Abstract
Despite remarkable advances in our understanding of a genetic basis of cancer, the precise molecular definition of the phenotypically relevant genetic features associated with human epithelial malignancies remains a significant and highly relevant challenge. Here we performed a systematic analysis of the chromosomal positions of cancer-associated transcripts for prostate, breast, ovarian, and colon tumors, and identified short segments of human chromosomes that appear to represent a common target for transcriptional activation in major epithelial malignancies in human. These cancer-associated transcriptomeres correspond well to the regions of transient transcriptional activity on chromosomes 1q21-q23 (144–160 Mbp), 12q13 (52–63 Mbp), 17q21 (38–50 Mbp), 17q23-q25 (72–82 Mbp), 19p13 (1–16 Mbp), and Xq28 (132–142 Mbp) during human cell cycle, suggesting a common epigenetic mechanism of transcriptional activation. Consistent with this idea, two of these transcriptomeres (12q13 and 17q21) seemed to be related to the p53-regulated transcriptional clusters, and some of the cancer-associated transcriptomeres appeared to correspond well to the recently identified regions of increased gene expression on human chromosomes.
Keywords: epithelial cancers, transcriptional activation, chromosomal domains, expression profiling; microarray
Introduction
During malignant progression, genomic instability leads to continuously emerging phenotypic diversity, clonal evolution, and clonal selection, resulting in the remarkable cellular heterogeneity of tumors. The phenotypic diversity of cancer cells is associated with significant mutation-driven changes in gene expression, although not all mutations and differences in gene expression are crucial to the malignant phenotype. Important goals are to identify mutations and gene expression changes that are highly relevant and characteristic of malignant phenotypes and progression pathways, more than one of which may exist [1]. At least some of the phenotypically relevant changes in the mRNA abundance levels characteristic of malignancy are mutationdriven and associated with the recurrent genetic alterations. Recent parallel comparisons of the alterations in DNA copy number and gene expression in human breast cancer cell lines [2,3] revealed that most differentially expressed genes were not amplified or deleted, nor did all regions of DNA amplifications or deletions cause gene expression changes. However, both groups reported that several genes highly overexpressed in the multiple human breast cancer cell lines were involved in recurrent DNA amplifications, suggesting that these genes are more likely to represent important mediators of the breast cancer progression. Collectively, these data support the idea that the systematic analysis of the recurrent transcriptional aberrations in cancer may be useful in the identification of the recurrent phenotypically relevant genomic changes.
Completion of the draft sequence of the human genome allowed identification of the chromosomal positions of human genes with unprecedented accuracy. Integration of these mapping data with genome-wide messenger RNA expression profiles as provided by serial analysis of gene expression for 12 tissue types resulted in generation of the human transcriptome map [4]. The map reveals an apparently nonrandom pattern of chromosomal distribution of transcriptionally active regions along human chromosomes reflected in a clustering of highly expressed genes to specific chromosomal segments. Therefore, we thought to take advantage of these novel analytical tools and determine whether a systematic analysis of the chromosomal positions of the cancer-associated genes with increased mRNA abundance levels would reveal recurrent chromosomal regions of transcriptional activations characteristic of cancer. Global gene expression monitoring in cancer cell lines and clinical tumor samples showed that genes with increased mRNA abundance levels exhibited largely nonoverlapping cancer type-specific patterns of expression [5–8], consistent with the concept of multiple independent pathways of tumor progression [1]. In this paper, we performed an analysis of chromosomal positions of cancer-associated transcripts identified in multiple independent data sets, including oligonucleotide microarray data generated by the Affymetrix gene expression profiling of human prostate cancer cell lines (this study) and previously published microarray data of clinical samples [5–8]. Surprisingly, analysis of the chromosomal positions of the cancer-associated genes revealed several recurrent malignancy-associated regions of transcriptional activation (MARTAs) common for human prostate, breast, ovarian, and colon cancers.
