Abstract
To optimize the sensitivity of imaging HSV1-tk/GFP reporter gene expression, a series of HSV1-tk/GFP mutants was developed with altered nuclear localization and better cellular enzymatic activity, compared to that of the native HSV1-tk/GFP fusion protein (HSV1-tk/GFP). Several modifications of HSV1-tk/GFP reporter gene were performed, including targeted inactivating mutations in the nuclear localization signal (NLS), the addition of a nuclear export signal (NES), a combination of both mutation types, and a truncation of the first 135 bp of the native hsv1-tk coding sequence containing a “cryptic” testicular promoter and the NLS. A recombinant HSV1-tk/GFP protein and a highly sensitive sandwich enzyme-linked immunosorbent assay for HSV1-tk/GFP were developed to quantitate the amount of reporter gene product in different assays to allow normalization of the data. These different mutations resulted in various degrees of nuclear clearance, predominant cytoplasmic distribution, and increased total cellular enzymatic activity of the HSV1-tk/GFP mutants, compared to native HSV1-tk/GFP when expressed at the same levels. This appears to be the result of improvedmetabolic bioavailability of cytoplasmically retargeted mutant HSV1-tk/GFP enzymes for reaction with the radiolabeled probe (e.g., FIAU). The analysis of enzymatic properties of different HSV1-tk/GFP mutants using FIAU as a substrate revealed no significant differences from that of the native HSV1-tk/GFP. Improved total cellular enzymatic activity of cytoplasmically retargeted HSV1-tk/GFP mutants observed in vitro was confirmed by noninvasive imaging of transduced subcutaneous tumor xenografts bearing these reporters using [131I]FIAU and a γ-camera.
Keywords: molecular imaging, herpes virus type one thymidine kinase, green fluorescent protein, FIAU, PET
Introduction
Recent advances in noninvasive molecular imaging have provided new research tools for monitoring the expression of different genes and activities of various signal transduction pathways. Reporter systems have been developed for tracking transgene expression in vitro as well as in vivo, and in transgenic animals. Reporter gene systems that are based on enzymatic signal amplification include luciferase [10], herpes virus 1 thymidine kinase (hsv1-tk) [45,47], cytosine deaminase (CD) [24,44], uracil phosphoribosyl transferase (UPRT) [16], xanthine phosphoribosyl transferase (XPRT) [39], and human truncated mitochondrial thymidine kinase type two (ΔhTK2) [38]. The fluorescent protein-based reporter gene systems include Aequorea victoria green fluorescent protein (GFP) and different spectral-shifted GFP variants [11,17,18,25], humanized version (hrGFP) [31,32], and red fluorescent proteins (Red1 and Red2) [7,35]. Several bioluminescent reporter genes are being widely used for whole body bioluminescent imaging in animals such as Renilla and Firefly luciferase enzymes in combination with their substrates coelenterazine and luciferine, correspondingly [2,41]. The receptor-based systems include dopamine receptor type 2 (D2R) [33], somatostatin receptor (SSTr2) [42], and sodium iodide symporter (NIS) [23].
HSV1-tk is one of the most studied reporter genes for noninvasive radionuclide imaging [21,26,45–47]. HSV1-tk can phosphorylate different radiolabeled purine and pyrimidine nucleoside analogs such as 2′-fluoro-2′-deoxy-1-β-d-arabinofuranosyl-5-iodo-uracil (FIAU), 9-(4-[18F]fluoro-3-hydroxymethylbutyl) guanine (FHBG), and other radiolabeled probes [1,6,40,47]. Noninvasive radionuclide imaging of hsv1-tk expression provides the opportunity to determine the location, magnitude, and temporal dynamics of transgene expression using γ-camera, SPECT, or positron emission tomography (PET) [21,45,47]. In previous reports, we described the HSV1-tk/GFP fusion reporter gene, which combines the advantages of hsv1-tk and GFP for monitoring gene transfer and expression in vitro using fluorescent microscopy and FACS, as well as in vivo using γ-camera, SPECT, and PET [26]. Recently, we extended the application of HSV1-tk/GFP reporter gene to imaging the expression of various endogenous genes and the activity of signal transduction pathways [15,36].
The use of tissue-specific hsv1-tk gene expression in transgenic mice for sensitization of these tissues to ganciclovir (GCV) treatment had limited success. This is due to the presence of a putative cryptic testis-specific promoter within the coding sequence of the hsv1-tk gene [9,43], which renders the transgenic males sterile. The latter occurs because of spermatozoal toxicity induced by high nuclear concentrations of the HSV1-tk enzyme. The N-terminal truncated variant of hsv1-tk has been shown to have no spermatozoal toxicity in transgenic mice in the absence of the cryptic testis-specific promoter. However, it was demonstrated that N-terminal truncation of hsv1-tk results in the attenuation of its enzymatic activity [9,43]. Degreve et al. [13,14] described nuclear localization signal (NLS), which targets the native HSV1-tk protein or its fusion with GFP to the cell nucleus. The NLS in HSV1-tk protein is a nonapeptide (25-RRTALRPRR-33). In addition, the amino acids R236–R237 and K317–R318 in the C-terminal portion of HSV1-tk have also been shown to mediate the nuclear tropism of HSV1-tk protein, its cell toxicity [12], and male sterility in transgenic mice [9,43].
The aim of this study was to develop and evaluate a series of HSV1-tk/GFP mutants with altered nuclear localization and with better cellular enzymatic activity compared to that of the native HSV1-tk/GFP fusion protein. We performed several modifications of the native HSV1-tk/GFP reporter gene, including targeted mutations in the NLS, the addition of a nuclear export signal (NES), the combination of both mutations, and the truncation of the first 135 bp of the native hsv1-tk coding sequence that contains the cryptic testicular promoter and the N-terminal NLS. These different mutations resulted in various degrees of nuclear clearance and predominant cytoplasmic distribution, as well as increased total cellular enzymatic activity of the mutant HSV1-tk/GFP variants compared to native HSV1-tk/GFP expressed at the same levels. The improved total cellular enzymatic activity of cytoplasmically retargeted HSV1-tk/GFP mutants was also observed by noninvasive imaging of transduced subcutaneous tumor xenografts bearing these reporters.
Materials and Methods
Hsv1-tk/GFP Mutations and Expression Vectors
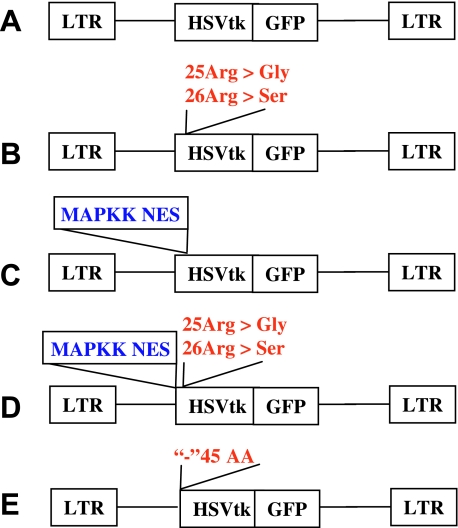
The schematic structures of the retroviral vectors used in this study are shown in Figure 1. The retroviral vector SFGwt-HSV1-tk/GFP has been described previously [26]. A mutation of two arginine residues (Arg25 and Arg26) into corresponding glycine-25 and serine-26 residues within the NLS of the native hsv1-tk sequence of the fusion gene was achieved using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, CA). To inactivate the NLS signal of hsv1-tk, the codons for Arg25–Arg26 amino acids were replaced by the BamHI restriction site, 5′-GGATCC-3′ (to encode Gly25-Ser26), by polymerase chain reaction (PCR) using the sense primer 5′-GCAACGGATCCACGGCGTTGCGCCCTC-3′ and the antisense primer 5′-TGGCCGCGAGAACGCGCAGCCTGGTCG-3′. PCR amplification of the mutant primers was performed with the Pfu DNA polymerase (Stratagene) and SFG-wt-HSV1-tk/GFP as a template using a temperature cycle program (30 seconds at 95°C followed by 12 cycles of 30 seconds at 95°C, 1 minute at 55°C, and 12 minutes at 72°C). The native plasmid was inactivated by digestion with the DpnI restriction enzyme (Stratagene). The Gly25-Ser26-containing mutant vector was transfected into the competent DH5α Escherichia coli (Invitrogen, Carlsbad, CA). Ampicillin-resistant colonies were screened for mutant plasmids by BamHI digestion. The resulting vector was termed SFG-mNLS-HSV1-tk/GFP.
