Abstract
Insertional mutagenesis techniques, including transposon- and T-DNA-mediated mutagenesis, are key resources for systematic identification of gene function in the model plant species Arabidopsis thaliana. We have developed a database (http://atidb.cshl.org/) for archiving, searching and analyzing insertional mutagenesis lines. Flanking sequences from approximately 10 500 insertion lines (including transposon and T-DNA insertions) from several tagging programs in Arabidopsis were mapped to the genome sequence through our annotation system before being entered into the database. The database front end provides World Wide Web searching and analyzing interfaces for genome researchers and other biologists. Users can search the database to identify insertions in a particular gene or perform genome-wide analysis to study the distribution and preference of insertions. Tools integrated with the database include a graphical genome browser, a protein search function, a graphical representation of the insertion distribution and a Blast search function. The database is based on open source components and is available under an open source license.
INTRODUCTION
The plant Arabidopsis thaliana has many advantages as an experimental system, including a complete genome sequence (1), that have increased the rate of gene discovery. Because <10% of the more than 25 000 predicted genes have an experimentally determined function, systematic approaches in defining gene function have been initiated recently (2; National Science Foundation, Program Solicitation NSF-01-162). These strategies use either maize transposable elements (3) or Agrobacterium tumefaciens T-DNA (4), a portion of a tumor-inducing plasmid that is transferred to plant cells, as mutagens. Inserted DNA can disrupt the expression of the gene into which it is inserted and can also be used as a marker for subsequent identification of the mutation. An insertion may reveal gene function via a gene knock-out and a gene knock-up (overexpression and misexpression) or through expression patterns revealed by modified insertion elements (5).
Recent reports (6–8) demonstrate the feasibility of using transposable elements and T-DNA to generate libraries of Arabidopsis lines in which each individual line carries a single, tagged gene disruption. Several large-scale T-DNA and transposon insertional mutagenesis projects using these strategies are underway (Table 1) to support gene discovery projects. Modifications to transposable elements and T-DNAs to create gene traps (GT) and enhancer traps (ET) (9), activation tagging (AT) (10–12) and promoter traps (13) provide information on enhancer and promoter function, gene expression patterns and phenotypes resulting from putative gain-of-function mutations. Transposon and T-DNA mutagenesis has created the opportunity to rapidly identify gene disruption mutants through the Arabidopsis genome by amplifying and sequencing genomic DNA flanking insertion sites (14–17). The precise location of sequenced insertion sites provides information about both the likely effect of the insertion on gene function and a unique tag for database searches. T-DNA and transposon insertion sites in the Arabidopsis genome are being sequenced in several large-scale projects and these are being integrated with genome databases (see Table 1). Users can access this information through a variety of databases, described in Table 1. These vary in utility.
Table 1. Gene disruption resources in Arabidopsis.
| Laboratory | Insertion element | Number | Source | Sequenced insertions |
|---|---|---|---|---|
| Salk Institute | T-DNA | 70 000 | http://signal.salk.edu/cgi-bin/tdnaexpress | ∼94 947 |
| TMRI | T-DNA | 100 000 | http://www.tmri.org/pages/collaborations/garlic_files/ | ∼100 000 |
| CSHL | Ds GT and ET | 27 000 | http://genetrap.cshl.org/ | ∼26 800 |
| GABI-KAT | T-DA | 23 000 | http://www.mpiz-koeln.mpg.de/GABI-Kat/ | ∼20 764 |
| FLAG | T-DNA | 50 000 | http://flagdb-genoplante-info.infobiogen.fr/ | ∼11 500 |
| John Innes Centre | DSpm and Ds GT | 50 000 | http://atidb.cshl.org/ | ∼13 000 |
Here we describe a new database system (ATIDB) to integrate A.thaliana transposon and T-DNA insertion data with genome features such as gene models. We have also developed World Wide Web interfaces conveniently integrating a suite of search and display tools for users to access the insertion information. In our demonstration system, users can search for transposon or T-DNA insertions of interest and order the corresponding lines/material directly from Nottingham Arabidopsis Stock Center and the Arabidopsis Biological Resource Center, where the seed collections are maintained. Users can also analyze and compare insertion features within genes or in the entire genome sequence of A.thaliana. This paper reports the design of the software in detail.