Materials and Methods
Cell Culture
Cell lines used in this study are described in Table 1. The PC3-derived and LNCaP-derived cell lines were developed by consecutive serial orthotopic implantation, either from metastases to the lymph node (for the LN series), or reimplanted from the prostate (Pro series). This procedure generated cell variants with differing tumorigenicity, frequency, and latency of regional lymph node metastasis [9]. The LNCaP and PC-3 panels of human prostate carcinoma cell lines of graded metastatic potential were provided by Dr. C. Pettaway (M.D. Anderson Cancer Center, Houston, TX) and described earlier [9]. A third progression model is represented by the P69 cell line, an SV40 large T-antigen-immortalized prostate epithelial line, and M12, a metastatic derivative of P69 [10–12]. The P69 and M12 cell lines [11–13] were obtained from Dr. S. Plymate and Dr. J. Ware. Two primary human prostate epithelial and one primary human prostate stromal cell line were obtained from Clonetics/BioWhittaker (San Diego, CA ) and grown in complete prostate epithelial and stromal growth medium provided by the supplier. Except where noted, other cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum and gentamicin (Gibco BRL, Gaithersburg, MD) to 70% to 80% confluence and subjected to serum starvation as described [13,14], or maintained in fresh complete media, supplemented with 10% FBS.
Table 1.
Divergent Evolution During Experimentally Extended Tumor Progression In Vivo in Nude Mice of Human Prostate Carcinoma Cell Lines Derived from Androgen-Dependent (LNCaP) and Androgen-Independent (PC3) Lineages (See Text for Details and References).
| Cell Lines | Cycles of Progression | Site of Transplantation/Recovery | Orthotopic Tumorigenicity | Metastatic Potential | RNA Sources Used in This Study |
| Normal epithelia | 0 | None | None | None | In vitro; triplicate samples |
| PC3 | 0 | None | High | Intermediate | |
| PC3M | 1 | Prostate/liver | High | High | |
| PC3M-LN4 | 4 | Prostate/lymph nodes | High | Very high | In vitro; duplicate samples |
| PC3M-Pro4 | 4 | Prostate/prostate | High | Intermediate | |
| LNCaP | 0 | None | Intermediate | Low | |
| LNCaP-LN3 | 3 | Prostate/lymph nodes | High | High | In vitro; duplicate samples |
| LNCaP-Pro5 | 5 | Prostate/prostate | High | Low | |
| P69 | 0 | None | Very low | None | |
| M12 | 3 | Subcutaneous/prostate | High | High | |
RNA from all conditions was prepared twice from independent experiments to assure reproducibility.
RNA Extraction
For gene expression analysis, cells were harvested in lysis buffer 2 hours after the last media change at 70% to 80% confluence, and total RNA or mRNA was extracted using the RNeasy (Qiagen, Chatsworth, CA) or FastTract kits (Invitrogen, Carlsbad, CA). Cell lines were not split more than five times, except where noted.
Affymetrix Arrays
The protocol for mRNA quality control and gene expression analysis was that recommended by Affymetrix (http://www.affymetrix.com). In brief, approximately 1 µg of mRNA was reverse-transcribed with an oligo(dT) primer that has a T7 RNA polymerase promoter at the 5′-end. Second strand synthesis was followed by cRNA production incorporating a biotinylated base. Hybridization to Affymetrix Hu6800 arrays representing 7129 transcripts overnight for 16 hours was followed by washing and labeling using a fluorescently labeled antibody. The arrays were read and data were processed using Affymetrix equipment and software [16]. Detailed protocols for data analysis and documentation of the sensitivity, reproducibility, and other aspects of the quantitative microarray analysis using Affymetrix technology have been reported [16]. To determine the quantitative difference in the mRNA abundance levels between two samples, in each individual sample for each gene, the average expression differences were calculated from intensity measurements of perfect match (PM) probes minus corresponding control probes representing single nucleotide mismatch (MM) oligonucleotides for each gene-specific set of 20 PM/MM pairs of oligonucleotides, after discarding the maximum, minimum, and any outliers beyond 3SD. The averages of pairwise comparisons for each individual gene were made between the samples and the corresponding expression difference calls (see below) were made with Affymetrix software. Microsoft Access was used for other aspects of data management and storage. For each gene, a matrix-based decision concerning the difference in the mRNA abundance level between two samples was made by the software and reported as a “difference call” [No change (NC), Increase (I), Decrease (D), Marginal increase (MI), and Marginal decrease (MD)], and the corresponding fold change ratio was calculated. The results of 7 array experiments are presented in this paper. Forty to 50% of the surveyed genes were called present by the Affymetrix software in these experiments. The concordance analysis of differential gene expression across the data set was performed using Microsoft Access and Affymetrix MicroDB software. Three of the normal prostate epithelial (NPE) microarrays are used as controls and referred to as the NPE expression profile. Thus, when a gene is required to show a two-fold or greater change relative to NPE, this must occur in all three microarrays, for either positive or negative changes. These stringent criteria exclude genes for which one of the three microarrays is in error. The strategy in this study is based on the idea that expression differences will not be called by chance in the same direction in multiple arrays (see Statistical Analysis and Quality Performance Criteria section for statistical justification). Each gene in the final list of the 165 differentially expressed genes was required to be called exclusively as either concordantly upregulated or downregulated in 12 separate comparisons (2 prostate cancer cell lines x 2 experimental serum conditions x 3 NPE controls).