Figure 1.
Schematic structures of retroviral vectors for mammalian expression of the native and different HSV1-tk/GFP mutants: (A) native HSV1-tk/GFP; (B) mNLS-HSV1-tk/GFP with mutations in the N-terminal nuclear localization signal; (C) NES-HSV1-tk/GFP with the addition of the nuclear export signal from MAPKK; (D) NESmNLS-HSV1-tk/GFP with a combination of mutations in vectors (B) and (C); and (E) Δ45HSV1-tk/GFP with truncation of N-terminal sequence encoding the first 45 amino acids of the native protein.
In another HSV1-tk/GFP mutant, the leucine-rich NES sequence of the MAPKK cDNA of Xenopus encoding 20 amino acids (32–51) was inserted into the N-terminus of HSV1-tk/GFP between His9 and Ala10 using MluI restriction site, resulting in the SFG-NES-HSV1-tk/GFP vector.
A third HSV1-tk/GFP mutant was produced by a combination of the mutagenic procedures in NLS and the addition of NES as described above. The resulting vector was termed SFG-NES/mNLS-HSV1-tk/GFP vector.
The truncated HSV1-tk/GFP mutant was obtained as follows: the Δ45HSV1-tk cDNA lacking the first 45 amino acids (135 nucleotides) of the original HSV1-tk was amplified using primers 5′-ACTGCCATGGCTCCCACGCTAC-3′ and 5′-CCATGGTGGCGACCGGTG-3′ and subsequently cloned between NcoI sites of the SFG-wt-HSV1-tk/GFP vector replacing native HSV1-tk cDNA. The resultant vector was named SFG-Δ45HSV1-tk/GFP.
For the production of the retroviral vectors for transduction of tumor cells, 10 µg of the DNA of each vector-encoding plasmid was transfected into the GPG29 packaging cell line using the calcium phosphate method as previously described [19]. The vesicular stomatitis virus G-protein (VSV-G)-pseudotyped retroviral particles were used as cell-free viral stocks for transduction.
The pET-29 vector (Novagen, Madison, WI) was used to establish HSV1-tk/GFP protein expression in prokaryotic cells. Briefly, HSV1-tk/GFP cDNA was amplified using primers 5′-ACTGACTGCATATGGCTTCGTACCCCTG-3′ and 5′-ACTGACTGGGATCCTTACTTGTACAGCTC-3′ and cloned into the pET-29 vector between NdeI and BamHI restriction sites. The resultant vector was named pET-29 HSV1-tk/GFP.
Transduction of Tumor Cells
The RG2 rat glioma cell line was kindly provided by Dr. D. Bigner (Duke University Medical Center, Durham, NC). RG2 cells were grown as monolayers in MEM supplemented with 10% fetal calf serum (FCS) in at 37°C in humidified atmosphere with 5% CO2. The in vitro transduction of RG2 cells with the retroviral vectors was accomplished by exposing the cell monolayers to a filtered (0.45 µm) culture medium obtained from the vector producer cells for 8 hours in the presence of polybrene (8 µg/ml; Sigma, St. Louis, MO).
Flow Cytometry and Fluorescent Microscopy
Retrovirally transduced RG2 cells were grown as bulk cultures for 48 hours and subsequently sorted for positive HSV1-tk/GFP expression using FACS (FACSVantage; Becton Dickinson, CA); the 488-nm excitation beam and 510-nm emission filters were used. Subcellular localization of the HSV1-tk/GFP protein in transduced tumor cells was visualized by fluorescence microscopy (Nikon, Osaka, Japan) using similar excitation and emission parameters.
Assessment of HSV1-tk/GFP mRNA Expression
The levels of HSV1-tk/GFP mRNA in transduced tumor cells expressing different HSV1-tk/GFP protein mutants were assessed using RP-PCR. Total RNA was isolated from the transduced tumor cells using the RNAeasy kit (Qiagen, CA). The HSV1-tk/GFP mRNA amplification was performed using specific primer 5′-ATGGCTTCGTACCCCTG-3′ (for all mutants except Δ45HSV1-tk/GFP) or 5′-ATGGCTCCCACGCTACTG-3′ (for Δ45HSV1-tk/GFP) and reverse primer 5′-AAGGTCGGCGGGATGAG-3′, and 2 µg of total RNA in a Single-Tube RT-PCR System (Stratagene). These sets of primers amplify a 578-bp fragment of the wild-type and mNLS-bearing HSV1-tk coding sequence (1–578), a 670-bp fragment of the NES-containing mutant hsv1-tk, and a 450-bp fragment of the truncated Δ45HSV1-tk. The parameters of RT-PCR were: RT at 45°C for 30 minutes, then 25 cycles of denaturing at 94°C for 1 minute, primer annealing at 58°C for 30 seconds, and extension at 72°C for 1 minute, using the DNA Engine thermal cycler (MJ Research, Waltham, MA). To normalize the level of TKGFP mRNA expression in different cell populations, rat GAPDH mRNA was measured using the 5′-CAAGTTCAATGGCACAGTCAAG-3′ and 5′-TTGGCAGGTTTCTCCAGG-3′ primers, which produced a 600-bp fragment.
HSV1-tk/GFP Protein Production and Purification
The plasmid pET-29HSV1-tk/GFP was transfected into a strain of E. coli BL21(DE3) (Invitrogen) according to the manufacturer's protocol; bacteria in 800 ml of inoculated media (with 100 µg/ml carbenicillin) were incubated at 37°C with continuous shaking until an OD of 0.6 was reached. Expression of HSV1-tk/GFP protein was induced by adding IPTG (Sigma, St. Louis, MO) to a final concentration of 1 mM. The incubation was continued until an OD of 1.2 was reached, and cells were harvested by centrifugation at 15,000g for 20 minutes at +4°C. The bacterial pellet was frozen at -80°C and stored until use. The cell pellet was thawed on ice and homogenized in 35 ml of lysis buffer (100 mM NaH2PO4, 0.5 M NaCl, 8 M urea, pH 8.0). Lysozyme (1 mg/ml; Sigma) was added to the lysate and the suspension was incubated at +4°C for 30 minutes with shaking, which was followed by sonication (6 x 10 seconds, 80 W, on ice). Benzoase (1 µl/ml; Novagen) and Protease Inhibitor Cocktail III (2 µl/ml; Novagen) were added and the lysate was incubated at room temperature for 3 hours, followed by centrifugation at 15,000g for 30 minutes at +4°C. The supernatant was filtered through a 0.45-µM filter (Pall, East Hills, NY). Purification of the recombinant HSV1-tk/GFP protein was performed using FPLC (BioLogic LP; Bio-Rad Laboratories, Hercules, CA) and HiTrap affinity columns loaded with Ni2+ (1 ml; Amersham Pharmacia Biotech, Piscataway, NJ) as follows: the bacterial cell lysate was applied to the column and eluted at a flow rate of 1 ml/min. The column was washed with the lysis buffer (see above) using a gradient pH from 8.0 to 4.0. Different elution fractions were collected and analyzed by Western blot using anti-HSV1-tk monoclonal antibody (kindly provided by Dr. W. Summers, Yale University, New Haven, CT) and fractions containing pure HSV1-tk/GFP protein were identified and collected.