MATERIALS AND METHODS
Insertion data
Our demonstration set of insertion data as of November 2002 consisted of more than 100 000 insertion lines. As shown in Table 2, 196 defective Suppressor/mutator (dSpm) pool, 6113 dSpm single copy (SM), 119 AT, 1586 GT, 1289 ET and 94 947 T-DNA lines came from several sources. Raw insertion site sequences were pre-screened to confirm the presence of a sequence tag from the transposon or T-DNA preceding the genomic sequence. This ensured that the precise insertion site was identified in the subsequent data pipeline. Sequence data were formatted into a FASTA file, in which the head line of each sequence includes data source, line type, the end of the element from which the sequence was derived and the accession number given by the sequencing center, before being entered into the data pipeline.
Table 2. Testing datasets for the demonstration ATIDB.
| Insertion type | No. of lines | No. of flanking sequences | Data source |
|---|---|---|---|
| dSpm (dSpm pool) | 196 | 1058 | John Innes Centre, UK |
| SM (dSpm single copy) | 6113 | 6113 | John Innes Centre, UK |
| AT | 119 | 159 | John Innes Centre, UK |
| GT | 1586 | 1998 | Cold Spring Harbor Laboratory, USA and Institute of Molecular Agrobiology, Singapore |
| ET | 1289 | 1383 | Cold Spring Harbor Laboratory, USA and Biotechnology Institute, Pennsylvania State University, USA |
| TN (T-DNA) | 94 947 | 94 947 | SALK Institute, USA |
The datasets consist of six insertion types with approximately 10 500 lines from five different resources.
Genome data
The Arabidopsis genome and its annotation data were downloaded from the FTP site of the TIGR Arabidopsis genome annotation database (The Institute for Genomic Research, ftp://ftp.tigr.org/pub/data/a_thaliana). Genomic DNA sequences corresponding to entire chromosomes (pseudo-chromosomes) were formatted into FASTA-format nucleotide database files for WU-BLAST (W. Gish, blast. wustl.edu). The genome annotations were also parsed into three files containing clone, gene and exon information based on the database design described later. These three files were then used for the insertion mapping and as input files of the database.
Insertion mapping
We used BLASTN of WU-BLAST to align the flanking sequences against the pseudo-chromosomes. Both parameters V and B were set to 5. The no gap case was chosen and other parameters were left in default.
If the Blast result of a flanking sequence had score of 90 or higher, we considered that this flanking sequence had a hit on the Arabidopsis genome. If the flanking sequence had one or more hits on the genome, we chose the hit with the highest score and took the starting position of the hit on the chromosome as the insertion position on the genome of Arabidopsis. The mapped insertions were then assigned to clones and, when appropriate, to genes, based on the insertion map locations relative to the TIGR genome annotations. For our purposes, a gene was defined as beginning 900 bp 5′ of the initiation codon and to the end of the 3′-UTR, where known.
Database design
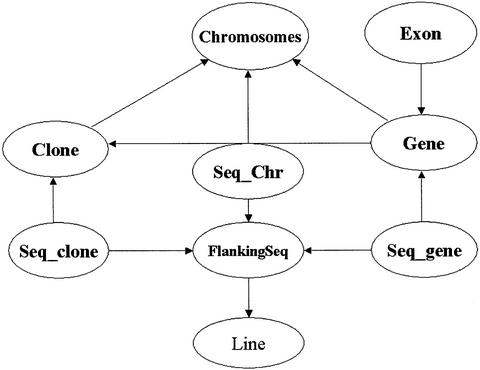
The insertion database has nine top level objects shown in Figure 1. Tables Clone, Gene and Exon store clone, gene and exon information separately. A raw insertion line usually consists of flanking sequence, the end of the sequence insertion, the pedigree and source. Therefore, the FlankingSeq table contains the flanking sequences and the Line table takes the other information on the insertion lines. Tables Seq_Chr, Seq_Clone and Seq_Gene store the insertion mapping positions on chromosomes, clones and genes, respectively. The relationships among these core object tables or relations are shown in Figure 1.
Figure 1.
Diagram of the relational database schema. The circles represent object tables or relations and the arrows show that foreign keys in the tables point back to the reference tables. Non-core object tables are not shown in the graphics for simplification.
Programming and database implementation
The data parsing, insertion mapping and data loading scripts of ATIDB were coded in Perl (18). The World Wide Web interfaces were programmed as Perl-CGI scripts (19). The database was implemented based on MySQL (20), a relational database management system.