Statistical Analysis and Quality Performance Criteria
We used stringent analytical approach to test the hypothesis that there are common genes with altered mRNA abundance levels, which appear to be significantly associated with the prostate cancer phenotype in PC3/LNCaP model systems of human prostate cancer. The Affymetrix GeneChip gene expression analysis software identifies in any given comparison of two chips only genes that are determined to be expressed at the levels of difference in the expression values determined to be statistically significant (P<.05). These transcripts are called differentially expressed. To be included in our final differentially regulated gene class, the given transcript was required to be determined differentially regulated in the same direction (up or down) at the statistically significant levels (P<.05) in 12 independent comparisons (2 experimental cell linesx2 experimental conditionsx3 control cell lines). Despite that an identified set of 165 upregulated genes has been differentially expressed in described experimental systems with an extremely high level of confidence, we carried out Q-PCR confirmation analysis for a subset of identified genes and confirmed their differential expression in all instances using an additional independent normal human prostate epithelial cell line as a control.
Quality Performance Criteria Adopted for the Affymetrix GeneChip System and Applied in This Study
Forty to 50% of the surveyed genes were called present by the Affymetrix software in these experiments. This is at the high end of the required standard adopted in many peer-reviewed publications using the same experimental system. Transcripts that are called present by the Affymetrix software in any given experiment were determined to have the signal intensities higher in the PM probe sets compared to single-nucleotide MM probe sets and background at the statistically significant level. This analysis was performed for each individual transcript using a unique set of 20 PM probes vs 20 single nucleotide MM probes. In our final list of 165 genes, all transcripts were called present in at least one experimental setting.
The inclusion error associated with two mRNA samples from identical cell lines was 2.7% for a difference called by the Affymetrix software. Thus, two independently obtained mRNA from the same cell lines will have 2.7% false positives. When a third independently derived epithelial cell line was included, only 4 (0.06%) of 7129 genes were called differentially expressed. The expression profiles of the NPE cell lines used in our experiments were determined to be indistinguishable. Therefore, controls are not likely sources of errors in gene expression analysis performed in this study. This is particularly important because the strategy adopted in this study is based on the idea that expression differences will not be called statistically significant by chance in the same direction in multiple arrays and during multiple independent comparisons of different phenotypes and variable experimental conditions. To impose additional stringent restrictions on the possibility of a gene to be detected as concordantly differentially regulated by chance, we apply the use of multiple experimental models and vastly variable experimental settings such as in vitro and in vivo growth and varying growth conditions. A similar strategy for the identification of consistent gene expression changes based on a concordant behavior of the differentially regulated genes using the Affymetrix GeneChip system and software was applied and validated in several peer-reviewed published papers (e.g., see Refs. [17,18]). We applied more stringent criteria in our study, requiring a concordance in at least 12 of 12 experiments compared to six of six comparisons in Ref. [17] and four of six comparisons in Ref. [18]. Ishida et al. [18] provided a formal statistical justification that four or more concordant calls out of six comparisons cannot be explained by chance, with the probability in the range of 10-4.