Assessment of the Integrity of Different HSV1-tk/GFP Mutant Proteins
The protein integrity of different HSV1-tk/GFP mutants was assessed using Western blot analysis. Total protein was extracted from tumor cells transduced with native HSV1-tk/GFP or with different HSV1-tk/GFP mutants using the lysis buffer 10 mM Tris-HCl (pH 7.5), 1 mM DDT, 1 mM EDTA, 20% glycerol, protease inhibitor cocktail (cat no. 8340; Sigma) 150 µL/100 mL buffer, and ultrasonication at 4°C. The lysate was centrifuged for 10 minutes at 5000 rpm, and the protein concentration in supernatants was assayed spectrophotometrically at 595 nm (UV-1201S; Shimadzu, Columbia, MD) using protein assay kit (cat no. 5000006; Bio-Rad, Hercules, CA). The proteins were resolved by electrophoresis in 10% acrylamide gel and transferred to the Immobilon-P membrane (Millipore, Bedford, MA) using a dry transfer apparatus Mini-PROTEAN 3 (Bio-Rad Laboratories). The HSV1-tk/GFP proteins were detected using a monoclonal anti-GFP antibody (cat no. 8362; Clontech, Palo Alto, CA) at a 1:500 dilution or an anti-HSV1-tk monoclonal antibody at 1:500 dilution and a secondary peroxidase antimouse antibody (cat no. PI-2000; Vector Laboratories, Burlingame, CA) at a 1:4000 dilution. Detection was performed using an ECL Western blot detection kit (cat no. RPN 2106; Amersham Pharmacia Biotech).
Sandwich enzyme-linked immunosorbent assay (ELISA) for HSV1-tk/GFP Proteins
Anti-GFP Coated Clear Strip Plate (Pierce, Rockford, IL) was used for HSV1-tk/GFP fusion protein detection in cell lysates. The plate was washed with PBS containing 0.05% Tween 20 (PBST) five times. Then, the plate was incubated for 1 hour at room temperature either with 100 µl of recombinant HSV1-tk/GFP protein serially diluted in PBS with 1% BSA (to produce a reference standard curve), or with 100 µl of sample (cell lysate, diluted in PBS with 1% BSA). Then, the plate was washed five times with PBST and incubated with 100 µl of anti-HSV1-tk mAb (diluted at 1:200 in PBS with 1% BSA, kindly provided by Dr. W. Summers, Yale University) for 1 hour at room temperature. Then, the plate was washed five times with PBST and incubated with 100 µl of HRP-conjugated goat antimouse IgG+A+M (H+L) secondary antibody (Zymed Laboratories, South San Francisco, CA) diluted at 1:2000 in PBS with 1% BSA for 1 hour at room temperature, washed with PBST five times, and incubated for 20 minutes with 100 µl of substrate reagent (1:1 mixture of H2O2 and tetramethylbenzidine; R&D Systems, Minneapolis, MN). The fluorochrome development reaction was stopped with 50 µl of 2NH2SO4 and the absorbance was measured at 450 nm with wavelength correction at 570 nm using plate reader Saphire (TECAN, Salzburg, Austria). The concentration of HSV1-tk/GFP protein in samples was calculated based on the standard curve obtained with the purified recombinant HSV1-tk/GFP protein and expressed as micrograms per milligram of total protein in transduced cells.
HSV1-tk In Vitro Enzymatic Assay
Different tumor cell lines stably transduced with various mutant forms of HSV1-tk/GFP were grown until confluence in 15-cm tissue culture dishes (Nunc, Naperville, IL) in MEM supplemented with 10% FCS in at 37°C in humidified atmosphere with 5% CO2. The cells were wash with 10 ml of sterile PBS and harvested by scraping in 10 to 15 ml of PBS, and the cell pellet was obtained by centrifugation at 4°C for 10 minutes. The cell pellet was transferred into a preweighed 1.5-ml vial to determine cell pellet weight (usually ∼20–30 mg/plate) and resuspended in 500 µl of lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM DTT, 1 mM EDTA, 20% glycerol, 150 µl of Sigma protease inhibitor cocktail in 100 ml of buffer). The cell pellet was lysed by ultrasonication on wet ice and centrifuged at 4°C for 10 minutes, and the supernatant was transferred to another vial.
The assay was initiated by mixing 150 µL of cell lysate with 750 µL of assay buffer (Tris-HCl 150 mM, pH 7.5, ATP 10 mM, MgCl210 mM, NaF 25 mM, β-mercaptoethanol 10 mM) containing 0.01 mM [14C] FIAU (Moravek Biochemicals, Brea, CA) adjusted to 0.1 µCi/ml, and incubated at 37°C. After 20, 40, 60, 90, 120, and 150 minutes, a 50-µl aliquot of the reaction mixture was removed and spotted on a filter paper DE81 (Whatman, Newton, MA), which was air-dried for 2 minutes and then washed with 10 ml of H2O followed by 10 ml of 95% ethanol. The phosphorylated nucleoside (e.g., [14C]FIAU) was selectively retained on the filter paper after these washes. The FIAU-MP radioactivity retained on the filters was measured by β-spectroscopy (TriCarb 1600; Packard, Billerica, MA) after the addition of solubilizer Soluene (Packard) and scintillation cocktail Instafluor III (Packard). To determine the total radioactivity in the aliquot (FIAU+ FIAUMP), 50 µl of the reaction mixture was spotted onto the filter paper and the radioactivity was measured without washing the filter. The nonspecific binding of [14C]FIAU to the filter paper was assessed by spotting the reaction mixture containing a nontransduced tumor cell lysate or no lysate at all followed by the regular washes. The FIAU-MP-specific fraction of radioactivity in the reaction mixture was calculated as: [(washed sample -nonspecific binding)/total reaction sample] multiplied by the initial FIAU concentration (0.01 mM). The MPspecific fraction was plotted versus time and the apparent rate of the reaction was assessed by fitting the data to single-site binding equation using PRIZM v. 3.02 software (GraphPad Software, San Diego, CA). The reaction rate (µM FIAU/sec) was normalized to enzyme concentration (µM HSV1-tk), which was measured by ELISA in a sample obtained from the corresponding cell lysate.
FIAU Accumulation Assay
The FIAU accumulation assays were performed as previously described [47]. The cells were seeded in 150x25-mm tissue culture plates (Nunc) at a concentration of 1 x 106 cells/plate and grown until 50% to 60% confluent. The incubation medium was replaced with 14 ml of medium containing 0.01 µCi/ml (56 mCi/mmol) [2-14C]FIAU and 0.2 µCi/ml (60 Ci/mmol) [met-3H]TdR (Moravek Biochemicals). Radiochemical purity of each compound was checked in our laboratory using HPLC and found to be >98%. The cells were harvested by scraping after various periods of incubation (10, 30, 60, 90, and 120 minutes) and centrifuged, then cell pellets were weighed and assayed for radioactivity concentration using a TriCarb 1600 β-spectrometer (Packard) using standard 3H and 14C dual-channel counting techniques. The medium was also counted before and after incubation. The data were expressed as a harvested cellto-medium concentration ratio: (dpm/g cells)/(dmp/ml medium). The rates of accumulation (Ki) for FIAU and TdR were determined from the slope of the cell-to-medium ratios versus incubation time plots and have units of tracer clearance from the medium (ml medium/min per g cells). The ratio of Ki values (the FIAU/TdR ratio) is a measure of HSV1-tk activity and correlates with independent measures of the gene expression [47].
Subcutaneous Tumors and Study Groups
The experimental protocol involving animals was approved by the Institutional Animal Care and Use Committee of the Memorial Sloan Kettering Cancer Center. The wild-type RG2 cells and transduced RG2 cells expressing different types of mutant HSV1-tk/GFP gene were released from the culture plates by treatment with 0.5% trypsin in PBS for 5 to 10 minutes, resuspended in the growth media to neutralize the trypsin, centrifuged to obtain the cellular pellet, and then resuspended in MEM (without FCS) at a concentration of 106 viable cells in 100 µl. Tumor cells (106 cells in 100 µl) were injected subcutaneously into rnu-rnu rats (Harlan Sprague-Dawley, Indianapolis, IN), weighing 200 to 250 g.
Five subcutaneous tumors were produced in each rat, including the wild-type RG2 tumor and four different transduced TKGFP/RG2 tumors (wt-HSV1-tk/GFP/RG2, NESH-SV1-tk/GFP/RG2, mNLS-HSV1-tk/GFP/RG2, and NESmNLS-HSV1-tk/GFP/RG2), by injection of 106 cells subcutaneous into five different sites. The transduced tumors were produced in the dorsal aspects of both shoulders, and into the right thigh and flank. The native RG2 tumor was produced in the left flank and served as a negative control.