Operating environments and availability of ATIDB
The software uses Unix as the operating system, Apache (21) 1.3.6 or newer as the Web server and MySQL 3.23 or higher as the database management system. It requires installing Perl 5.004 or higher, Bioperl (22) 1.1.1 or later and a WU-BLAST 2.0 or newer (or current NCBI-BLAST) server. The Perl modules DBI, DBD::MySQL (20), Digest::MD5, Text::Shellwords and GD (23) are also required. The source code is downloadable from the website at URL http://www.gmod.org/.
RESULTS
The data pipeline and web interfaces are the major components of the ATIDB software.
Data pipeline
The system begins with an automated data pipeline that maps insertion sites to the genome. First, Arabidopsis genome annotations (The Institute for Genomic Research, ftp://ftp.tigr.org/pub/data/a_thaliana) are retrieved and parsed into three files containing clone, gene and exon information. Batch flanking sequences are Blasted one by one against the chromosomal sequences. The Blast results are analyzed to determine the insertion sites in the genome. Finally, all of the information about the insertion sites, clones, genes and exons is entered into the insertion database.
For our test data set, the insertion mapping results (Table 3) show a success rate of >96% for insertion mapping. A total of 57–72% of the insertions fall within genes.
Table 3. Success ratio of insertions mapping to the Arabidopsis genome.
| Insertion type | Ratio of mapping to entire genome (%) | Ratio of mapping to genes (%) |
|---|---|---|
| dSpm | 98.68 | 71.46 |
| SM | 98.95 | 66.60 |
| AT | 96.86 | 70.80 |
| GT | 96.95 | 66.42 |
| ET | 100 | 64.70 |
| TN | 100 | 57.26 |
See Table 2 for explanations about the insertion type abbreviations.
World Wide Web interfaces
Based on user requirements, we have designed graphical user-friendly interfaces for examining the distribution of insertions, for browsing and searching for an insertion on the genome of A.thaliana and for retrieving information on gene knock-outs and gene knock-ups.
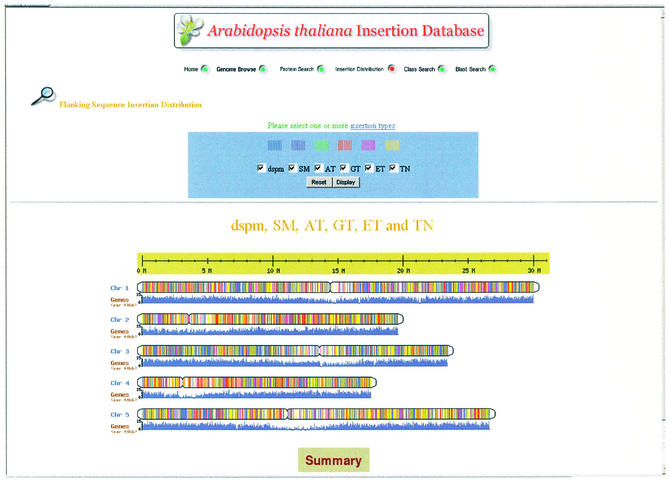
A web-based graphical user interface provides a view of the insertion distribution (Fig. 2). It allows users to select one or more insertion types to see the distribution on the five Arabidopsis chromosomes for comparison. The distribution of genes is also shown. Clicking on a location on a chromosome will display a page describing the genes and insertions mapped to the selected region. If the user clicks on the ‘Summary’ button, a page listing insertions on all five chromosomes in tabular form appears.
Figure 2.
Interface for the distribution of insertions. The figure shows the distribution of all the insertions in the demonstration ATIDB on Arabidopsis genome. The lines with different colors represent different insertion types as shown on the upper part of the graphic. The histogram below each chromosome shows the distribution of genes per 40 kb of the genome.
