Abstract
Hepatitis B x antigen (HBxAg) is a trans-activating protein that may be involved in hepatocarcinogenesis, although few natural effectors of HBxAg that participate in this process have been identified. To identify additional effectors, whole cell RNA isolated from HBxAg-positive and HBxAg-negative HepG2 cells were compared by polymerase chain reaction select cDNA subtraction, and one clone, upregulated gene, clone 11 (URG11), was chosen for further characterization. Elevated levels of URG11 mRNA and protein were observed in HBxAg-positive compared to HBxAg-negative HepG2 cells. Costaining was observed in infected liver (P < .01). URG11 stimulated cell growth in culture (P < .01), anchorage-independent growth in soft agar (P < .001), and accelerated tumor formation (P < .01), and yielded larger tumors (P < .02) in SCID mice injected subcutaneously with HepG2 cells. These data suggest that URG11 is a natural effector of HBxAg that may promote the development of hepatocellular carcinoma.
Keywords: hepatitis B virus, hepatitis B x antigen, hepatocellular carcinoma, oncogene, cell cycle
Introduction
Chronic hepatitis B virus (HBV) infection is a major public health problem worldwide because it is associated with the development of hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [1–3]. The mechanism(s) whereby HBV causes HCC remains to be worked out, but increasing evidence suggests that the viral contribution to HCC involves persistently high levels of the virus-encoded X antigen, or hepatitis B x antigen (HBxAg), in the liver of chronically infected patients [4–7]. High levels of intrahepatic HBxAg expression directly correlate with the intensity of liver disease [8,9]. HBxAg transforms cells in vitro [10–12], whereas sustained high levels of HBxAg in transgenic mice are associated with the development of HCC [13–15], suggesting that HBxAg plays an important role in the pathogenesis of this tumor type.
HBxAg has been identified as a potentially promiscuous trans-activating protein [16,17]. HBxAg binds to and alters the function of transcription factors such as OCT-1 [18], ATF-2 [19], CREB [19], TBP [20], a subunit common to RNA polymerases [21], and other elements of the transcriptional machinery [22,23]. HBxAg may also repress gene expression by binding to and inactivating the tumor suppressor, p53 [15,24], and the senescence-related factor p55sen [25], as well as by downregulating the expression of p21WAF1/CIP1/SDI1 [26] and the translation initiation factor, sui1 [27], all of which negatively regulate hepatocellular growth and survival. In addition, HBxAg stimulates several cytoplasmic signal transduction pathways, such as those involving NF-κB [28,29], AP-1 [30], MAPK [31,32], and PI3K [33], that promote cell growth and/or promote cell survival in the face of apoptotic stimuli. Since little is known about the natural effectors of HBxAg that contribute to the development of HCC, experiments were designed to identify and characterize such effectors. In this report, it is shown that HBxAg upregulates the expression of a unique gene (upregulated gene, clone 11; URG11) that promotes hepatocellular growth and tumorigenesis.
Materials and Methods
Polymerase Chain Reaction (PCR) Select cDNA Subtraction of RNA From HepG2X and HepG2CAT Cells
HepG2X and HepG2CAT cells were constructed, as described [27]. HBxAg and CAT expressions were verified prior to conducting this study [27]. RNA from each cell line was then isolated and subjected to PCR select cDNA subtraction using a commercial kit according to instructions provided by the manufacturer (Clontech, Palo Alto, CA), as described [27]. The PCR fragments corresponding to differentially expressed mRNAs were cloned, sequenced, and compared to existing sequences within GenBank. One of these differentially expressed PCR fragments, designated as URG11, was chosen for further characterization.
In Situ Hybridization (ISH)
To verify that URG11 was differentially expressed in HepG2X compared to HepG2CAT cells, the URG11 cDNA fragment obtained from PCR select cDNA subtraction was used as a probe for ISH. ISH was carried out using the Oncor ISH and digoxigenin/biotin detection kits (Oncor, Gaithersburg, MD), as described [27,34]. ISH was also carried out using fresh frozen samples from tumor (HCC) and nontumor livers collected from HBV carriers (see below).
Patient Samples
The HCC and surrounding nontumor liver tissues used for analyses were obtained from three sets of patients. Twenty-three paired tumor/nontumor samples came from as many Chinese patients who had undergone surgery for the removal of their tumors. Many patients lived in and around Xi'an and were treated at the Fourth Military Medical University. Other Chinese patients underwent surgery at the Hepatobiliary Hospital of the Second Military Medical University in Shanghai. Fourteen additional paired tumor/nontumor samples were obtained from South African patients. These tissues were usually obtained at necropsy, although some were obtained as a result of surgical resection at the University of Witwatersrand in Johannesburg. Additional characteristics of these patients have been published [27]. Formalin-fixed, paraffinembedded tissues, fresh frozen blocks, and -80°C snap-frozen paired liver and tumor samples were collected from most patients, used for diagnostic purposes, and then made available for these studies. Analogous pieces of uninfected human liver from two individuals were available to serve as controls. Other normal tissues from uninfected individuals available for staining were from the gastrointestinal mucosa, heart, placenta, ovary, brain, kidneys, prostate, pancreas, and spleen. In addition, tumor samples from 14 Chinese patients with colon cancer, from 10 patients with gastric cancer, from 7 patients with lung cancer, and from 4 patients with breast cancer were used as tumor controls. The use of all tissues for this work was approved by the Institutional Review Board at the Thomas Jefferson University.
Northern Blotting
Northern blotting was conducted as previously reported [27]. Briefly, total RNA was extracted using the Qiagen RNeasy RNA Kit (Qiagen, Valencia, CA). Twenty micrograms of total RNA from each sample was electrophoresed on formaldehyde-denaturing agarose gels, and transferred to nylon membranes (Schleicher and Schuell, Keene, NH). The URG11 cDNA fragment obtained from PCR select cDNA subtraction was radiolabeled with α[32P]dCTP (NEN, Boston, MA) by random priming using the Prime-A-Gene labeling kit (Promega, Madison, WI) and used for hybridization under stringent conditions. The results were detected by autoradiography and quantitated by phosphoimaging. A probe for glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used for the normalization of results in each lane.
Cloning and Sequencing of Full-Length URG11 cDNA
To obtain the full-length clone of URG11, two genespecific primers were designed according to the sequence of the URG11 cDNA fragment. The sense primer was URG11-1p (5′-GGAGCTGGAGGAGATGAAGCACCGG-3′) whereas the antisense primer was URG11-2p (5′-GGCTCTCCCCTCGCAGAAATGTGGC-3′). These primers were used in 5′ and 3′ RACE PCR with the Marathon cDNA Amplification Kit according to enclosed instructions (Clontech). Human placental cDNA was used as the template. The PCR products were cloned into pT7blue vector (Novagen, Madison, WI) and sequenced. The appropriate 3′ and 5′ gene-specific fragments were then digested with EcoRI and NotI and cloned into pcDNA3 (Invitrogen, San Diego, CA), and the integrity of the full-length clone was verified by DNA sequencing. The full-length sequence was analyzed for homology to entries in GenBank.
Subcloning of HBxAg
The HBx gene obtained from the ayw sequence was subcloned into pcDNA3, as described [27,34].