No-Carrier-Added Synthesis [131I]FIAU and γ-Camera Imaging
No-carrier-added [131I]FIAU with >97% radiochemical purity was prepared by the iododestanylation reaction using the tin precursor and carrier-free 131I, followed by isolation of the product by column chromatography as previously described [47]. The specific activity of [131I]FIAU was estimated to be more than 30 Ci/µmol. One day before [131I]FIAU administration, the animals received an intraperitoneal injection of 0.9% NaI solution (1 ml) to block the thyroid uptake of radioactive iodide (low amounts of which develop by radiolysis). [131I]FIAU (300 µCi per animal) was injected intravenously and 24 hours later, γ-camera imaging was performed on a Vertex dual-headed γ-camera (ADAC, Milpitas, CA) equipped with a high-energy high-resolution (HEHR) collimator. Planar images were obtained and reconstructed to 512 x 512 matrix with 1.2 x 1.2-mm pixel size.
Statistics
Descriptive statistics of group data were performed using univariate analysis. Group data were compared using ANOVA analysis, regression analysis, and Student's t-test; a P value of <.05 was considered significant. Statistical analysis of data was performed using the Stat-View 4.57 (Abacus Concepts, Piscataway, NJ) and KaleidaGraph 3.5 (Synergy, Reading, PA).
Results
Subcellular Localization of Different Mutant HSV1-tk/GFP Fusion Proteins
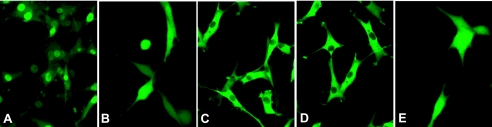
Fluorescent microscopic analysis of transduced tumor cells demonstrated distinct differences in subcellular localization of native and mutant HSV1-tk/GFP proteins. The wildtype HSV1-tk/GFP localized predominantly to the cell nucleus (Figure 2A). The mNLS-HSV1-tk/GFP and Δ45HSV1-tk/GFP proteins exhibited pancellular distribution with no visible differences between the nucleus and cytoplasm (Figure 2, B and E). The distribution of NES-HSV1-tk/GFP and NESmNLS-HSV1-tk/GFP proteins was predominantly cytoplasmic, and the cell nuclei were dark and nonfluorescing, indicating that the NES is functional (Figure 2C). Notably, the double mutant NESmNLS-HSV1-tk/GFP protein distribution exhibited an improved nuclear clearance compared to NES-HSV1-tk/GFP protein (Figure 2D).
Figure 2.
Fluorescent photomicrographs of RG2 cells expressing the native and different HSV1-tk/GFP mutants: (A) native HSV1-tk/GFP; (B) mNLS-HSV1-tk/GFP with mutations in the N-terminal nuclear localization signal; (C) NES-HSV1-tk/GFP with the addition of the nuclear export signal from MAPKK; (D) NESmNLSHSV1-tk/GFP with a combination of mutations in vectors (B) and (C); and (E) Δ45HSV1-tk/GFP with truncation of N-terminal sequence encoding the first 45 amino acids of the native protein.
HSV1-tk/GFP Protein Expression in Transduced Cells by RT-PCR and Western Blot
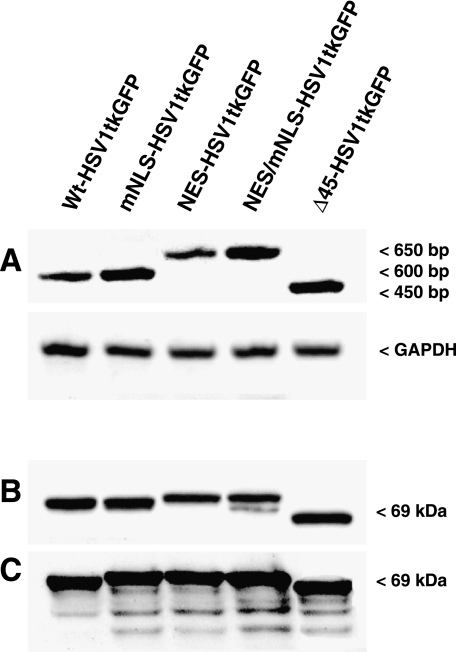
The RT-PCR-amplified fragments of wt-HSV1-tk/GFP and mNLS-HSV1-tk/GFP mRNA had lengths of 578 bp and the NES-HSV1-tk/GFP and NESmNLS-HSV1-tk/GFP mRNA fragments had lengths of 648 bp. The length of Δ45HSV1-tk/GFP was expectedly shorter, 443 bp. The larger size of the NES-HSV1-tk/GFP and NESmNLSHSV1-tk/GFP mRNA fragments reflects the addition of a sequence encoding the NES (Figure 3A). The native and fulllength mutant HSV1-tk/GFP proteins had the predicted molecular mass of about 69 to 72 kDa, whereas the truncated Δ45HSV1-tk/GFP was about 64 kDa, as determined by Western blot analysis. The native and mutant HSV1-tk/GFP proteins had similar stability and exhibited only a minimal degree of degradation (Figure 3, B and C). These results may be somewhat different in other tumor cell lines.
Figure 3.
Detection and analysis of the native and different HSV1-tk/GFP mutants in transduced RG2 cells: (A) RT-PCR analysis; (B) Western blot using a monoclonal anti-HSV1-tk antibody; and (C) Western blot using a monoclonal anti-GFP antibody. Native—wt-HSV1-tk/GFP; with mutations in the N-terminal nuclear localization signal—mNLS-HSV1-tk/GFP; with the addition of the nuclear export signal from MAPKK—NES-HSV1-tk/GFP; with both the NLS mutations and the addition of NES from MAPKK—NESmNLSHSV1-tk/GFP; with truncation of N-terminal sequence encoding the first 45 amino acids of the native protein—Δ45HSV1-tk/GFP.
Enzymatic Properties of Different HSV1-tk/GFP Mutant Proteins
The enzymatic properties of different HSV1-tk/GFP proteins were assessed in vitro using whole cell lysates of transduced tumor cells and the results are summarized in Table 1. The enzyme concentration-normalized apparent reaction rates observed with FIAU and different HSV1-tk/GFP mutants were fairly similar.
Table 1.
Comparison of Enzymatic Activities of Different HSV1-tk/GFP Mutants In Vitro.
| Reporter Protein Type | Enzyme Activity (sec-1)* |
| NES-HSV1-tk/GFP | 4.44±0.58 |
| Δ45HSV1-tk/GFP | 1.94±0.13 |
| mNLS-HSV1-tk/GFP | 2.19±0.10 |
| NESmNLS-HSV1-tk/GFP | 1.22±0.89 |
| Native HSV1-tk/GFP | 2.04±0.70 |
The apparent rate of the reaction normalized by the enzyme concentration (µM FIAU/sec per µM HSV1-tk or sec-1). The concentration of FIAU was 0.01 mM.
FACS and Radiotracer Assessment of Mutant HSV1-tk/GFP Protein Expression
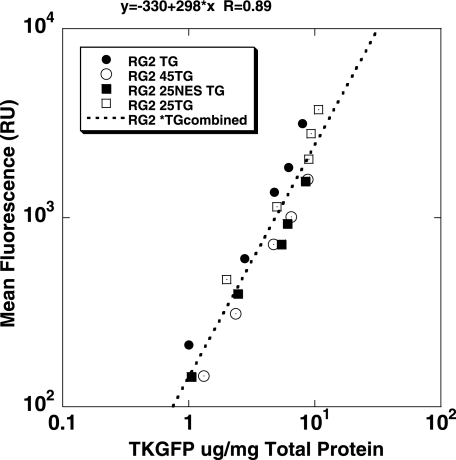
The levels of enzymatic activity of the cytoplasmically retargeted mutant HSV1-tk/GFP proteins (mNLS-HSV1-tk/GFP, NES-HSV1-tk/GFP, NESmNLS-HSV1-tk/GFP, and Δ45HSV1-tk/GFP) were assessed by FACS and the FIAU accumulation assay in populations of transduced RG2 cells with different levels of expression for each of the mutant TKGFP variants. These different populations of cells were selected on the basis of different levels of fluorescence as measured by FACS. Different levels of native HSV1-tk/GFP expression in tumor cell populations were achieved by mixing transduced and nontransduced tumor cells in various proportions; the native HSV1-tk/GFP fluorescence in the resulting populations was also measured by FACS and was found to correlate with the percentage of native HSV1-tk/GFP cells in the population. The levels of HSV1-tk/GFP fluorescence in different native HSV1-tk/GFP/nontransduced cell mixtures and in various transduced cell lines expressing different HSV1-tk/GFP mutant proteins correlated with the concentrations of HSV1-tk/GFP (HSV1-tk/GFP, µg/mg total cellular protein) in corresponding cell populations as measured with ELISA (Figure 4; R= 0.89, P < .0001, paired Student's t-test).