Calculation of a Clustering Effect
For every member of individual sets of genes with increased mRNA abundance levels, we identified the precise chromosomal position by retrieving the RH mapping data using the LocusLink database (http://www.ncbi.nlm.nih.gov). Within each gene set for all combinations of the three nearest neighbors distributed along the length of the individual chromosomes, we calculated the average distance between three nearest neighboring genes (the experimental clustering distance). A clustering effect analysis was performed for each individual data set (set of transcripts differentially regulated in the prostate cancer cell lines as well as sets of transcripts identified in published papers for corresponding type of human cancer, human cell cycle, p53 response, and dsRNA response). As a first step of this analysis, we identified a precise chromosomal position (in Mb) for every gene listed in individual data sets. We were able to calculate the average distance between three nearest neighbors only for genes with chromosomal position defined in Mb. Typically, the fraction of genes with chromosomal positions defined with this precision constitutes ∼65% to 80% of total genes included in data sets based on a gene expression analysis. To account for random pseudo-clustering effect, we performed a similar analysis for a randomly selected set of 165 genes from the list of 7129 transcripts comprising Affymetrix Hu6800 probe set (the random gene set). To determine the expected random density of gene distribution, we calculated the average distance between three nearest neighbors within a random gene set from a total of 102 individual measurements (the average random clustering distance). The clustering effect in the experimental data set was calculated as a ratio of the average random clustering distance to the individual measurements of the experimental clustering distance within a given class of differentially regulated transcripts. To account for the effect of random chromosomal distribution of transcripts present on the array, we generated two independent random lists of genes derived from genes present on the array. We utilize one random gene list as a control set to generate the expected density of transcript distribution and a second random gene list was used as mock experimental transcript set. The cutoff value for the identification of the transcription activation clusters in the experimental data sets was set to exceed the expected random density of gene distribution by at least 10-fold. A higher ratio due to a shorter experimental clustering distance was interpreted as a more significant clustering effect. The random distribution of the individual clustering distances was obtained by performing a similar analysis for the second random gene set (a total of 105 individual measurements). There were no random pseudo-clusters exceeding the cutoff value that was set for the identification of the transcription activation clusters. The data were plotted for genome-wide visualization of distribution of transcription activation clusters (Figure 2 and Figures 1S-7S, Supplement).
Figure 2.
Genome-wide representation of distribution of transcription activation clusters within a PC3MLN4/LNCaPLN3 consensus class of 165 genes with increased mRNA abundance levels. The clustering effect in the experimental data set was calculated as a ratio of the average random clustering distance to the individual measurements of the experimental clustering distance within a given class of differentially regulated transcripts. Higher ratio due to a shorter experimental clustering distance was interpreted as more significant clustering effect. The cutoff value for identification of the transcription activation clusters was set to exceed the expected random density of gene distribution by at least 10-fold. The random distribution of the individual clustering distances (d) was obtained by performing similar analysis for the random gene set (a total of 105 individual measurements). There were no random pseudo-clusters exceeding the cutoff value that was set for identification of the transcription activation clusters. Note that for more accurate visual comparisons of the clustering effects within experimental and random gene sets, (a) and (b) scaled to the different Y-axis values, and (c) and (d) scaled to the same Y-axis value. A similar analysis of a genome-wide distribution of transcription activation clusters was performed for genes upregulated in ovarian (Figure 1S), breast (Figure 2S), and colon (Figure 3S) cancers as well as genes activated during human cell cycle (Figures 4S and 5S), dsRNA-induced transcripts (Figure 6S), and p53-regulated genes (Figures 7S). These data are presented in the supplement.