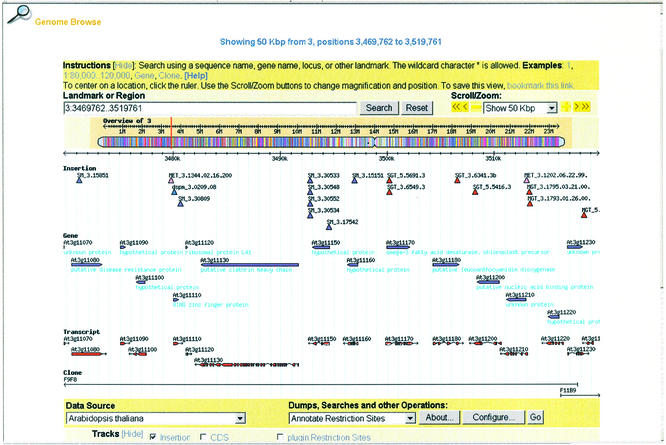
The genome browser, adopted from the generic genome browser (24), displays the sites of insertion integration, the location of genes and their transcripts and the clones used in the assembly of the genome (Fig. 3). A user can click the chromosome overview to browse the insertions within a region or, for fine adjustment, click on the scale that appears at the top of the detailed view to center the displayed region at that point in the chromosomal assembly. Figure 3 shows the distribution of insertions within a 50 kb region of chromosome 3, between positions 3 533 295 and 3 583 294. One dSpm and eight SM transposons, represented by blue and purple triangles, respectively, are inserted into the left hand side of the region. Seven GT transposons (red) are inserted into the right hand side of the region and two ET transposons (pink) are inserted into the left and right hand sides of the region, respectively. Fifteen genes, represented by purple arrowheads, appear within this region. Corresponding to the genes, there are 15 transcripts (red) displayed below the genes. Two genomic clones are also shown in this region. Clicking the gene representation brings up an expanded view (Fig. 4).
Figure 3.
Insertions within a 50 kb region of chromosome 3. Purple, blue, red and pink triangles represent SM, dSpm, GT and ET transposon insertions, respectively. Genes, cognate transcripts and sequenced BAC clones underlie the insertion data and are clickable.
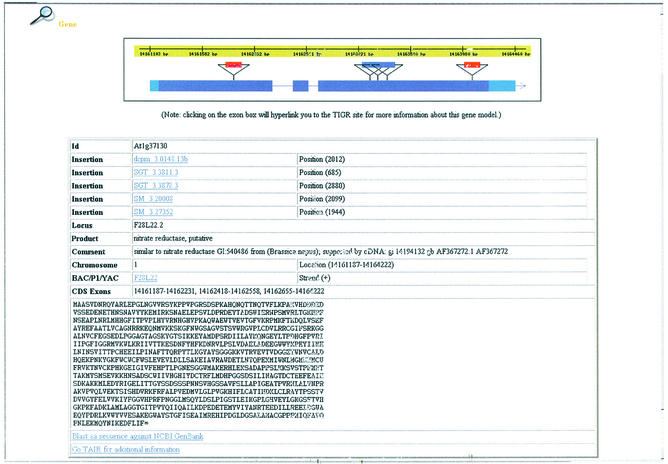
Figure 4.
Insertions in gene structure. Purple, blue and red triangles represent SM, dSpm and GT transposon insertions, respectively. The dark blue rectangles represent exons and the light blue rectangles represent untranslated regions of the mRNA, derived from cDNA sequence.
The protein search lets users retrieve insertion lines that may affect particular genes or proteins. The resulting genes or proteins are graphically displayed on the chromosomes according to their location on the genome. More information about these genes or proteins is provided in a detail table. The users can click each marked point on the chromosomes or a gene id listed in the table to check detail information about insertions within a gene via the gene graphical interface (Fig. 4). The graphic at the top shows that two GT, one dSpm and two SM insertions, represented by red, blue and purple rectangles, respectively, are inserted in the first and third exons of gene At1g37130. The table below the graphic lists information about the id, locus, product, location, exons and translated protein sequence of the gene as well as the insertion sequence identifiers and their insertion positions downstream from the initiation codon of the gene.
The Blast search allows users to search for insertions and genes by running WU-BLAST against the DNA or protein database of A.thaliana and then following the hits to the nucleotide or protein sequences within the Arabidopsis genome to locate nearby insertion lines. For example, if a user has a favorite gene and wants to find what insertion lines may lead to a knock-out for this gene, they can paste the protein sequence to the data receiving field and run the BLASTP program against the protein database. The resulting pages will display that gene with insertion information. If there are insertions within this gene, the user can order the insertion lines directly from the seed stock centers by clicking the insertion line link and then the stock code link in the flanking sequence information page.
DISCUSSION
ATIDB is a comprehensive insertion database for A.thaliana. It provides insertional mutagenesis projects in A.thaliana with a depository to store insertion data from a variety of transposon and T-DNA populations. The suite of graphical user-friendly interfaces we have developed in ATIDB provides users with useful tools to search for the insertions of interest and to order them directly from the seed stock centers of A.thaliana. Furthermore, ATIDB provides researchers with an effective environment to track the progress of insertional mutagenesis projects.