Preparation and Use of URG11 Antibodies
The full-length cDNA of URG11 was used to deduce the corresponding amino acid sequence using the TRANSLATE program. The amino acid sequence was then subjected to analysis in the PEPTIDESTRUCTURE and PLOTSTRUCTURE programs to identify hydrophilic peptides that would be suitable candidates for solid-phase peptide synthesis [35]. Three peptides were made at the Kimmel Cancer Center of the Thomas Jefferson University. Each of these peptides was then coupled to keyhole limpet hemocyanin, and New Zealand white rabbits were immunized (two rabbits per peptide), as described [36]. Corresponding antibodies were measured by specific enzyme-linked immunosorbent assays [37]. A mixture of antibodies to peptide 2 (amino acids 568–591, inclusive) and peptide 3 (amino acids 611–630, inclusive) was used at a dilution of 1:6000 each for staining formalin-fixed, paraffin-embedded tissues, and at a dilution of 1:10,000 each for staining fresh frozen samples. Staining was otherwise carried out, as described [34]. Controls included staining with preimmune serum in place of primary antibodies, and preincubation of primary antibodies with an excess (25 µg) of the corresponding synthetic peptide(s) prior to staining [37,38].
Western blot analysis was carried out, as described [27]. One hundred micrograms of protein from each sample was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 10% running gels and transferred to PVDF membranes (Millipore, Bedford, MA). The primary antibody consisted of a mixture of anti-URG11 from the three peptides (each at 1:600 dilution). For some membranes, anti-HBx was used as the primary antibody, as described [37,38]. The secondary antibody was horseradish peroxidase goat antirabbit Ig (Santa Cruz Biotechnology, Santa Cruz, CA). As a negative control, preimmune serum was used in place of anti-URG11 or anti-HBx. As a positive control, the peptides used for immunization were spotted near the edge of the PVDF membranes following the transfer step. The results were visualized using the enhanced chemiluminescence (ECL) detection system (Amersham, Uppsala, Sweden). β-Actin was used as internal control for loading using monoclonal mouse antihuman β-actin (Sigma, St. Louis, MO) as primary antibody at a dilution of 1:10,000.
Construction of URG11 Over-expressing HepG2 and Control Cells
Separate cultures of 1 x 106 HepG2 cells were transfected with 10 µg of pcDNA3-URG11, pcDNA3-HBx, or pcDNA3 vector using the SuperFect Transfection Reagent (Qiagen) as previously reported [27]. Cells were selected in G418 (800 µg/ml complete medium) for 4 weeks, and resistant cultures were passaged without selecting colonies. URG11 and HBxAg protein expressions were verified by Western blotting using anti-URG11 or anti-HBx, respectively.
HBxAg Mutant Expressing HepG2 Cells
pcDNA3-HBx was used to make HBxAg mutants lacking the carboxy-terminal 10 amino acids (HBx1–135), the carboxy-terminal 40 amino acids (HBx1–105), the amino-terminal 10 amino acids (HBx10–145), or the aminoterminal 40 amino acids (HBx41–145), as previously described [27]. Additional HBxAg mutants lacking amino acid residues 42 to 77, inclusive (HBxML), or residues 78 to 115, inclusive (HBxMR), were also constructed, as described [27]. The mutant cDNA were verified by sequence analysis. The polypeptides made from these various mutants were verified by in vitro translation of each construct, and then each construct was stably transfected into HepG2 cells. Cultures were selected in G418 and passaged without selection of individual colonies in each case. Mutant expression was assayed in cell lysates by Northern and Western blotting.
Transient Transfection of HepG2 Cells
To determine whether HBxAg upregulates the expression of URG11, 1 x 106 HepG2 cells were seeded overnight into each of three 60-mm-diameter plates, and then transiently transfected with 5 µg of pcDNA3, pcDNA3-HBx, or pcDNA3-URG11 using Superfect (Qiagen, Santa Clara, CA) according to instructions provided by the manufacturer [27]. Cell lysates were prepared 48 hours post-transfection and analyzed by SDS-PAGE. The levels of URG11 were then determined by Western blotting using anti-URG11 antibodies.
Additional transient transfection experiments were performed to determine the trans-activation function of each HBxAg mutant. Five micrograms of each mutant was cotransfected into separate cultures of HepG2 cells along with 2 µg of the reporter plasmid, pGL2-HIV-1-LTR, in which expression of the luciferase gene is under the control of the HBxAg-responsive HIV-LTR promoter. Luciferase reporter gene activity was determined 48 hours post-transfection. Additional details of this assay have been previously published [27].
Growth Curves in Medium Containing 10% or 0% FCS
HepG2 cells stably transfected with pcDNA3, pcDNA3-HBx, or pcDNA3-URG11 were seeded into six-well plates in duplicate and grown in complete or serum-free medium. The number of viable cells was determined at daily intervals for up to 5 days by trypan blue staining. Cell viability was independently determined using the modified tetrazolium salt (MTT) assay (Cell Titer 96 Nonradioactive Cell Proliferation Assay; Promega). Growth curves from HepG2.2.15 cells [39] were generated in parallel for comparison.
Flow Cytometry
To assess the effect of URG11 on cell cycle, 1 x 105 HepG2-pcDNA3, HepG2-pcDNA3-HBx, or HepG2-pcDNA3-URG11 cells were seeded into 60-mm-diameter plates in complete medium overnight, placed in serum-free medium for 48 hours to synchronize the cells, and then again in complete medium. At 24, 48, and 72 hours, cells were recovered, fixed, stained with propidium iodide, and subjected to flow cytometry analysis for DNA content in the FACS facility at the Thomas Jefferson University.
Growth of Cells in Soft Agar and Tumorigenicity in Nude Mice
HepG2 cells stably transfected with pcDNA3, pcDNA3-URG11, or pcDNA3-HBx were selected in G418 for 4 weeks. To test for growth in soft agar, 1 x 104 cells/well were seeded in triplicate into six-well plates, allowed to grow for 21 days, and counted under code using an inverted microscope. Colonies that were at least 0.5 mm in diameter were scored as positive.
For tumorigenicity assays, three groups of 10 mice each were injected subcutaneously at a single site with 5 x 106 cells (HepG2-pcDNA3, HepG2-pcDNA3-URG11, or HepG2-pcDNA3-HBx) under code. Tumor onset was scored visually and by palpitation at the sight of injection by two trained laboratory personnel at different times on the same day. Average tumor size was estimated by physical measurement of the excised tumor at the time of sacrifice. With the exception of mice with large tumor burdens, animals were sacrificed 6 weeks postinjection. These tumors were verified as being HCCs by hematoxylin and eosin (H&E) staining. Blocks were available for further analysis.
Statistics
The relationship between HBxAg and URG11 signals obtained by ISH and immunohistochemistry was determined using 2 x 2 comparisons in the Fisher's exact test. Statistical significance was observed when P < .05. The mean difference between cell numbers (in culture), colonies (in soft agar), or cell cycle phase for HepG2X, HepG2-pcDNA3-URG11, and HepG2 vector (control) cells was determined by the Student's t-test. A significant relationship was indicated when P < .05.