Q-PCR Confirmation Analysis of the Differentially Regulated Genes
To confirm the differential regulation of the transcripts comprising a PC3/LNCaP consensus class using an independent method, a sample of 14 genes (12 upregulated and 2 downregulated genes) was tested using Q-PCR (quantitative polymerase chain reaction) on an ABI7900 according to the vendor's recommended protocols (http://www.appliedbiosystems.com/support/tutorials/). This PCR experiment used a further new batch of RNA from a third normal human prostate epithelial cell line and human transcript-specific pairs of PCR primers. In addition, for seven genes (two upregulated, three downregulated, and two controls), we carried out a semiquantitative reverse transcription (RT) PCR confirmation analysis (Figure 1 and data not shown). For confirmation of array results, RNA expression levels were quantified by semiquantitative PCR. An amount of 0.5 Ag of total RNA from NPE cells or prostate tumor cells was reverse-transcribed into cDNA using Superscript II RNase H- Reverse Transcriptase Kit (Invitrogen) according to the manufacturer's instructions. Semiquantitative PCR primer sequences were selected for each cDNA with the aid of commercial software: chromosome 18—oligo 3 (CYB5) forward: 5′ AAA TTA CAC ATT AAG GAA ACA TCA A 3′, reverse: 5V′GAA GAG CCT GCT TTG GAC AC 3′, product size: 216 bp; oligo 4 (maspin) forward: 5′ AGA CATT CTC GCT TCC CT 3′, reverse: 5′ AAT TTT GAC CCC TTA TGG GC 3′, product size: 333 bp; oligo 5 (serpin B3) forward: 5′ CAG ATG TTC TGG TAA ACT GAT TGC 3′, reverse: 5′ AAA GAA ATG TGT GTT TCT AGG TTG C 3′, product size: 330 bp; oligo 8 (serpin B2) forward: 5′ TGCTCT TCT GAA CAA CTT CTG C 3′, reverse: 5′ ATA GAA GGG CAT GGA GGG AT 3′, product size: 339 bp; chromosome X—oligo m12 (mage A12) forward: 5′ GGT GGA AGT GGT CCG CAT CG 3′, reverse: 5′ GCC CTC CAC TGA TCT TTA GCA A 3′, product size: 392 bp; oligo 13 (SYBL1) forward: 5′ GCA ATC CAT GTG ACT CAA G 3′, reverse: 5′ GCA ATG AAT GGT TCA ATC TG 3′, product size: 161 bp; oligo 14 (mage A3) forward: 5′TGA GTC TGA GCA CGA GTT GC 3′, reverse: 5′ TTA AAA GGA ACA TTT GAA CAA CTC C 3′, product size: 224 bp. PCR reactions were performed with HotStarTaq DNA Polymerase Kit (Qiagen, Valencia, CA) according to the manufacturer's instruction. An amount of 1 µl of RT product was amplified by using 1.25 U of polymerase in a final volume of 50 µl containing 1.5 mM MgCl2, 0.2 mM dNTP, and 0.3 µM of each primer. The polymerase was activated by incubation at 95°C for 15 minutes, and the reactions were cycled 30 to 40 times at 95°C for 30 seconds, 56°C for 30 seconds (chromosome 18) or 57°C for 30 seconds (chromosome X), and 72 for 40 seconds, followed by a final extension at 72°C for 7 minutes. PCR products at cycle 30, 35, or 40 were analyzed by electrophoresis through 2% agarose gels containing ethidium bromide.
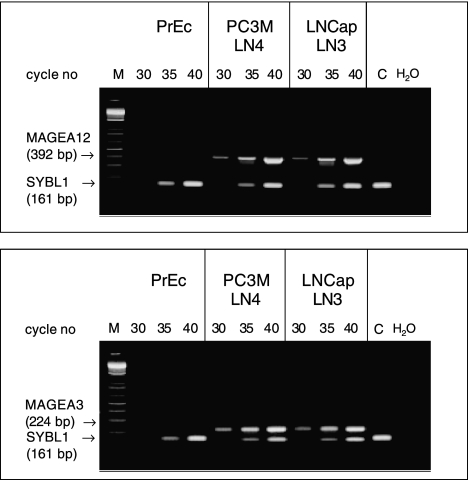
Figure 1.
RT-PCR confirmation analysis of the upregulation of two genes representing Xq28 transcription activation cluster in human prostate carcinoma cell lines [MAGEA12 (top panel) and MAGEA3 transcripts (bottom panel)]. Standard RT-PCR protocol was used to amplify fragments of corresponding genes from mRNA of the normal human prostate epithelial cells (PrEc) and highly metastatic PC3MLN4 and LNCaPLN3 human prostate carcinoma cell lines. To control PCR amplification efficiency and loading, the experiments were carried out using coamplification in the same tube with each experimental gene of a fragment of control gene (SYBL1) that was selected to have a similar chromosomal location but distinct amplification product size and regulation pattern. In the control experiments (C), PCR amplification was carried out only for corresponding control genes. H2O—negative control of PCR amplification; M—molecular weight markers.