Figure 4.
Relationship between two measurements of HSV1-tk/GFP expression—FACS measurement of GFP fluorescence (mean value) and ELISA assay of HSV1-tk/GFP protein levels. Strong linear relationship was observed in corresponding cell populations (paired t-test, P < .001).
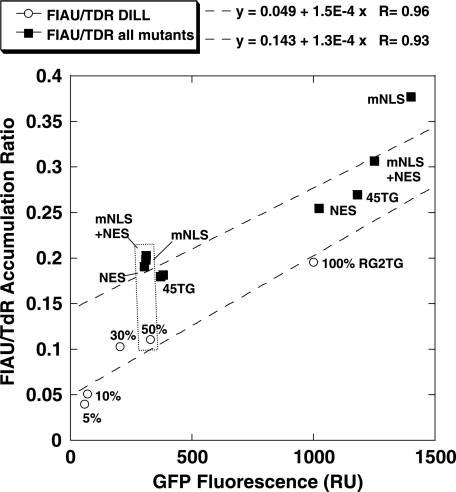
The relationship between GFP fluorescence and FIAU accumulation is shown in Figure 5. The FIAU/TdR normalized accumulation ratios (enzymatic activity of the HSV1-tk subunit) were similar in tumor cell populations expressing different mutant HSV1-tk/GFP proteins. To increase the statistical significance of the analysis, the data from tumor cell populations expressing different mutant HSV1-tk/GFP proteins were grouped together and the slope of the relationship (1.3E-4±3.8E-5) was determined. This was very similar to the value obtained in cell populations expressing different levels of the native HSV1-tk/GFP protein (1.5E-4±2.4E-5). Based on the ratio of Y-intercepts of the slopes in Figure 5, the magnitude of the FIAU/TdR accumulation ratio in various populations of tumor cells expressing different levels of mutant HSV1-tk/GFP proteins was roughly two-fold higher than that measured in populations of cells expressing different levels of native HSV1-tk/GFP protein.
Figure 5.
A comparison of expression levels of the native (open circles) and different HSV1-tk/GFP mutants (closed squares) based on GFP domain fluorescence with their ability to accumulate FIAU in transduced RG2 cells. To achieve different levels of the native HSV1-tk/GFP expression levels, the transduced RG2 cells were mixed with nontransduced cells in percentages indicated on the plot above open circles. The data for HSV1-tk/GFP mutants were grouped together and reflect multiple populations of transduced RG2 cells with different levels of mutant HSV1-tk/GFP expression. The data sets for the native and different HSV1-tk/GFP mutants were fitted with a linear function to assess the relationship. The cell populations with similar levels of transgene expression (dotted rectangle) were later used to produce subcutaneous tumors for the in vivo imaging studies.
Imaging Expression of Different HSV1-tk/GFP Proteins by γ-Camera Imaging In Vivo
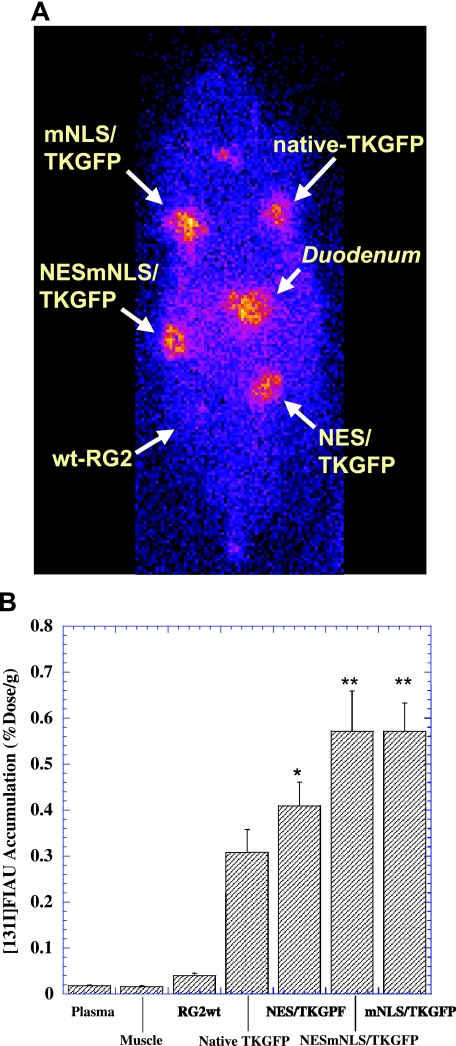
γ-Camera imaging studies were performed in rats bearing multiple subcutaneous tumors grown from transduced RG2 cell populations expressing the wt-HSV1-tk/GFP protein or different mutant HSV1-tk/GFP proteins at similar levels. These populations are identified as the outlined (dotted cluster) in Figure 6. The nontransduced RG2 tumor served as a negative control. Highly specific localization of [131I]FIAU-derived radioactivity in tumors expressing the native and mutant TKGFP proteins was seen (Figure 6A). The levels of [131I]FIAU radioactivity in control (nontransduced) RG2 tumors were very low and similar to body background.
Figure 6.
γ-Camera imaging of [131I]FIAU accumulation in rats bearing multiple subcutaneous tumors produced from RG2 cells expressing native HSV1-tk/GFP or different HSV1-tk/GFP mutants at similar levels based on GFP expression (see dotted cluster box in Figure 5) and from nontransduced RG2 cells (panel A). The levels of [131I]FIAU accumulation measured in tissue samples are expressed as percent injected dose per gram of tissue (panel B). Paired t-test of FIAU accumulation in tumors expressing different mutant HSV1-tk/GFP proteins versus tumors expressing native HSV1-tk/GFP protein (n = 6, *P < .05, **P < .01).
The highest levels of [131I]FIAU accumulation were observed in tumors expressing the mNLS-HSV1-tk/GFP and NES/mNLS-HSV1-tk/GFP protein -0.57±0.06% ID/g and 0.57±0.09% dose/g, respectively (Figure 6B). Somewhat lower levels of [131I]FIAU radioactivity were observed in NES-HSV1-tk/GFP tumors, 0.41±0.05% ID/g, and lower levels of [131I]FIAU radioactivity were observed in tumors expressing the wt-HSV1-tk/GFP protein, 0.31±0.05% ID/g. Overall, the levels of [131I]FIAU accumulation in tumors expressing mutant HSV1-tk/GFP proteins were about two-fold higher than in those expressing the wt-HSV1-tk/GFP protein.