Results
Discovery and Cloning of the HBxAg Upregulated Gene, URG11
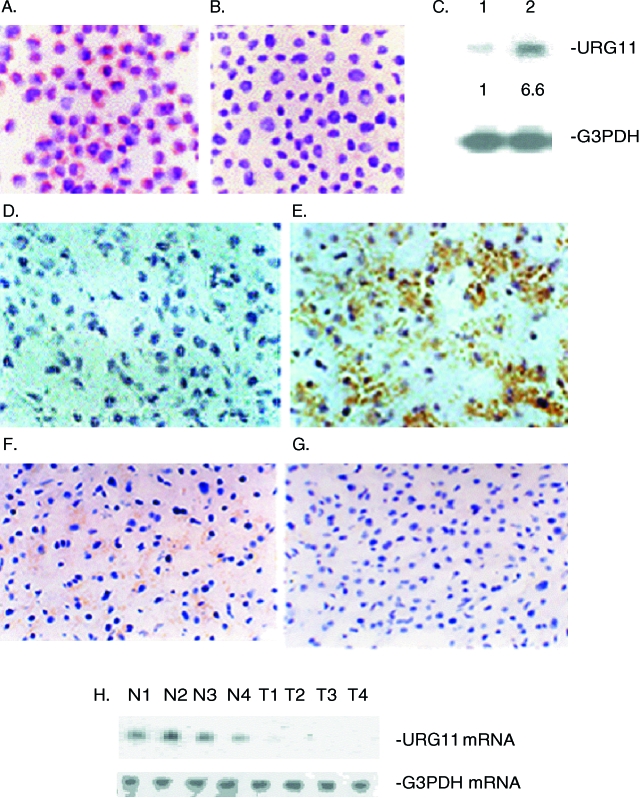
Previous work established HepG2X and HepG2CAT cells, and verified the expression of HBxAg or CAT, respectively, in these cell lines [27]. Whole cell RNA was extracted from each of these cell lines, and subjected to PCR select cDNA subtraction, as described [27,34]. One of the cDNA fragments upregulated in HepG2X compared to HepG2CAT cells was 580 bases long but, on sequencing and GenBank analysis, did not show homology with any known gene. To check whether the RNA corresponding to the 580-bp fragment was differentially expressed in HepG2X compared to HepG2CAT cells, ISH was performed. The results show hybridization in the cytoplasm of HepG2X cells (Figure 1A, red-brown color) but little or no signal in HepG2CAT cells (Figure 1B). Northern blot analysis showed only a single band of about 3 kb in both cell lines (Figure 1C). However, the levels of this RNA were 6.6 ± 0.4-fold higher in HepG2X compared to HepG2CAT cells. Together, these results verify the PCR select cDNA subtraction, and suggest that HBxAg is associated with increased steady state levels of RNA from this cellular gene.
Figure 1.
Expression of URG11 in tissue culture cells and liver tissues was evaluated by ISH. ISH was performed in HepG2X (A) and HepG2CAT (B) cells. (C) Northern blot hybridization was conducted with RNA isolated from HepG2CAT cells (lane 1) or HepG2X cells (lane 2). The relative amounts of URG11 mRNA are indicated below each lane, and are based on normalization with G3PDH mRNA from the same lanes. (D–G) ISH for URG11 was performed in fresh frozen sections from HCC (D), from infected liver surrounding the tumor (E), and from an uninfected liver (F). In panel G, ISH was performed on a consecutive section of tissues from panel E with an irrelevant (SV40 DNA) probe. (H) Northern blot analysis was performed on RNA extracted from the nontumor liver of four patients (N1–N4) and from the corresponding tumor from these same patients (T1–T4). Again, G3PDH in the same RNA samples was used for normalization.
To determine whether this gene is differentially expressed in vivo, ISH was performed on fresh frozen liver and tumor sections from HBV carriers, and in sections from uninfected livers. In HCC tissues from 14 South African patients, ISH signals were observed in nine (64%), whereas 12 of 23 (52%) Chinese patients had detectable signals in tumors (Table 1). The great majority of patients from both groups had faint signals in less than 10% of HCC cells. An example of the ISH results in a tumor from one of these patients, indicated by the brown color in the cytoplasm, is presented in Figure 1D. In contrast, when surrounding nontumor liver tissues were analyzed by ISH in these same carriers, 13 of 14 South African patients (93%) and all 23 Chinese patients (100%) analyzed had readily detectable signals. These signals were observed in more than 30% of the hepatocytes in most of the samples from each group, suggesting that higher and more widespread expression of this cellular gene occurred in nontumor livers compared to HCC. Interestingly, when ISH was performed on liver sections from two uninfected individuals, faint ISH signals were observed in less than 10% of hepatocytes in both cases (Figure 1F). When a consecutive section of nontumor liver tissues from the patient presented in Figure 1E was analyzed by ISH using an irrelevant probe, no signal was detected (Figure 1G), verifying the specificity of hybridization. The relatively strong ISH signal in nontumor compared to tumor tissues was independently verified by Northern blot analysis. In four cases, where large-enough pieces of tumor and nontumor liver tissues were available and intact RNA was obtained, the levels of RNA in nontumor tissues were roughly two- to six-fold higher than in tumors when normalized to the levels of G3PDH mRNA in the same tissue extracts (Figure 1H). Hence, a cDNA fragment identified by PCR select cDNA subtraction in HBxAg-positive compared to HBxAg-negative cell lines identified a unique RNA whose expression is upregulated in the liver of HBV carriers, but not in the tumor tissues from these same patients, nor from the liver sections of two uninfected individuals.
Table 1.
ISH Results for URG11 in Tumor/Nontumor Pairs from HCC Patients.
| South African patients | Uninfected livers | |||||||||||||||
| Case Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1 | 2 |
| T | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | - | - |
| NT | 2 | 1 | 1 | 2 | 3 | 2 | 2 | 2 | 3 | 3 | 3 | 0 | 2 | 3 | 1 | 1 |
| Chinese patients | |||||||||||||||||||||||
| Case Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 |
| T | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| NT | 2 | 2 | 3 | 2 | 3 | 2 | 3 | 3 | 2 | 1 | 3 | 3 | 2 | 2 | 2 | 3 | 1 | 2 | 2 | 1 | 2 | 2 | 3 |
ISH staining is estimated as follows: 0 = no signal; 1 = ISH signal in <10% of cells; 2 = ISH signal in 10% to 25% of cells; 3 = ISH signal in 25% to 50% of cells; and 4 = ISH signal in >50% of cells. T = tumor; NT = nontumor.
Based on these observations, the full-length cDNA clone of URG11 was obtained using the RACE approach. An NCBI blast search of the human genome showed that URG11 cDNA is located within human chromosome 11q11. A Gen-Bank Fasta search showed that URG11 cDNA had 99.5% homology over a 3044-bp overlap with an existing entry (AK056571) and an 87.7% homology in a 673-amino-acid residue overlap within the same clone (Figure 2A). A more thorough analysis of URG11 cDNA showed that it was 3074 bp long, with an open reading frame potentially encoding a protein of 673 amino acids in length (spanning nucleotides 634–2652, inclusive) (Figure 2B). The putative protein product is 70,463.48 Da and contains five von Willebrand factor type-C (VWFC) repeats and one C-type lectin domain, as revealed by GCG motifs analysis (Figure 2B). No other structural features were identified.
Figure 2.
(A) Alignment of amino acid sequences for URG11 (bottom sequence) and AK056571 (top sequence). (B) cDNA sequence and predicted protein structural features of URG11. The five VWFC repeat sequences are indicated in bold italic and span amino acid residues 121 to 157, 256 to 295, 306 to 342, 366 to 401, and 424 to 459. The C-type lectin domain is underlined and spans residues 366 to 387, which partially overlaps with the fourth VWFC repeat. Synthetic peptides used to generate URG11-specific antisera span amino acids 254 to 269 (16 residues, peptide 1), 568 to 591 (24 residues, peptide 2), and 611 to 630 (20 residues, peptide 3) and are indicated by a double underline.