Results and Discussion
To define precise chromosomal positions of the genes overexpressed in human prostate, breast, ovarian, and colon cancers, we retrieved the radiation hybrid (RH) mapping data of the individual genes using the LocusLink database (http://www.ncbi.nlm.nih.gov). Our initial analysis was focused on 165 genes overexpressed in vitro in two highly metastatic human prostate carcinoma cell lines PC3MLN4 and LNCaPLN3 compared to normal human prostate epithelial cells (see Table 1S, Supplement for a complete gene list). Genome-wide visualization of the chromosomal positions of the 165 genes of PC3LN4/LNCaPLN3 consensus set appears to indicate a clustering pattern of chromosomal distribution of the UniGene and Gene Bank hits corresponding to these genes (Figure 2a). To test this assumption, we calculated a clustering effect within an experimental gene set compared to a random gene set selected from the list of genes subjected to a gene expression analysis. Interestingly, we found that, in contrast to a random gene set, a significant fraction (∼40%) of the upregulated human prostate cancer-associated genes appears to reside in small continuous chromosomal regions comprising dense transcriptional transcriptional islands of at least three coregulated genes and exceeding the expected random density of gene distribution by at least 10-fold and often >100-fold (Figures 2, b–d and Tables 1S and 9S, Supplement). We propose to call these discrete continuous transcriptional islands of coregulated genes the transcriptomeres. We performed clustering effect analysis for genes upregulated in human tumors from patients with breast [5], ovarian [6], colon [7], and prostate [8] cancers and found that genes with increased transcript abundance levels exhibited a similar clustering pattern of chromosomal distribution (Figures 1S-3S, and Tables 2S-4S, Supplement). Remarkably, when we compared the results of an independent analysis of the chromosomal distribution of the cancer-associated genes identified by the global gene expression monitoring of the human prostate cancer cell lines as well as clinical samples of breast [5], ovarian [6], colon [7], and prostate [8] tumors, we found that there are several shared chromosomal regions that appear to be commonly targeted for transcriptional activation in different types of human cancer (Table 2). It should be pointed out, however, that a majority of the cancerassociated transcriptomeres appear to be nonoverlapping and, thus, cancer type-specific (Figures 1S-3S and Tables 1S-4S, Supplement).
Table 2.
Common MARTAs for Human Prostate, Breast, Ovarian, and Colon Cancers.
| Type of Cancer Cytobands | Prostate Cancer; Cell Lines | Breast Cancer; Cell Lines | Prostate Cancer; Clinical Samples | Breast Cancer; Clinical Samples | Ovarian Cancer; Clinical Samples | Colon Cancer; Clinical Samples | Human Cell Cycle Genes | |||||||
| Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | Gene ID | RH map (kbp) | |
| 12q13 | X78136 | 56,673,798 | TEGT | 50,807,778 | U04810 | 52,354,304 | X74929 | 54,644,765 | HNRPA1 | 57,707,744 | X78136 | 56,673,798 | ||
| X79536 | 57,702,941 | KRT8 | 54,634,170 | KRT7 | 55,931,470 | X12876 | 54,723,961 | M22382 | 59,928,471 | X06256 | 57,824,703 | |||
| M19483 | 60,050,913 | Hs.199067 | 59,463,438 | KRT18 | 54,727,618 | KRT5 | 56,272,873 | Hs.23881 | 55,931,470 | P23 | 60,123,762 | U48707 | 58,009,594 | |
| X94754 | 62,585,109 | ERBB3 | 59,486,794 | Hs.5181 | 59,516,056 | M34309 | 59,463,438 | M22919 | 59,368,893 | |||||
| U37022 | 62,777,307 | Hs.25 | 60,050,913 | X70991 | 60,773,574 | |||||||||