Discussion
The objective of this study was to show that specific mutations in the HSV1-tk/GFP reporter gene result in improved imaging characteristics and improved sensitivity using established radiolabeled probes. We demonstrate that targeted mutations of HSV1-tk/GFP reporter protein designed to overcome or disrupt its NLS lead to a wider subcellular distribution of the mutant HSV1-tk/GFP proteins, and result in improved total cellular enzymatic activity. The observed improvements in functional imaging characteristics of mutant HSV1-tk/GFP proteins did not result from an enhancement of their enzyme kinetic properties with respect to phosphorylation of radiolabeled probe, FIAU, which was the preferred substrate for our studies [46]. The enzyme kinetic studies performed with extracts of cells transduced with either the native or different HSV1-tk/GFP mutants demonstrated similar apparent rates of FIAU phosphorylation by these different enzymes at a concentration of FIAU that was significantly greater than the ∼2 µM Km of FIAU for HSV1-tk (Dr. K. Watanabe, personal communications). The mutation in NLS results in pancellular and proportionally more cytoplasmic localization of the mNLS-TG protein compared to the wild-type TG. Further addition of NES into the NESmNLS-TG was intended to completely clear the cell nucleus from this protein. Therefore, the amount of residual mNLS-TG protein in the nucleus is relatively low, as documented by virtually no gain of total cellular enzymatic activity upon its clearance into the cytoplasm (in form of NESmNLSTG protein). Our data suggest that the increase in accumulation of FIAU in cells transduced with cytoplasmically retargeted HSV1-tk/GFP mutants occurs because cytoplasmic retargeting results in improved bioavailability of these mutant enzymes for FIAU compared to the native HSV1-tk/GFP enzyme (which has a nuclear localization). This explanation is also supported by in vitro data (Figure 5), which demonstrate a higher accumulation of FIAU in cells transduced with cytoplasmically retargeted HSV1-tk/GFP mutants compared to native HSV1-tk/GFP transduced cells when the expression levels of the gene product (amount of enzyme) are similar or normalized. It is noteworthy that the expression levels of HSV1-tk/GFP in transduced cells were assessed using two independent methods—FACS (GFP subunit fluorescence) and a newly developed sandwich ELISA for HSV1-tk/GFP fusion protein measurement.
Analysis of the crystal structure of native HSV1-tk enzyme bound to different substrates [5,8,48] has demonstrated that the N-terminal pole of the parallel β-sheet (containing the N-terminus of the polypeptide) forms a part of the molecular surface, but is not involved in the formation of the enzyme catalytic site. As demonstrated in the current study, mutations within the N-terminus or removal of a large portion of the N-terminus (the first 45 amino acids) of the native HSV1-tk did not impact on the enzymatic characteristics of cytoplasmically retargeted mutant HSV1-tk/GFP proteins. These results are confirmatory of previous reports, which also demonstrated that truncation of the first 34 N-terminal amino acids or mutation of different Arg residues within this region resulted in pancellular distribution of HSV1-tk/GFP chimeras [14]. The retention of enzymatic activity of HSV1-tk after truncation of the first 45 amino acids was demonstrated previously using a GCV sensitivity assay in tumor cells transduced with the N-terminally truncated HSV1-tk gene [43], although no enzyme kinetic studies had been performed in these studies [14,43].
Enzyme kinetic studies were performed in Q125, T63, A168, R176, and C336 site-directed mutants of HSV1-tk to clarify the contribution of these residues to the binding of deoxyribose-containing or acycloanalogues of thymidine, Mg2+, and ATP [30]. It was demonstrated that the Q125L mutant is able to phosphorylate only thymidine, whereas the Q125N mutant accepts both thymidine and acyclovir (ACV) as substrates, and the Q125E mutation results in an abolition of phosphorylation activity of the enzyme. T63 appears to be essential for the binding of Mg2+ and thus the catalytic activity of the enzyme, whereas A168 limits steric accessibility and, if mutated to a bulkier residue, will exclude binding of larger substrate analogues. R176 appears to be essential for electrostatic balance within the active site, and C336, which is located at the surface of TK and directed toward the ATP binding site, disrupts the three-dimensional structure of the whole active site by shifting the LID domain.
Seminal studies by Black et al. [3,4], Kokoris and Black [27], and Kokoris et al. [28,29] further identified the role of different amino acids in substrate specificity of HSV1-tk. Using random sequence and targeted mutagenesis approaches, these researchers have produced numerous mutant HSV1-tk enzymes with improved specificity towards different acycloguanosine analogues, including GCV, ACV, and pencyclovir (PCV) [3,4,27–29]. One of such mutant enzymes, sr39TK, was extensively studied as a reporter gene for noninvasive imaging with PET using different radiolabeled acycloguanosine analogues as reporter probes, including 8-[18F]GCV [20], [18F]FHPG [6,46], and [18F]FHBG [46,49]. The results of these studies are complimentary to those reported here because the mutations we introduced into the HSV1-tk sequence of the HSV1-tk/GFP fusion reporter gene were quite distant from the catalytic site of the enzyme and, therefore, did not significantly impact on substrate specificity or enzyme kinetics.
Following our initial reports on HSV1-tk/GFP mutagenesis described in the current paper [37], specific mutations in sr39HSV1-tk were performed in arginines (R) 25 and 26 to glycine (G) and serine (S), respectively, to create mNLS-sr39HSV1-tk/GFP reporter gene to image two-protein interactions, p53 and SV40, in living animals using [18F]FHBG and PET [34]. The sr39HSV1-tk/GFP was reported to localize predominantly in the nucleus of transiently transfected HeLa cells. Conversely, the mNLS-sr39HSV1-tk/GFP was distributed more uniformly in both the nucleus and cytosol, consistent with disruption of one NLS. In the latter study, it was also reported that the accumulation of a radiolabeled nucleoside analog [8-3H]PCV in HeLa cells transiently transfected with mNLS-sr39HSV1-tk was more than two-fold greater than HSV1-sr39HSV1-tk and 12-fold greater than native HSV1-tk, which is in accord with the results obtained in our current studies.
It is known that the nuclear tropism of native HSV1-tk causes cell toxicity when the enzyme is expressed at high levels [9,43] and limits the expression level of this transgene. In contrast, bulk populations of cells transduced with cytoplasmically retargeted HSV1-tk/GFP mutants consistently exhibited higher levels of the transgene expression compared to cells transfected with the native HSV1-tk/GFP using the same vector backbone and promoter sequences. The latter phenomenon is consistent with reduced nuclear toxicity of cytoplasmically retargeted HSV1-tk/GFP mutants and the absence of a nonspecific feedback inhibition of transgene expression. Furthermore, it has been demonstrated that the N-terminal truncated variant of HSV1-tk does not cause spermatozoal toxicity in transgenic mice due to the removal of cryptic testis-specific promoter sequences and of nuclear localization-associated toxicity [12]. Therefore, if should be feasible to use cytoplasmically retargeted or N-terminal-truncated HSV1-tk/GFP reporter gene mutants for generation of transgenic mice bearing various reporter systems. It is noteworthy that noninvasive imaging of the albumin promoter-driven expression of the wild-type HSV1-tk reporter gene (AL-HSV1-tk) in heterozygous transgenic mice has been reported using 18F-FHBG and PET [22]. The ALHSV1-tk transgenic mice had been developed from FVB/N mice by pronuclear injection, although no information was provided regarding the fertility status of these AL-HSV1-tk transgenic male mice.
Conclusions
The nuclear clearance and cytoplasmic retargeting of HSV1-tk/GFP were achieved with different site-directed and N-terminally truncated HSV1-tk/GFP mutants. The retargeted HSV1-tk/GFP mutant proteins are expressed at higher levels compared to the native HSV1-tk/GFP in bulk populations of transduced cells due to the absence of nuclear toxicity. The observed increase in total cellular enzymatic activity of the mutant HSV1-tk/GFP variants compared to native HSV1-tk/GFP expressed at the same protein level is probably due to improved bioavailability of the retargeted mutant HSV1-tk/GFP enzymes in the cytoplasm. Cytoplasmic retargeting of mutant HSV1-tk/GFP is likely to facilitate reaction with exogenous substrates and radiolabeled probes (e.g., FIAU), with no significant change in enzyme kinetic properties. The improved total cellular enzymatic activity of retargeted HSV1-tk/GFP mutants confers higher levels of [131I]FIAU accumulation in transduced subcutaneous tumor xenograftbearing rats, and this should result in a higher sensitivity of these mutant HSV1-tk/GFP reporter systems.