Expression of URG11 and HBxAg in HCC and Surrounding Nontumor Liver Tissues
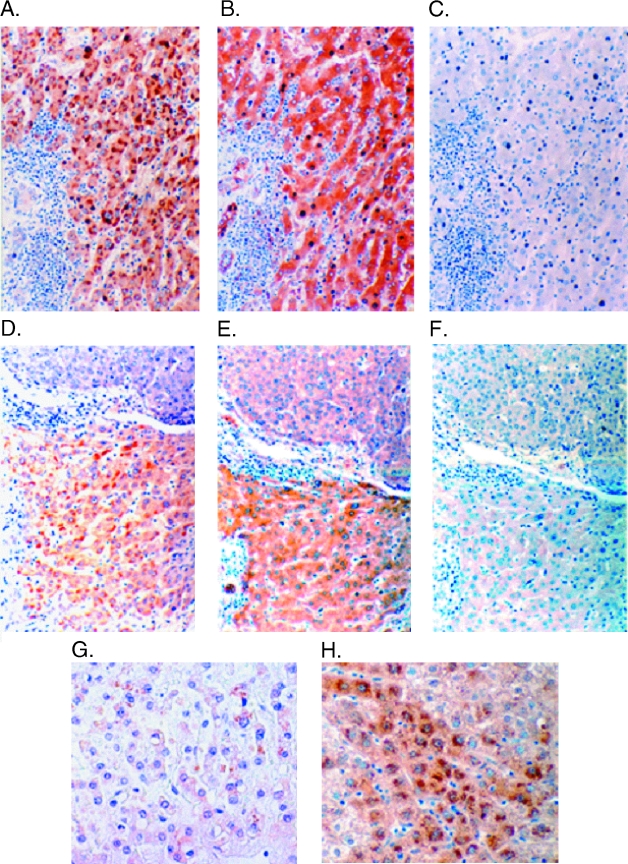
If URG11 is upregulated by HBxAg in natural infection, then there should be considerable costaining in tumor and/or nontumor liver cells. To test this, URG11 antisera were raised in rabbits against three peptides within the URG11 sequence (Figure 2B). Consecutive sections of formalinfixed, paraffin-embedded tissues from the South African and Chinese populations were then stained with anti-URG11, anti-HBx, or preimmune serum. Among South African patients, 10 of 14 (71%) had detectable URG11 protein staining in tumor cells, as did 13 of 23 (57%) Chinese patients (Table 2). When compared to the results of ISH using fresh frozen samples from the same patients, both assays were tightly correlated (P < .001). ISH and immunostaining likewise correlated in nontumor liver tissues from these two groups of patients (Tables 1 and 2) (P < .001), indicating cross-validation and suggesting that the upregulated expression of URG11 was at both the RNA and protein levels. When the protein staining patterns of URG11 and HBxAg were compared in consecutive sections from the same patients, costaining was observed in 6 of 14 (42%) tumor tissues from African patients, and in 8 of 23 (35%) tumor tissues from Chinese patients. These relationships were not statistically significant (P >.2). However, when URG11 and HBxAg staining patterns were compared in surrounding nontumor liver tissues from these patients, costaining was seen in 11 of 14 (79%) nontumor liver tissues from African patients, and in 21 of 23 (91%) nontumor liver tissues from Chinese patients (P < .01). URG11 staining was cytoplasmic Figure 3, A and D) and costaining with HBxAg (Figure 3, B and E) was common in liver samples from Chinese (Figure 3, A and B) and South African (Figure 3, D and E) patients. Staining with preimmune rabbit serum yielded no detectable brown color (Figure 3, C and F). In addition, no brown colorwasobservedwhenstaining wasconducted in the absence of secondary antibody, or when staining was conducted after preincubation of the primary antibodies with the synthetic peptides used for immunization (data not shown), indicating that the staining was specific. URG11 staining was present in the cytoplasm of hepatocytes from uninfected liver, but this staining was often weak and only detectable in 10% to 20% of the cells (Figure 3G). This staining pattern was similar to that observed in HCC cells. In contrast, a higher magnification of an infected liver from an HCC patient with chronic hepatitis showed cytoplasmic URG11 staining in most hepatocytes (Figure 3H), especially in those surrounding tumor nodules. Collectively, these results demonstrate considerable costaining between URG11 and HBxAg in vivo.
Table 2.
Immunohistochemistry for URG11 and HBxAg in Tumor/Nontumor Pairs from HCC Patients.
| South african patients | Uninfected livers | ||||||||||||||||
| Case number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1 | 2 | |
| T | URG11 | 1 | 1 | 0 | 2 | 1 | 0 | 2 | 1 | 2 | 0 | 2 | 0 | 2 | 2 | ||
| HBxAg | 0 | 1 | 0 | 2 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| NT | URG11 | 4 | 3 | 1 | 2 | 4 | 3 | 3 | 2 | 2 | 1 | 3 | 0 | 3 | 4 | 1 | 1 |
| HBxAg | 1 | 2 | 1 | 3 | 3 | 0 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | |
| Chinese patients | ||||||||||||||||||||||||
| Case number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
| T | URG11 | 1 | 1 | 2 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| HBxAg | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |
| NT | URG11 | 3 | 2 | 3 | 1 | 3 | 2 | 2 | 3 | 2 | 1 | 3 | 3 | 2 | 3 | 3 | 2 | 2 | 0 | 3 | 2 | 3 | 3 | 4 |
| HBxAg | 3 | 1 | 1 | 2 | 1 | 2 | 1 | 3 | 1 | 0 | 2 | 1 | 1 | 1 | 2 | 3 | 2 | 0 | 1 | 2 | 1 | 2 | 2 | |
Staining is estimated as follows: 0 = no signal; 1 = IHC signal in <10% of cells; 2 = IHC signal in 10% to 25% of cells; 3 = IHC signal in 25% to 50% of cells; and 4 = IHC signal in >50% of cells.
Figure 3.
Immunohistochemical staining for URG11 (A and D) and HBxAg (B and E) in liver sections from a Chinese (A–C) and a South African (D–F) patient with HCC. Consecutive sections were also stained with preimmune serum in place of primary antibody (C and F) (original magnification, x 100). URG11 staining in uninfected liver (G) and at higher power of infected liver adjacent to tumor (H) (original magnification of G and H, x 200).
Extrahepatic Distribution of URG11
Immunohistochemical staining was conducted in several tumor types other than HCC and in a variety of normal tissues from uninfected patients that were available from archival paraffin blocks. Staining was observed in 10% to 50% of the cells from 13 of 14 patients (93%) who had colon cancer, in 9 of 10 patients (90%) who had gastric cancer, in 6 of 7 patients (86%) with lung cancer, and in 4 of 4 patients (100%) with breast cancer (data not shown). Relatively weak staining was observed in surrounding nontumor cells in 9 of 14 patients (64%) with colon cancer, in 7 of 10 patients (70%) with gastric cancer, in 3 of 4 patients (75%) with lung cancer where nontumor lung tissues were available, and in 3 of 4 patients (75%) with breast cancer (data not shown). Weak or trace amounts of staining were observed in the normal gastrointestinal mucosa, heart, placenta, ovaries, brain, kidneys, prostate, pancreas, and spleen (data not shown). These results show that URG11 expression is upregulated in a variety of tumors compared to peritumor and normal, uninfected tissues, suggesting that it plays a role in the pathogenesis of tumor types, in addition to HCC.