| U41635 | 62,804,248 | D79989 | 62,783,643 | |||||||||||
| 17q21 | U81599 | 47,868,482 | Hs.156346 | 41,032,491 | HSD17B1 | 41,950,889 | GRB7 | 38,774,557 | Y00503 | 39,734,738 | X64330 | 41,335,169 | X55954 | 37,792,038 |
| D13118 | 48,038,113 | D12765 | 43,796,455 | ERBB2 | 38,820,725 | L47276 | 41,032,491 | L38951 | 44,678,610 | X72632 | 39,478,751 | |||
| S85655 | 48,572,139 | KRT13 | 39,757,417 | X17620 | 50,311,072 | X90763 | 39,781,598 | |||||||
| D87989 | 48,869,441 | Hs.69563 | 40,927,738 | L47276 | 41,032,491 | |||||||||
| X17620 | 50,311,072 | Hs.156346 | 41,032,491 | J04088 | 41,032,491 | |||||||||
| Hs.118638 | 50,311,072 | L19527 | 42,423,499 | |||||||||||
| U18018 | 43,796,455 | |||||||||||||
| X82895 | 44,096,205 | |||||||||||||
| 17q23-q25 | X81788 | 75,432,555 | SOX9 | 72,334,148 | Hs.1050907 | 78,231,494 | Z46629 | 72,334,148 | ||||||
| M15205 | 78,231,494 | SYNGR2 | 78,245,850 | ITGB4 | 76,115,990 | J02783 | 81,727,734 | X81788 | 75,432,555 | |||||
| M32304 | 79,217,687 | Hs.79339 | 79,555,927 | P4HB | 81,727,879 | Hs.1578 | 78,438,000 | M77836 | 81,750,633 | D90209 | 76,644,082 | |||
| D21853 | 80,464,033 | PYCR1 | 81,750,633 | M32304 | 79,217,687 | |||||||||
| U30894 | 80,565,151 | M77836 | 81,750,161 | |||||||||||
| M77836 | 81,750,161 | |||||||||||||
| 19p13 | U75370 | 1,186,071 | Hs.24879 | 991,821 | UQCR | 469,828 | M63904 | 2,026,745 | ||||||
| U49070 | 9,818,575 | Hs.168383 | 10,214,410 | ATP5D | 824,197 | Hs.76084 | 7,124,136 | X12492 | 4,003,943 | |||||
| X69819 | 10,320,179 | Hs.110837 | 11,105,937 | Hs.77462 | 10,115,002 | X63692 | 10,115,059 | U40343 | 10,553,783 | |||||
| D50922 | 10,473,454 | J04430 | 13,739,065 | CNN1 | 13,703,253 | X81479 | 12,056,463 | |||||||
| Z50853 | 10,914,862 | Hs.25292 | 14,563,116 | K02765 | 11,821,766 | U20734 | 14,548,010 | |||||||
| U41804 | 12,994,354 | Hs.3107 | 15,936,572 | Hs.78202 | 13,123,023 | X51345 | 14,548,010 | |||||||
| M60459 | 13,538,712 | Hs.118110 | 19,412,795 | Hs.180455 | 14,702,432 | U76764 | 15,936,572 | |||||||
| U07424 | 14,679,005 | U90426 | 15,963,940 | U90426 | 15,963,940 | |||||||||
| U33053 | 15,988,578 | U61263 | 17,076,547 | U33053 | 15,988,578 | |||||||||
| MIC-1 | 20,610,730 | X12794 | 18,865,648 | X79439 | 17,001,239 | |||||||||
| L37033 | 20,786,013 | |||||||||||||
| Xq28 | U36341 | 132,416,389 | Z69043 | 132,523,092 | L22206 | 132,633,367 | ||||||||
| Z69043 | 132,523,092 | X78817 | 132,635,828 | X53416 | 132,965,702 | |||||||||
| X77588 | 132,658,377 | Hs.3109 | 132,635,828 | Hs.182018 | 132,738,957 | X12458 | 133,104,381 | |||||||
| X79353 | 133,054,257 | Hs.18212 | 133,094,859 | |||||||||||
| Xq28 | D83260 | 133,061,242 | Hs.87225 | 133,228,507 | ||||||||||
| X92896 | 133,094,859 | |||||||||||||
| L18920 | 138,297,657 | Hs.36980 | 138,297,657 | |||||||||||
| L18877 | 138,311,892 | Hs.169246 | 138,311,892 | |||||||||||
| U03735 | 138,347,200 | Hs.36978 | 138,347,200 | |||||||||||
| M77481 | 138,702,845 | U47105 | 138,411,489 | |||||||||||
| X92396 | 141,641,489 | U46023 | 140,466,991 | |||||||||||
| M34677 | 142,020,376 | |||||||||||||
The genes with increased mRNA abundance levels in human prostate, breast, ovarian, and colon cancers as well as genes induced during the human cell cycle were identified as described in the legend to Figure 3. RH mapping data for each individual gene were retrieved using the LocusLink database and utilized to identify common malignancy-associated chromosomal regions of transcriptional activations. The genes commonly induced in human cancer and during the human cell cycle are in bold.