Many major genome information databases implement insertion modules. For example, the Berkeley Fly Database (25,26) provides users with searches of Drosophila genomic and cDNA clones, sequences, STSs and P insertion lines. This database contains more than 6000 P transposon insertion lines. However, it only provides users with a simple text-based interface to search for insertions. WormBase (27) has graphical user-friendly interfaces with many search functions. However, it does not provide a proper tool for users to access insertion information and there are only a small number of transposons in the database. Two Arabidopsis T-DNA insertion databases presently support public searches for gene disruptions. The SAIL Web interface (the Torrey Mesa Research Institute, www.tmri.org/pages/collaborations/garlic_files/) provides a Blast service for single sequences of less than 10 000 characters against the Torry Mesa Research Institute’s collection of 100 000 T-DNA insertions. The T-DNA Express database (J. Ecker, signal.salk.edu/cgi-bin/tdnaexpress) maps T-DNA insertions from the Salk Institute’s large collection of publicly available lines relative to gene models and full-length c-DNAs. This database allows text-based searches and direct links from the map viewer to insertion site sequence. However, the map viewer displays a limited region of the genome each time and users are unable to overview the insertions across the entire genome in one window. The FLAGdb/FST (28) is a database of mapped flanking insertion sites (FSTs) of Arabidopsis T-DNA, collected from the Institute Nationale Research Agronomie (INRA), Versailles, France. The Arabidopsis Information Resource (TAIR) (29) is a comprehensive database and Web-based information retrieval, analysis and visualization system for Arabidopsis currently linked out to third party insertion, knock-out and mutation resources. Lastly, the GeneTrap database (R. Martienssen, genetrap.cshl.org) stores ET and GT transposon insertion line sequences, images of gene expression patterns and phenotypes from the Cold Spring Harbor Laboratory Arabidopsis collection and allows users to access the insertion data via text-based queries. However, each of these insertion databases has been developed for specific laboratory use. To our knowledge, none have been distributed as open source software.
The technique of functional genomic analysis via insertional mutagenesis is a general technique that extends to many plant, invertebrate and vertebrate species. Therefore we designed ATIDB to be a generic insertion database that can be used for any experimental organism. To set up a new insertion database, researchers need to customize a configuration file and the homepage HTML file. The researchers then must also retrieve and parse the genome annotations and replace the Blast database used for insertion mapping with one appropriate for their organism. The rest of the software, including the mapping pipeline, the user interfaces and the query services, do not need to be altered. To update the genome annotations of the insertion database, researchers only need to retrieve and parse the new genome annotations, then re-map the insertion data to the new genome and, finally, re-populate the database with the new genome annotations and the newly mapped data. We have developed a series of scripts to retrieve and parse Arabidopsis genome annotations from TIGR for ATIDB.
Several developments of ATIDB are underway. The first is to populate it with all available sequenced insertion sites to provide users with a useful experimental tool to search for insertions in genes of interest and to track progress of the multinational effort to obtain insertions in each gene (2). We are also developing modules to represent phenotypes resulting from altered gene function and images of gene expression patterns and to establish links to microarray data sets. Finally, we are developing phenotype description modules based on the emerging trait ontology system (30) to facilitate phenotypic comparisons.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Institute of Molecular Agrobiology, Singapore, Joe Ecker at the SALK Institute, CA, the Biotechnology Institute at Pennsylvania State University and Robert Martienssen’s group at Cold Spring Harbor Laboratory, NY, for part of the insertion data. We are grateful to Ravi Sachidanandam for a DBI utility module, Marco Mangone, Steven Schmit and Peter Vanburen for computer technical help and Bruce May and Mark Crowe for data supply assistance. This project was partly supported by a grant from the NSF BDI-0110143, the BBSRC Investigating Gene Function Programme (M.B. and J.C.) and the EC PlaNet Project (contract QRL1-CT-2001-0006) (M.B.).
REFERENCES
- 1.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Chory J., Ecker,J.R., Briggs,S., Caboche,M., Coruzzi,G.M., Cook,D., Dangl,J., Grant,S., Guerinot,M.L., Henikoff,S. et al. (2000) National Science Foundation-Sponsored Workshop Report: “The 2010 Project” functional genomics and the virtual plant. A blueprint for understanding how plants are built and how to improve them. Plant Physiol., 123, 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedoroff N. (1989) Maize transposable elements. In Howe,M. and Ber,D. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 375–411.
