar]
- 13.Anderson R., Gao,X.M., Papakonstatinopoulou,A., Roberts,M. and Dougan,G. (1996) Immune response in mice following immunization with DNA encoding fragment C of tetanus toxin. Infect. Immun., 64, 3168–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda H. and Maliga,P. (2001) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res., 29, 970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda H. and Maliga,P. (2001) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol., 125, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svab Z. and Maliga,P. (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA, 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murashige T. and Skoog,F. (1962) A revised medium for the growth and bioassay with tobacco tissue culture. Physiol. Plant., 15, 473–497. [Google Scholar]
- 18.Silhavy D. and Maliga,P. (1998) Mapping of the promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet., 33, 340–344. [DOI] [PubMed] [Google Scholar]
- 19.Carrer H., Hockenberry,T.N., Svab,Z. and Maliga,P. (1993) Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet., 241, 49–56. [DOI] [PubMed] [Google Scholar]
- 20.Douce G., Fontana,M., Pizza,M., Rappuoli,R. and Dougan,G. (1997) Intranasal immunogenicity and adjuvanticity of site-directed mutant derivatives of cholera toxin. Infect. Immun., 65, 2821–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueiredo D., Turcotte,C., Frankel,G., Li,Y., Dolly,O., Wilkin,G., Marriott,D., Fairweather,N. and Dougan,G. (1995) Characterization of recombinant tetanus toxin derivatives suitable for vaccine development. Infect. Immun., 63, 3218–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- 23.Komine Y., Kikis,E., Schuster,G. and Stern,D. (2002) Evidence for in vivo modulation of chloroplast RNA stability by 3′-UTR homopolymeric tails in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA., 99, 4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlak F.J., Fuchs,R.L., Dean,D.A., McPherson,S.L. and Fischhoff,D.A. (1991) Modification of the coding sequence enhances plant expression of insect control protein genes. Proc. Natl Acad. Sci. USA, 88, 3324–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride K.E., Svab,Z., Schaaf,D.J., Hogan,P.S., Stalker,D.M. and Maliga,P. (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology, 13, 362–365. [DOI] [PubMed] [Google Scholar]
- 26.Lutz K.A., Knapp,J.E. and Maliga,P. (2001) Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol., 125, 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makrides S.C. (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev., 60, 512–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kane J.F. (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol., 6, 494–500. [DOI] [PubMed] [Google Scholar]
- 29.Shimada H. and Sugiura,M. (1991) Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res., 19, 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye G.N., Hajdukiewicz,P.T.J., Broyles,D., Rodriquez,D., Xu,C.W., Nehra,N. and Staub,J.M. (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J., 25, 261–270. [DOI] [PubMed] [Google Scholar]
- 31.Franklin S., Ngo,B., Efuet,E. and Mayfield,S.P. (2002) Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J., 30, 733. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda H. and Maliga,P. (2002) Over-expression of the clpP 5′-UTR in a chimeric context causes a mutant phenotype suggesting competition for a clpP-specific RNA maturation factor in tobacco chloroplasts. Plant Physiol., 129, 1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardor M., Faye,L. and Lerouge,P. (1999) Analysis of the N-glycosylation of recombinant proteins produced in transgenic plants. Trends Plant Sci., 4, 376–380. [DOI] [PubMed] [Google Scholar]