Abbreviations
- HSV1-tk
herpes simplex virus type 1 thymidine kinase
- GFP
green fluorescent protein
- PET
positron emission tomography
- FIAU
2′-fluoro-2′-deoxy-1-β-d-arabinofuranosyl-5-iodo-uracil
- FHBG
9-(4-[18F]fluoro-3-hydroxymethylbutyl)guanine
- FACS
fluorescence-activated cell sorting
- DMEM
Dulbecco's modified Eagle's medium
- ELISA
enzyme-linked immunosorbent assay
- VSV-G
vesicular stomatitis virus G-protein
- FCS
fetal calf serum
References
- 1.Bengel FM, Anton M, Avril N, Brill T, Nguyen N, Haubner R, Gleiter E, Gansbacher B, Schwaiger M. Uptake of radiolabeled 2′-fluoro-2′-deoxy-5-iodo-1-beta-D-arabinofuranosyluracil in cardiac cells after adenoviral transfer of the herpesvirus thymidine kinase gene: the cellular basis for cardiac gene imaging. Circulation. 2000;102:948–950. doi: 10.1161/01.cir.102.9.948. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black ME, Kokoris MS, Sabo P. Herpes simplex virus-1 thymidine kinase mutants created by semi-random sequence mutagenesis improve prodrug-mediated tumor cell killing. Cancer Res. 2001;61:3022–3026. [PubMed] [Google Scholar]
- 4.Black ME, Rechtin TM, Drake RR. Effect on substrate binding of an alteration at the conserved aspartic acid-162 in herpes simplex virus type 1 thymidine kinase. J Gen Virol. 1996;77(Pt 7):1521–1527. doi: 10.1099/0022-1317-77-7-1521. [DOI] [PubMed] [Google Scholar]
- 5.Brown DG, Visse R, Sandhu G, Davies A, Rizkallah P, Melitz C, Summers W, Sanderson MR. Crystal structures of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and ganciclovir. Nat Struct Biol. 1995;2:876–881. doi: 10.1038/nsb1095-876. [DOI] [PubMed] [Google Scholar]
- 6.Brust P, Haubner R, Friedrich A, Scheunemann M, Anton M, Koufaki ON, Noll S, Noll B, Haberkorn U, Schackert G, Schackert HK, Avril N, Johannsen B. Comparison of [18F]FHPG and [124/125I]FIAU for imaging herpes simplex virus type 1 thymidine kinase gene expression. Eur J Nucl Med. 2001;28:721–729. doi: 10.1007/s002590100526. [DOI] [PubMed] [Google Scholar]
- 7.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champness JN, Bennett MS, Wien F, Visse R, Summers WC, Herdewijn P, de Clerq E, Ostrowski T, Jarvest RL, Sanderson MR. Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands. Proteins. 1998;32:350–361. doi: 10.1002/(sici)1097-0134(19980815)32:3<350::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Cohen JL, Boyer O, Salomon B, Onclerco R, Depetris D, Lejeune L, Dubus-Bonnet V, Bruel S, Charlotte F, Mattei MG, Klatzmann D. Fertile homozygous transgenic mice expressing a functional truncated herpes simplex thymidine kinase delta TK gene. Transgenic Res. 1998;7:321–330. doi: 10.1023/a:1008893206208. [DOI] [PubMed] [Google Scholar]
- 10.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 11.Contag CH, Jenkins D, Contag PR, Negrin RS. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2:41–52. doi: 10.1038/sj.neo.7900079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowsill C, Southgate TD, Morrissey G, Dewey RA, Morelli AE, Maleniak TC, Forrest Z, Klatzmann D, Wilkinson GW, Lowenstein PR, Castro MG. Central nervous system toxicity of two adenoviral vectors encoding variants of the herpes simplex virus type 1 thymidine kinase: reduced cytotoxicity of a truncated HSV1-TK. Gene Ther. 2000;7:679–685. doi: 10.1038/sj.gt.3301147. [DOI] [PubMed] [Google Scholar]
- 13.Degreve B, Esnouf R, De Clercq E, Balzarini J. Characterization of multiple nuclear localization signals in herpes simplex virus type 1 thymidine kinase. Biochem Biophys Res Commun. 1999;264:338–342. doi: 10.1006/bbrc.1999.1485. [DOI] [PubMed] [Google Scholar]
- 14.Degreve B, Johansson M, De Clercq E, Karlsson A, Balzarini J. Differential intracellular compartmentalization of herpetic thymidine kinases (TKs) in TK gene-transfected tumor cells: molecular characterization of the nuclear localization signal of herpes simplex virus type 1 TK. J Virol. 1998;72:9535–9543. doi: 10.1128/jvi.72.12.9535-9543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doubrovin M, Ponomarev V, Beresten T, Balatoni J, Bornmann W, Finn R, Humm J, Larson S, Sadelain M, Blasberg R, Gelovani Tjuvajev J. Imaging transcriptional regulation of p53-dependent genes with positron emission tomography in vivo. Proc Natl Acad Sci USA. 2001;98:9300–9305. doi: 10.1073/pnas.161091198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrovin M, Ponomarev V, Beresten T, Matei C, Koutcher J, Tjuvajev J. In Vivo 19F Nuclear Magnetic Resonance Measurements of Enhanced 5FU Conversion to Fluoronucleotides after UPRT Gene Transduction; Abstract, 3rd Annual Meeting of the ASGT, Denver, CO, May 31–June 4, 2000; 2000. [Google Scholar]
- 17.Ellenberg J, Lippincott-Schwartz J, Presley JF. Dual-colour imaging with GFP variants. Trends Cell Biol. 1999;9:52–56. doi: 10.1016/s0962-8924(98)01420-2. [DOI] [PubMed] [Google Scholar]
- 18.Falk MM, Lauf U. High resolution, fluorescence deconvolution microscopy and tagging with the autofluorescent tracers CFP, GFP, and YFP to study the structural composition of gap junctions in living cells. Microsc Res Tech. 2001;52:251–262. doi: 10.1002/1097-0029(20010201)52:3<251::AID-JEMT1011>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Gallardo HF, Tan C, Ory D, Sadelain M. Recombinant retroviruses pseudotyped with the vesicular stomatitis virus G glycoprotein mediate both stable gene transfer and pseudotransduction in human peripheral blood lymphocytes. Blood. 1997;90:952–957. [PubMed] [Google Scholar]
- 20.Gambhir SS, Barrio JR, Wu L, Iyer M, Namavari M, Satyamurthy N, Bauer E, Parrish C, MacLaren DC, Borghei AR, Green LA, Sharfstein S, Berk AJ, Cherry SR, Phelps ME, Herschman HR. Imaging of adenoviral-directed herpes simplex virus type 1 thymidine kinase reporter gene expression in mice with radiolabeled ganciclovir. J Nucl Med. 1998;39:2003–2011. [PubMed] [Google Scholar]
- 21.Gambhir SS, Herschman HR, Cherry SR, Barrio JR, Satyamurthy N, Toyokuni T, Phelps ME, Larson SM, Balatoni J, Finn R, Sadelain M, Tjuvajev J, Blasberg R. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green L, Yap C, Nguyen N, Barrio J, Namavari M, Satyamurthy N, Phelps M, Sandgren E, Herschman H, Gambhir S. Indirect monitoring of endogenous gene expression by positron emission tomography (PET) imaging of reporter gene expression in transgenic mice. Mol Imaging Biol. 2002;4:71–81. doi: 10.1016/s1095-0397(01)00071-1. [DOI] [PubMed] [Google Scholar]
- 23.Haberkorn U. Gene therapy with sodium/iodide symporter in hepatocarcinoma. Exp Clin Endocrinol. 2001;1:60–62. doi: 10.1055/s-2001-11010. [DOI] [PubMed] [Google Scholar]
- 24.Hackman TDM, Balatoni J, Beresten T, Ponomarev V, Beattie B, Finn R, Bornmann W, Blasberg R, Gelovani Tjuvajev J. Imaging expression of cytosine deaminase-herpes virus thymidine kinase fusion gene (CD/TK) expression with [124I]FIAU and PET. Mol Imaging. 2002;1:36–42. doi: 10.1162/15353500200200003. [DOI] [PubMed] [Google Scholar]