URG11 is Upregulated by HBxAg in HepG2 cells
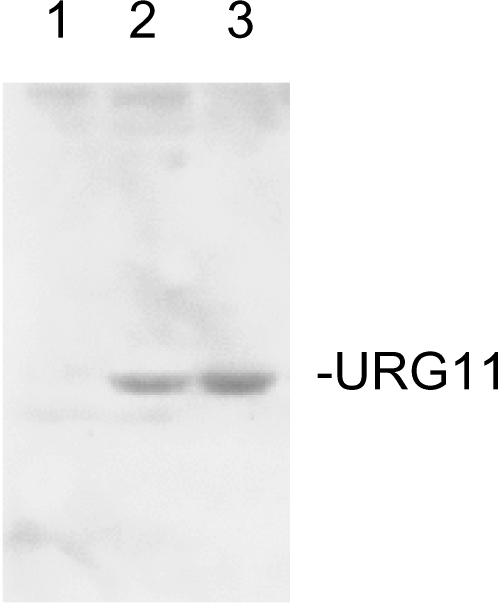
Although the mechanisms whereby elevated URG11 expression in extrahepatic tissues remains to be explored, the higher levels of URG11 in HepG2X compared to HepG2-CAT cells (Figure 1), combined with HBxAg-URG11 costaining in infected liver (Figure 3), suggest that URG11 expression is upregulated by HBxAg. To test this hypothesis,HepG2 cells were transiently transfected with pcDNA3, pcDNA3-HBx, or pcDNA3-URG11. The results (Figure 4) show that the introduction of HBxAg stimulates the expression levels of URG11 protein (lane 2), whereas pcDNA3 does not (lane 1). The band in lane 2 has both the size and immunoreactivity of URG11 in lysates from HepG2 cells transiently transfected with pcDNA3-URG11. These findings are consistent with the hypothesis that URG11 is an effector of HBxAg.
Figure 4.
Western blot analysis of HepG2 cells transiently transfected with pcDNA3 (negative control, lane 1), pcDNA3-HBx (lane 2), or pcDNA3-URG11 (positive control, lane 3).
Relationship Between HBxAg Trans-Activation and Upregulated Expression of URG11
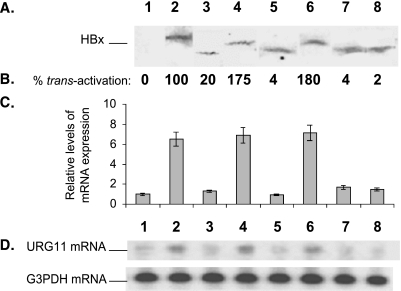
To address whether HBxAg trans-activation is responsible for the upregulated expression of URG11, HepG2 cells were stably transfected with pcDNA3, pcDNA3-HBx, or individual partial deletion mutants of HBxAg. Although cells were selected in G418, assays were performed on cultures that were passaged instead of on colonies chosen from each culture. Initially, expressions of wild type and mutant HBxAg polypeptides were verified by Western blotting in lysates prepared from each of the stably transfected cell lines (Figure 5A). The trans-activation function of each of these mutants was then assayed by transient transfection of pGL2-HIV-1-LTR [27]. The results verify that large deletions within the HBx gene result in loss of trans-activation activity (Figure 5B). When the levels of endogenous URG11 mRNA were assayed in each culture by Northern blot analysis, there was a close correlation between HBxAg trans-activation activity (Figure 5B) and elevated expression of URG11 mRNA (Figure 5C). The Northern blot of URG11 mRNA from one of three experiments is presented in Figure 5D. These results suggest that HBxAg transcriptionally trans-activates URG11.
Figure 5.
Relationship between HBxAg trans-activation and upregulated expression of URG11 mRNA. Separate cultures of HepG2 cells were stably transfected with pcDNA3, pcDNA3-HBx (HBx1–145), or one of the partially deleted mutants. (A) Western blot analysis of lysates prepared from HepG2 cells stably transfected with pcDNA3 (lane 1), with full-length HBx (HBx1–145) (lane 2), HBx41–145 (lane 3), HBx11–145 (lane 4), HBx1–105 (lane 5), HBx1–135 (lane 6), XML (lane 7), or XMR (lane 8). (B) Corresponding trans-activation activities of the various HBxAg polypeptides in HepG2 cells transiently transfected with pGL2-HIV-1-LTR. (C) Average of Northern blot analysis of endogenous URG11 mRNA in three experiments (done in duplicate) from each of the HBxAg-expressing cell lines in (A). (D) Actual Northern blot data of URG11 mRNA from one experiment. The numbered lanes in (A) correspond to the same numbered and aligned lanes in the subsequent panels.
URG11 Stimulates Cell Growth in Tissue Culture
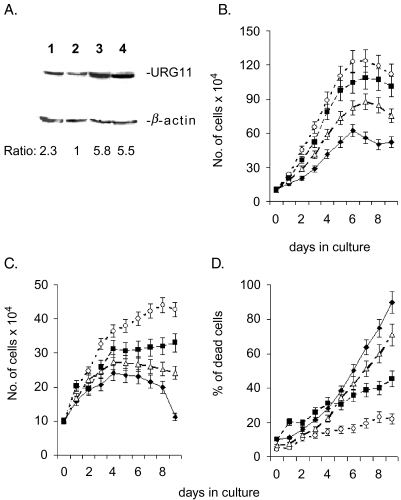
To study the effects of URG11 on cell growth, HepG2 cells were stably transfected with pcDNA3-URG11 and the characteristics of these URG11-over-expressing cells were compared to those of HepG2X, HepG2CAT, and HepG2.2.15 cells, the latter of which stably replicates HBV [39]. Preliminary experiments were carried out to assess the levels of URG11expression in these cell lines by Western blotting. The results show the presence of a single band reactive with URG11 antisera at the expected value of approximately 68 kDa (Figure 6A). Compared to HepG2 vector-transfected cells (lane 2), the levels ofURG11protein were approximately 2.3-fold higher in HepG2.2.15 cells (lane 1), 5.8-fold higher in HepG2X cells (lane 3), and 5.5-fold higher in HepG2-URG11-over-expressing cells (lane 4). These results not only verify URG11 expression, but also show that when HBxAg is expressed in the context of viral replication (as in HepG2.2.15 cells), URG11 expression is also elevated. When the growth curves of these cell lines were compared in medium containing 10% FCS, the curves for HepG2X and HepG2-pcDNA3-URG11 cells were significantly higher than control cells (P < .01 on days 4–8), whereas the growth curve for Hep2.2.15 cells was between that of HepG2-pcDNA3-URG11 and control cultures (Figure 6B). The same trends were observed when cells were grown in serum-free medium (Figure 6C). When the number of dead cells (attached + floating) for each culture was determined, there were significantly fewer cultures of HepG2X and HepG2-pcDNA3-URG11 cells compared to controls (P < .03 on days 6–8) (Figure 6D). Similar results were obtained using trypan blue (Figure 6D) and MTT assays (data not shown). Hence, the protection against serum-induced apoptosis documented for HBxAg [40] also seems to be a property of URG11.
Figure 6.
(A) Western blot of URG11 in HepG2.2.15 (lane 1), HepG2-pcDNA3 (lane 2), HepG2-pcDNA3-HBx (lane 3), and HepG2-pcDNA3-URG11 (lane 4) using anti-URG11 as the primary antibody. Equal amounts of cell lysate (25 µg) were loaded onto each lane. The numbers below the lanes are the relative amounts of URG11 in the Western blot based on gel scanning and corrected by comparison with the corresponding β-actin control shown below each sample. (B and C) Growth curves for HepG2-pcDNA3 (◆), HepG2-pcDNA3-HBx (○), HepG2-pcDNA3-URG11 (■), and HepG2.2.15 cells (△) in medium containing 10% serum (B) or 0% serum (C) for the days indicated. (D) Percent of cells in each culture from panel C that were dead at each time point. Adherent cells were stained with trypan blue at the indicated time points. The number of nonadherent (trypan blue [+]) cells was included in the count of dead cells at each time point. Experiments were done in triplicate, and the curves in each case represent the average values from these experiments.