- 4.Apiroz-Lenehan R. and Feldmann,K.A. (1997) T-DNA insertional mutagenesis in Arabidopsis: going back and forth. Trends Genet., 13, 152–156. [DOI] [PubMed] [Google Scholar]
- 5.Sundaresan V., Springger,P., Volpe,T., Haward,S., Jones,J.D., Dean,C., Ma,H. and Martienssen,R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev., 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- 6.Parinov S., Sevugan,M., Ye,D., Yang,W.-C., Kumaran,M. and Sundaresan,V. (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell, 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tissier A.F., Marillonnet,S., Klimyuk,V., Patel,K., Torres,M.A., Murphy,G. and Jones,J.D.G. (1999) Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell, 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krysan P.J., Young,J.C. and Sussman,M.R. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell, 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Springer P.S. (2000) Gene traps: tools for plant development and genomics. Plant Cell, 12, 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigel D., Ahn,J.H., Blazquez,M.A., Borevitz,J.O., Christensen,S.K., Fankhauser,C., Ferrandiz,C., Kardailsky,I., Malancharuvil,E.J., Neff,M.M. et al. (2000) Activation tagging in Arabidopsis. Plant Physiol., 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson K., Long,D., Swinburne,J. and Coupland,G. (1996) A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell, 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsch-Martinez N., Greco,R., Van Arkel,G., Herrera-Estrella,L. and Pereira,A. (2002) Activation tagging using the en-I maize transposon system in Arabidopsis. Plant Physiol., 129, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topping J.F. and Lindsey,K. (1997) Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell, 9, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parinov S. and Sundaresan,V. (2000) Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol., 11, 157–161. [DOI] [PubMed] [Google Scholar]
- 15.Martienssen R.A. (1998) Functional genomics: probing plant gene function and expression with transposons. Proc. Natl Acad. Sci. USA, 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsthoefel N.R., Wu,Y., Schulz,B., Bennett M.J. and Feldmann,K.A. (1992) T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust. J. Plant Physiol., 19, 353–366. [Google Scholar]
- 17.Azpiroz-Leehan R. and Feldmann,K.A. (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet., 13, 152–156. [DOI] [PubMed] [Google Scholar]
- 18.Christiansen T., Torkington,N. and Wall,L. (1998) Perl Cookbook. O’Reilly & Associates, Sebastopol, CA.
- 19.Stein L. (1998) The Official Guide to CGI.pm. John Wiley & Sons, New York, NY.
- 20.DuBois P. (1999) MySQL. New Riders, Indianapolis, IN.
- 21.Stein L. and MacEachern,D. (1999) Writing Apache Modules with Perl and C. O’Reilly & Associates, Sebastopol, CA.
- 22.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G.R., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verbruggen M. (2002) Graphics Programming with Perl. Manning Publications Co., Greenwich, CT.
- 24.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J., Harris,T.W., Arva,A. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin G.M. (1996) Around the genomes: the Drosophila genome project. Genome Res., 6, 71–79. [DOI] [PubMed] [Google Scholar]
- 26.Sprading A.C., Stein,D., Beaton A., Rhem,E.J., Laverty,T., Mozden,N., Misra,S. and Rubin,G.M. (1999) The BDGP gene distribution project: single P element insertions mutating 25% of vital Drosophila genes. Genetics, 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein L., Sternberg,P., Durbin,R. Thierry-Mieg,J. and Spieth,J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson F., Brunaud,V., Balzergue,S., Dubreucq,B., Lepiniec,L., Pelletier,G., Caboche,M. and Lecharny,A. (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res., 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huala E., Dickerman,A., Garcia-Hernandez,M., Weems,D., Reiser,L., LaFond,F., Hanley,D., Kiphart,D., Zhuang,J., Huang,W. et al. (2001) The Arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis and visualization system for a model plant. Nucleic Acids Res., 29, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaiswal P., Ware,D., Ni,J., Chang,K., Zhao,W., Schmidt,S., Pan,X., Clark,K., Teytelman,L., Cartinhour,S. et al. (2002) Gramene: development and integration of trait and gene ontologies for rice. Comp. Funct. Genomics, 3, 132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]