- 25.Hadjantonakis AK, Nagy A. The color of mice: in the light of GFP-variant reporters. Histochem Cell Biol. 2001;115:49–58. doi: 10.1007/s004180000233. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs A, Dubrovin M, Hewett J, Sena-Esteves M, Tan CW, Slack M, Sadelain M, Breakefield XO, Tjuvajev JG. Functional coexpression of HSV-1 thymidine kinase and green fluorescent protein: implications for noninvasive imaging of transgene expression. Neoplasia. 1999;1:154–161. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kokoris MS, Black ME. Characterization of herpes simplex virus type 1 thymidine kinase mutants engineered for improved ganciclovir or acyclovir activity. Protein Sci. 2002;11:2267–2272. doi: 10.1110/ps.2460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokoris MS, Sabo P, Adman ET, Black ME. Enhancement of tumor ablation by a selected HSV-1 thymidine kinase mutant. Gene Ther. 1999;6:1415–1426. doi: 10.1038/sj.gt.3300966. [DOI] [PubMed] [Google Scholar]
- 29.Kokoris MS, Sabo P, Black ME. In vitro evaluation of mutant HSV-1 thymidine kinases for suicide gene therapy. Anticancer Res. 2000;20:959–963. [PubMed] [Google Scholar]
- 30.Kussmann-Gerber S, Kuonen O, Folkers G, Pilger BD, Scapozza L. Drug resistance of herpes simplex virus type 1-structural considerations at the molecular level of the thymidine kinase. Eur J Biochem. 1998;255:472–481. doi: 10.1046/j.1432-1327.1998.2550472.x. [DOI] [PubMed] [Google Scholar]
- 31.Lalwani AK, Han JJ, Walsh BJ, Zolotukhin S, Muzyczka N, Mhatre AN. Green fluorescent protein as a reporter for gene transfer studies in the cochlea. Hear Res. 1997;114:139–147. doi: 10.1016/s0378-5955(97)00151-2. [DOI] [PubMed] [Google Scholar]
- 32.Levy JP, Muldoon RR, Zolotukhin S, Link CJ., Jr Retroviral transfer and expression of a humanized, red-shifted green fluorescent protein gene into human tumor cells. Nat Biotechnol. 1996;14:610–614. doi: 10.1038/nbt0596-610. [DOI] [PubMed] [Google Scholar]
- 33.Liang Q, Satyamurthy N, Barrio JR, Toyokuni T, Phelps MP, Gambhir SS, Herschman HR. Noninvasive, quantitative imaging in living animals of a mutant dopamine D2 receptor reporter gene in which ligand binding is uncoupled from signal transduction. Gene Ther. 2001;8:1490–1498. doi: 10.1038/sj.gt.3301542. [DOI] [PubMed] [Google Scholar]
- 34.Luker GD, Sharma V, Pica CM, Dahlheimer JL, Li W, Ochesky J, Ryan CE, Piwnica-Worms H, Piwnica-Worms D. Noninvasive imaging of protein-protein interactions in living animals. Proc Natl Acad Sci USA. 2002;99:6961–6966. doi: 10.1073/pnas.092022399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathieu S, El-Battari Monitoring E-selectin-mediated adhesion using green and red fluorescent proteins. J Immunol Methods. 2003;272:81–92. doi: 10.1016/s0022-1759(02)00434-9. [DOI] [PubMed] [Google Scholar]
- 36.Ponomarev V, Doubrovin M, Lyddane C, Beresten T, Balatoni J, Bornman W, Finn R, Akhurst T, Larson S, Blasberg R, Sadelain M, Tjuvajev JG. Imaging TCR-dependent NFAT-mediated T-cell activation with positron emission tomography in vivo. Neoplasia. 2001;3:480–488. doi: 10.1038/sj.neo.7900204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponomarev VDM, Beresten T, Balatoni J, Blasberg R, Tjuvajev JG. Functional coexpression of HSV1-tk and GFP and generation of mutants with different subcellular localization of TK/GFP fusion protein for gene therapy of gliomas. J Nucl Med. 2000;41:81. [Google Scholar]
- 38.Ponomarev VDM, Serganova I, Ageyeva L, Beresten T, Sagomonyan S, Balatoni J, Finn R, Blasberg R, Gelovani Tjuvajev JG. Human Thymidine Kinase Type 2—A Novel Non-Immunogenic Reporter Gene For Non-Invasive Imaging In Humans; AACR Meeting, Molecular Imaging in Cancer, Orlando, FL, January 23–27, 2002; 2002. [Google Scholar]
- 39.Ponomarev VSIALBT, Doubrovin M, Gelovani Tjuvajev JG. Xanthine Phosphoribosyl Transferase and Red Fluorescent Protein Fusion—A Novel Reporter Gene for Multi-Modality Imaging; Abstract, 9th Annual Meeting of the ESGT, Antalya, Turkey, November 2–4, 2001; 2001. [Google Scholar]
- 40.Qiao J, Doubrovin M, Sauter BV, Huang Y, Guo ZS, Balatoni J, Akhurst T, Blasberg RG, Tjuvajev JG, Chen SH, Woo SL. Tumorspecific transcriptional targeting of suicide gene therapy. Gene Ther. 2002;9:168–175. doi: 10.1038/sj.gt.3301618. [DOI] [PubMed] [Google Scholar]
- 41.Ray P, Bauer E, Iyer M, Barrio JR, Satyamurthy N, Phelps ME, Herschman HR, Gambhir SS. Monitoring gene therapy with reporter gene imaging. Semin Nucl Med. 2001;31:312–320. doi: 10.1053/snuc.2001.26209. [DOI] [PubMed] [Google Scholar]
- 42.Rogers BE, Zinn KR, Buchsbaum DJ. Gene transfer strategies for improving radiolabeled peptide imaging and therapy. Q J Nucl Med. 2000;3:208–223. [PubMed] [Google Scholar]
- 43.Salomon B, Maury S, Loubiere L, Caruso M, Onclercq R, Klatzmann D. A truncated herpes simplex virus thymidine kinase phosphorylates thymidine and nucleoside analogs and does not cause sterility in transgenic mice. Mol Cell Biol. 1995;15:5322–5328. doi: 10.1128/mcb.15.10.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stegman LD, Rehemtulla A, Beattie B, Kievit E, Lawrence TS, Blasberg RG, Tjuvajev JG, Ross BD. Noninvasive quantitation of cytosine deaminase transgene expression in human tumor xenografts with in vivo magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 1999;96:9821–9826. doi: 10.1073/pnas.96.17.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tjuvajev JG, Avril N, Oku T, Sasajima T, Miyagawa T, Joshi R, Safer M, Beattie B, DiResta G, Daghighian F, Augensen F, Koutcher J, Zweit J, Humm J, Larson SM, Finn R, Blasberg R. Imaging herpes virus thymidine kinase gene transfer and expression by positron emission tomography. Cancer Res. 1998;58:4333–4341. [PubMed] [Google Scholar]
- 46.Tjuvajev JG, Doubrovin M, Akhurst T, Cai S, Balatoni J, Alauddin MM, Finn R, Bornmann W, Thaler H, Conti PS, Blasberg RG. Comparison of radiolabeled nucleoside probes (FIAU, FHBG, and FHPG) for PET imaging of HSV1-tk gene expression. J Nucl Med. 2002;43:1072–1083. [PubMed] [Google Scholar]
- 47.Tjuvajev JG, Stockhammer G, Desai R, Uehara H, Watanabe K, Gansbacher B, Blasberg RG. Imaging the expression of transfected genes in vivo. Cancer Res. 1995;55:6126–6132. [PubMed] [Google Scholar]
- 48.Wild K, Bohner T, Aubry A, Folkers G, Schulz GE. The three-dimensional structure of thymidine kinase from herpes simplex virus type 1. FEBS Lett. 1995;368:289–292. doi: 10.1016/0014-5793(95)00680-8. [DOI] [PubMed] [Google Scholar]
- 49.Yaghoubi S, Barrio JR, Dahlbom M, Iyer M, Namavari M, Satyamurthy N, Goldman R, Herschman HR, Phelps ME, Gambhir SS. Human pharmacokinetic and dosimetry studies of [(18)F]FHBG: a reporter probe for imaging herpes simplex virus type-1 thymidine kinase reporter gene expression. J Nucl Med. 2001;42:1225–1234. [PubMed] [Google Scholar]