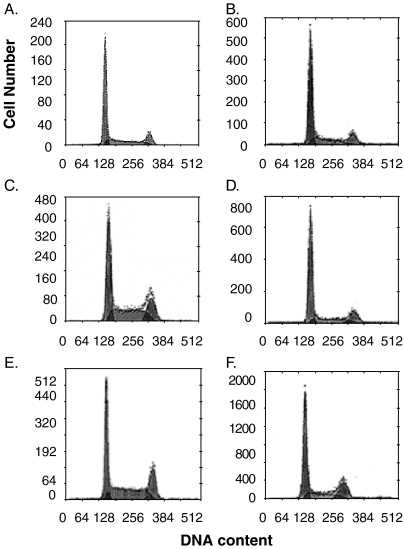
To test whether HBxAg-stimulated cell growth is associated with enhanced cell cycle, HepG2X, HepG2-pcDNA3-URG11, and HepG2 vector control cells were synchronized by serum starvation and then released by the addition of medium containing 10% serum. The results showed that at 24 hours after the release of synchronized cultures, 32.2% of HepG2-URG11 cells were in S-phase compared to 21.4% of HepG2-pcDNA3 cells (P < .01) (Figure 7). Differences were also observed in the fraction of cells in G2-phase, which was 24.2% for HepG2-pcDNA3-URG11 cells compared to 12.4% for HepG2-pcDNA3 cells (P < .005). For HepG2-pcDNA3-HBx cells, 36.6% of the cells were in S-phase at 24 hours after the release from serum starvation, whereas 30.1% were in G2-phase (Figure 7). These differences disappeared by 48 hours after the release from serum starvation. Hence, both URG11 and HBxAg stimulate cell cycle progression significantly more than control cells.
Figure 7.
Flow cytometry of HepG2-pcDNA (panels A and B), HepG2-pcDNA3-URG11 (panels C and D), and HepG2-pcDNA3-HBx (panels E and F) cells at 24 hours (panels A, C, and E) and 48 hours (panels B, D, and F) after synchronization, followed by the addition of 10% FCS. The results shown here illustrate one of the three independent analyses.
URG11 Promotes Growth in Soft Agar and Tumor Formation in SCID Mice
The observations that URG11 stimulated cell growth (Figure 6) and cell cycle progression (Figure 7) suggested that URG11 may contribute to tumor development. To directly test this hypothesis, HepG2-pcDNA3, HepG2-pcDNA3-HBx, and HepG2-pcDNA3-URG11 were seeded into soft agar, and anchorage-independent growth was determined after 3 weeks. HBxAg stimulated growth in soft agar more than five-fold above background, whereas URG11-overexpressing cells stimulated growth about threefold above background (P < .001) (Table 3). To test whether URG11 stimulated tumor formation, the HepG2 lines tested in soft agar were also evaluated for subcutaneous tumor formation in SCID mice. Both HBxAg and URG11 accelerated the onset of tumor and the size of tumors that were recovered 6 weeks after injection (Table 4). Hence, both URG11 and HBxAg stimulate anchorage-independent growth in soft agar and tumor formation in SCID mice.
Table 3.
Growth of URG11-Overexpressing HepG2 Cells in Soft Agar.
| Cell line | Average number of colonies | Student's t-test (P) |
| HepG2-pcDNA3 | 8.2 ± 3.3 | < .001 |
| HepG2-pcDNA3-URG11 | 24 ± 4.3 | |
| HepG2-pcDNA3-HBx | 43.7 ± 7.9 | |
The average number of colonies is from three independent experiments performed in triplicate.
Table 4.
Tumor Growth in HepG2 Cells Over-expressing URG11.
| Cell line | Onset of tumor (days) | Student's t-test (tumor onset) (P) | Average size of tumor (cm3) | Student's t-test (tumor size) (P) |
| HepG2 vector | 41 ± 4 | 0.85 ± 0.3 | ||
| HepG2-HBx | 31 ± 4 | < .03 | 1.5 ± 0.51 | <.02 |
| HepG2-URG11 | 23 ± 3 | < .01 | 1.8 ± 0.55 | <.02 |
The column to the right of “Onset of Tumor” lists the P values calculated from comparisons of HepG2-pcDNA3-HBx or HepG2-pcDNA3-URG11 cells culture to the HepG2 vector controls. The column to the right of “Average Size of Tumor” compares the tumor sizes for each cell line with that of the HepG2 vector cells.
Discussion
There is considerable evidence that sustained high levels of HBxAg expression in the liver are associated with the development of HCC [41]. The finding that HBxAg is a promiscuous trans-activator [16,17] implies that HBxAg may contribute to tumorigenesis by altering the patterns of host gene expression in chronically infected liver. To identify natural effectors of HBxAg, whole cell RNA from HepG2X and HepG2CAT cells were subjected to PCR select cDNA subtraction. One of the genes, URG11, whose expression is upregulated by HBxAg in HepG2 cells (Figure 1, A–C) verifies that the PCR select cDNA subtraction yielded a gene that is differentially expressed. The additional findings that URG11 is strongly expressed in the chronically infected liver by ISH (Figure 1E, Table 1) and by protein staining (Figure 3, Table 2) compared to tumor and uninfected liver tissues (Figures 1 and 3, Tables 1 and 2) strongly suggests that URG11 is upregulated in chronic HBV infection at the RNA and protein levels. The additional finding that HBxAg expression in liver closely resembles that of URG11 (Figure 3, Table 2) strongly suggests that URG11 is a natural effector of HBxAg in vivo. The finding that HBxAg transiently introduced into HepG2 cells stimulates expression of URG11 at the protein level (Figure 4) is consistent with the costaining data in tissues, and further suggests that URG11 is an effector of HBxAg. The observation that upregulated URG11 mRNA levels correlate with HBxAg trans-activation activity (Figure 5) provides at least a partial explanation for how HBxAg may upregulate URG11 in vivo. Hence, HBxAg trans-activation triggers the increased expression of a cellular protein in chronically infected liver that promotes hepatocellular growth and survival, both of which are important for tumor development.
If URG11 is an effector of HBxAg, it should demonstrate some of the properties of HBxAg when over-expressed in liver cells. The observation that URG11 stimulated HepG2 growth in serum-free medium (Figure 6C) suggests that URG11, like HBxAg, promotes serum-independent survival. This appears to be contributed by a URG11-mediated increase in cell cycle progression (Figure 7) and by an decrease in cell death (Figure 6D). These observations imply that URG11 stimulates cell cycle regulatory pathways and/or may be anti-apoptotic. Additional work will be necessary to identify the pathways whereby URG11 alters cell growth and survival and whether it is anti-apoptotic. Interestingly, other HBxAg-upregulated cellular proteins, such as URG7 [34] and URG4 [42], also stimulate cell growth and inhibit cell death. This suggests that during chronic infection, multiple upregulated genes are likely to cooperate in protecting infected hepatocytes from immune-mediated destruction, thereby promoting virus persistence and the carrier state. This cooperation may also promote the growth of infected over uninfected hepatocytes during liver regeneration, which may partially explain the close correlation between HBxAg staining and the intensity of chronic liver disease [6–9].
The observation that URG11 promotes colony formation in soft agar (Table 3) suggests that it plays an important role in tumorigenesis. However, the promotion of colony formation by URG11 is weaker than that of HBxAg, suggesting that other effectors of HBxAg, such as URG4 [42], may also contribute. When URG11-over-expressing cells were evaluated for tumor formation, URG11 accelerated both the onset of tumor and tumor size, as does HBxAg (Table 4). These combined results suggest that URG11 promotes tumor development. Given that HBxAg upregulates URG11 expression in preneoplastic tissues, URG11 likely mediates some of the properties of HBxAg during the early stages of hepatocarcinogenesis. As outlined above, elevated URG11 may directly contribute to the transformed phenotype, but it is also possible that its upregulated expression in peritumor tissues stimulates the release of growth factors that promote tumor growth. Another HBxAg effector, URG4, also accelerates tumor onset and size [42], suggesting that these two proteins cooperate in HBxAg-mediated transformation. Future work will focus on the molecular-based mechanisms whereby these URGs trigger transformation.
The finding of little HBxAg and URG11 in HCC cells, and of little costaining in this compartment (Table 2), implies that once autonomously growing tumors form, the expression of these proteins is no longer rate-limiting. This suggests a fundamental difference in the mechanisms that support hepatocellular growth and survival in preneoplastic and neoplastic tissues. Many of the changes in preneoplastic cells appear to be epigenetic and reversible in nature, whereas many of the changes observed in tumor cells are genetic, the latter of which include loss of heterozygosity and/or gene amplification at many loci. In this context, URG11 is located at chromosome 11q11, and that a gain in chromosome 11q13 is one of the changes documented in moderately to poorly differentiated HCC [43,44]. In addition, it has been proposed that gene amplification at 11q13 may involve the upregulated expression of the bcl-1 oncogene and fibroblast growth factor [44,45]. Interestingly, there is a recent report showing that mice transgenic for fibroblast growth factor develop HCC [46], suggesting that gene amplification in this region may have functional consequences in tumor development. For URG11, it is proposed that the HBxAg-independent expression of URG11 in some tumors may be associated with gene duplication involving URG11. This would not only provide an explanation for why there is little costaining in tumor cells, but would suggest that sustained URG11 expression may also be important after the formation of tumor nodules. The finding of a minority of HCCs with upregulated URG11 expression in the absence of detectable HBxAg (Table 2) is consistent with these ideas. This mechanism may not only be operative in HCCs, but also in many other tumor types.
When the full-length nucleic acid sequence of URG11 was obtained, the deduced polypeptide sequence revealed the presence of five von Willebrand domains and a single C-type lectin domain (Figure 2B). These domains have been observed in many other proteins and may participate in cell/cell recognition and cell/matrix interaction, both of which are important in cell fate decisions that distinguish normal liver cells from tumor cells [47–52]. Moreover, C-type lectin domains have been found in some proteins that appear to regulate cell growth [53]. In this context, it will be of great interest to see, through the mutation or deletion of one or more of these domains, how they contribute to the tumorigenic properties of URG11 in cells over-expressing mutated URG11 polypeptides.
Although HBxAg stimulates expression of URG11 and other cellular genes, the responsible mechanisms have not been elucidated. In this context, it has recently been shown that HBxAg triggers the release of intracellular calcium from the endoplasmic reticulum and/or mitochondria, which in turn activates the cytosolic calcium-dependent proline-rich tyrosine kinase-2 (Pyk2) [54]. Pyk2 is an src family kinase that, in turn, can activate ras signaling. Intracellular calcium release is also known to activate other signaling molecules, such as protein kinase C, calmodulin, calcineurin, nitric oxide synthase, and protein kinase A [55]. It will be interesting to see whether any of these pathways contributes to the HBxAg-triggered alterations in host gene expression that contribute to hepatocellular transformation on the molecular level.
Abbreviations
- AP-1
activating protein-1
- ATF-2
activating transcription factor-2
- CAT
chloramphenicol acetyltransferase
- CREB
cAMP-responsive element-binding protein
- ECL
enhanced chemiluminescence
- FACS
fluorescence-activated cell sorter
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
- H&E
hematoxylin and eosin
- HBV
hepatitis B virus
- HBxAg
hepatitis B x antigen
- HCC
hepatocellular carcinoma
- ISH
in situ hybridization
- MAPK
mitogen-activated protein kinase
- MTT
modified tetrazolium salt
- NF-κB
nuclear factor kappa B
- NT
nontumor
- OCT-1
octamer-binding factor-1
- PCR
polymerase chain reaction
- PI3K
phosphatidylinositol-3-kinase
- PVDF
porous poly(vinylidene fluoride) membrane RACE
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBP
TATA-binding protein
- T
tumor
- URG11
upregulated gene, clone 11
- SCID
severe combined immunodeficient
Footnotes
This work was supported by NIH grants CA48656 and CA66971 to M.F.
Current address: Department of Pathology, Pamukkale University School of Medicine, Kinikli-Denizli, Turkey.
References
- 1.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 2.Beasley RP, Hwang LY. Epidemiology of hepatocellular carcinoma. In: GN Vyas, JL Dienstag, JH Hoofnagle., editors. Viral Hepatitis and Liver Disease. New York: Grune and Stratton; 1984. pp. 209–224. [Google Scholar]
- 3.Szmuness W. Hepatocellular carcinoma and hepatitis B virus: evidence for a causal association. Prog Med Virol. 1978;24:40–69. [PubMed] [Google Scholar]
- 4.Paterlini P, Poussin K, Kew M, Franco D, Brechot C. Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology. 1995;21:313–321. [PubMed] [Google Scholar]
- 5.Diamantis ID, McGandy CE, Chen T-J, Liaw Y-F, Gudat F, Bianchi L. Hepatitis B x gene expression in hepatocellular carcinoma. J Hepatol. 1992;15:400–403. doi: 10.1016/0168-8278(92)90077-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, London WT, Lega L, Feitelson MA. HBxAg in the liver from carrier patients with chronic hepatitis and cirrhosis. Hepatology. 1991;14:29–37. doi: 10.1002/hep.1840140106. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, London WT, Feitelson MA. Hepatitis B x antigen in hepatitis B virus carrier patients with liver cancer. Cancer Res. 1991;51:4971–4977. [PubMed] [Google Scholar]
- 8.Feitelson MA, Lega L, Duan LX, Clayton M. Characteristics of woodchuck hepatitis X antigen in the livers and sera from chronically infected animals. J Hepatol. 1993;17(Suppl 3):S24–S34. doi: 10.1016/s0168-8278(05)80420-9. [DOI] [PubMed] [Google Scholar]
- 9.Jin YM, Yun C, Park C, Wang HJ, Cho H. Expression of hepatitis B virus X protein is closely correlated with the high periportal inflammatory activity of liver diseases. J Viral Hepatitis. 2001;8:322–330. doi: 10.1046/j.1365-2893.2001.00308.x. [DOI] [PubMed] [Google Scholar]
- 10.Koike K, Shirakata Y, Yaginuma K, Arii M, Takada S, Nakamura I, Hayashi Y, Kawada M, Kobayashi M. Oncogenic potential of HBV. Mol Biol Med. 1989;6:151–160. [PubMed] [Google Scholar]
- 11.Hohne M, Schaefer S, Seifer M, Feitelson MA, Paul D, Gerlich WG. Malignant transformation of immortalized hepatocytes by HBV DNA. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifer M, Hohne M, Schaefer S, Gerlich WH. In vitro tumor-igenicity of hepatitis B virus DNA and HBx protein. J Hepatol. 1991;13(Suppl 4):61–65. doi: 10.1016/0168-8278(91)90026-8. [DOI] [PubMed] [Google Scholar]
- 13.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of HBV induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 14.Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, Kurokawa K. High level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810–819. [PubMed] [Google Scholar]
- 15.Ueda H, Ullrich SJ, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 16.Henkler F, Koshy R. Hepatitis B virus transcriptional activators: mechanisms and possible role in oncogenesis. J Viral Hepatitis. 1996;3:109–121. doi: 10.1111/j.1365-2893.1996.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 17.Rossner MT. Hepatitis B virus X-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 18.Autunovic J, Lemieux N, Cromlish JA. The 17 kDa HBx protein encoded by hepatitis B virus interacts with the activation domains of Oct-1, and functions as a coactivator in the activation and repression of a human U6 promoter. Cell Mol Biol Res. 1993;39:463–482. [PubMed] [Google Scholar]
- 19.Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;52:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 20.Qadri K, Maguire HF, Siddiqui A. Hepatitis B virus trans-activator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheong JH, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:142–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XW, Forrester K, Yeh H, Feitelson MA, Gu J, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity and association with ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun BS, Zhu X, Clayton MM, Pan J, Feitelson MA. Identification and preliminary characterization of a protein involved in cellular senescence which binds to hepatitis B virus X antigen. Hepatology. 1988;27:228–239. doi: 10.1002/hep.510270135. [DOI] [PubMed] [Google Scholar]
- 26.Feitelson MA, Reis H, Pan J, Lian Z, Fang J, Liu J, Zhu X, Zhu M, Sun B. Abrogation of negative growth regulatory pathways by hepatitis B virus encoded X antigen in the development of hepatocellular carcinoma. In: WE Fleig., editor. Normal and Malignant Liver Cell Growth: FALK Workshop. Lancaster, UK: Kluwer Academic Publishing; 1999. pp. 156–170. [Google Scholar]
- 27.Lian Z, Pan J, Liu J, Zhu M, Arbuthnot P, Kew MC, Feitelson MA. The translation initiation factor, SUI1, may be a target of Hepatitis B x antigen in hepatocarcinogenesis. Oncogene. 1999;18:1677–1687. doi: 10.1038/sj.onc.1202470. [DOI] [PubMed] [Google Scholar]
- 28.Lucito R, Schneider RJ. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chirillo P, Falco M, Puri PL, Artini M, Balsano C, Levrero M, Natoli G. Hepatitis B virus pX activates NF-κB dependent transcription through a raf-independent pathway. J Virol. 1996;70:641–666. doi: 10.1128/jvi.70.1.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto E, Mitchell PJ, Yen TSB. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature. 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- 31.Benn J, Schneider RJ. Hepatitis B virus HBx protein activates ras-GTP complex formation and establishes a ras, raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nijhara R, Jana SS, Goswami SK, Rana A, Majumdar SS, Kumar V, Sarkar DP. Sustained activation of mitogen-activated protein kinases and activator protein 1 by the hepatitis B virus X protein in mouse hepatocytes in vivo. J Virol. 2001;75:10348–10358. doi: 10.1128/JVI.75.21.10348-10358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih WL, Kuo ML, Chuang SE, Cheng AL, Doong SL. Hepatitis B virus X protein inhibits transforming growth factor-beta-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J Biol Chem. 2000;275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- 34.Lian Z, Liu J, Pan J, Tufan NLS, Zhu M, Arbuthnot P, Kew M, Clayton MM, Feitelson MA. A cellular gene up-regulated by hepatitis B virus encoded X antigen promotes hepatocellular growth and survival. Hepatology. 2001;34:146–157. doi: 10.1053/jhep.2001.25545. [DOI] [PubMed] [Google Scholar]
- 35.Benson DA, Boguski MS, Lopman DJ, Ostell J. GenBank. Nucleic Acids Res. 1997;25:1–6. doi: 10.1093/nar/25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feitelson MA, Millman I, Duncan GD, Blumberg BS. Presence of antibodies to the polymerase gene product(s) of hepatitis B and woodchuck hepatitis virus in natural and experimental infections. J Med Virol. 1988;24:121–136. doi: 10.1002/jmv.1890240202. [DOI] [PubMed] [Google Scholar]
- 37.Feitelson MA, Clayton MM. X antigen polypeptides in the sera of hepatitis B virus infected patients. Virology. 1990;177:367–371. doi: 10.1016/0042-6822(90)90492-a. [DOI] [PubMed] [Google Scholar]
- 38.Feitelson MA, Clayton MM, Blumberg BS. X antigen/antibody markers in hepadnavirus infection: presence and significance of hepadnavirus X gene product(s) in serum. Gastroenterology. 1990;98:1071–1078. doi: 10.1016/0016-5085(90)90035-y. [DOI] [PubMed] [Google Scholar]
- 39.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottlob K, Fulco M, Levrero M, Greessmann A. The hepatitis B virus HBx protein inhibits caspase 3 activity. J Biol Chem. 1998;273:33347–33353. doi: 10.1074/jbc.273.50.33347. [DOI] [PubMed] [Google Scholar]
- 41.Feitelson MA, Duan XL. Hepatitis B virus x antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- 42.Tufan NLS, Lian Z, Liu J, Pan J, Arbuthnot P, Kew M, Clayton MM, Feitelson MA. Hepatitis B x antigen stimulates expression of a novel cellular gene, URG4, that promotes hepatocellular growth and survival. Neoplasia. 2002;4:355–368. doi: 10.1038/sj.neo.7900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858–1862. doi: 10.1002/hep.510290636. [DOI] [PubMed] [Google Scholar]
- 44.Feitelson MA, Sun B, Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 45.Knuutila S, Aalto Y, Autio K, Bjorkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. DNA copy number losses in human neoplasms. Am J Pathol. 1999;155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholes K, Guillet S, Tomlinson E, Hillan K, Wright B, Frantz GD, Pham TA, Dillard-Telm L, Tsai SP, Stephan JP, Stinson J, Stewart T, French DM. A mouse model of hepatocellular carcinoma. Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am J Pathol. 2002;160:2295–2307. doi: 10.1016/S0002-9440(10)61177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishiu J, Tanaka T, Nakamura Y. Identification of a novel gene (ECM2) encoding a putative extracellular matrix protein expressed predominantly in adipose and female-specific tissues and its chromosomal localization to 9q22. Genomics. 1998;52:378–381. doi: 10.1006/geno.1998.5455. [DOI] [PubMed] [Google Scholar]
- 48.Drickamer K. C-type lectin-like domains. Curr Opin Struct Biol. 1999;9:585–590. doi: 10.1016/s0959-440x(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 49.Tomasetto C, Masson R, Linares JL, Wendling C, Lefebvre O, Chenard MP, Rio MC. pS2/TFF1 interacts directly with the VWFC cysteine-rich domains of mucins. Gastroenterology. 2000;118:70–80. doi: 10.1016/s0016-5085(00)70415-x. [DOI] [PubMed] [Google Scholar]
- 50.Christian S, Ahorn H, Koehler A, Eisenhaber F, Rodi HP, Garin-chesa P, Park JE, Rettig WJ, Lenter MC. Molecular cloning and characterization of endosialin, a C-type lectin-like cell surface receptor of tumor endothelium. J Biol Chem. 2001;276:7408–7414. doi: 10.1074/jbc.M009604200. [DOI] [PubMed] [Google Scholar]
- 51.Llera AS, Viedma F, Sanchez-Madrid F, Tormo J. Crystal structure of the C-type lectin-like domain from the human hematopoietic cell receptor CD69. J Biol Chem. 2001;276:7312–7319. doi: 10.1074/jbc.M008573200. [DOI] [PubMed] [Google Scholar]
- 52.Olin AI, Morgelin M, Sasaki T, Timple R, Heinegard D, Aspberg A. The proteoglycans aggrecan and versican from networks with fibulin-2 through their lectin domain binding. J Biol Chem. 2001;276:1253–1261. doi: 10.1074/jbc.M006783200. [DOI] [PubMed] [Google Scholar]
- 53.Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 54.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 55.Nassal M. Ca2 +: the clue to hepatitis B virus X protein function? Hepatology. 2002;36:755–757. doi: 10.1002/hep.510360329. [DOI] [PubMed] [Google Scholar]