Next we attempted to determine whether a gene expression profile characteristic of a physiological but highly relevant to cancer process such as cell cycle would exhibit a similar discrete nonrandom pattern of chromosomal distribution. Using the LocusLink database, we retrieved the RH mapping data of the 378 genes comprising the human cell cycle transcriptome [19]. We found that coordinate transcriptional regulation of gene expression during human cell cycle seems to occur in a nonrandom fashion from discrete continuous chromosomal regions, suggesting an epigenetic regulatory nature of this phenomenon (Figures 4S and 5S and Table 5S, Supplement). Furthermore, several of the common MARTAs appear to be closely related to the human cell cycle-associated transcriptomeres (Table 2 and Figure 3, a and c; Tables 1S-5S, Supplement).
Figure 3.
Profiles of the chromosomal distribution of human breast cancer-associated transcripts (a), dsRNA-induced genes (b), and cell cycle-activated genes (c) residing on chromosome 17. A total of 132 estrogen receptor-negative breast cancer-associated transcripts was obtained from Ref. [5] by combining the lists of genes comprising basal epithelial cell clusters 1 and 2, Erb-B2 overexpression cluster, and a proliferative cluster. A total of 144 ovarian cancer-associated transcripts was derived from Ref. [6] as a sum of the top 100 biomarker genes, proliferative and tumor clusters. The redundant entries were eliminated from the final gene lists. A total of 165 prostate cancer-associated transcripts was identified by comparing gene expression profiles of two human prostate carcinoma cell lines (PC3MLN4 and LNCaPLN3) to the gene expression pattern of cultured normal human prostate epithelial cells using the Affymetrix GeneChip system. A concordant set of 165 genes upregulated in cancer cell lines was generated utilizing the Affymetrix software for pairwise comparisons of duplicate cancer mRNA samples from each cell line versus a triplicate normal mRNA samples derived from two different normal prostate epithelial cell lines. Thus, each differentially expressed gene was required to be called in the same direction in 12 pairwise comparisons. The list of 378 genes comprising the human cell cycle transcriptome was obtained from Ref. [19]. The list of the dsRNA-induced genes was derived from Ref. [20]. RH mapping data were retrieved using the LocusLink database and utilized to generate the chromosome-specific map of gene distribution. One unit value on the Y-axis corresponds to a single gene with a placement resolution of 1 Mb along the length of the chromosome. The complete lists of genes and RH mapping data are presented in the supplement (Tables 1S-8S).
Lastly, we thought to analyze whether a transcriptional response to the activation of certain signaling pathways, such as double-stranded (ds) RNA-triggered signaling [20] or p53-dependent transcription [21], would exhibit a nonrandom pattern of chromosomal distribution. dsRNA is thought to be the primary viral gene product that causes induction of type I interferon synthesis and interferon production by virusinfected cells. Activation of the interferon-inducible gene cluster was consistently found in the clinical cancer samples [5–7]. We analyzed the RH mapping data for genes activated in the type I interferon locus-deficient GRE cells in response to the dsRNA treatment [20]. We found that dsRNA-induced genes are distributed along human chromosomes in a nonrandom fashion with the multiple clusters of transcriptionally activated genes positioned at chromosomes 1, 2, 3, 4, 6, 7, 10, 12, 13, 14, 17, 19, and 20 (Figures 6S and Table 6S, Supplement). Interestingly, one of the dsRNA response clusters on chromosome 17 appears to be closely related to the breast cancer-associated and cell cycle-associated transcriptomeres on chromosome 17 (Figure 3, a and b), suggesting a potential overlap of corresponding transcription activation pathways. Consistent with this hypothesis, several others cancer type-specific transcriptomeres have overlapping chromosomal positions with the dsRNA response clusters (Figures 1S-6S and Tables 1S-6S, Supplement).
The p53-regulated genes [21] seem to exhibit a clustering pattern of chromosomal distribution represented by multiple transcriptional islands at chromosomes 1, 6, 10, 12, 16, 17, 19, and 22 (Figures 7S and Table 7S, Supplement). Two of the common cancer-associated transcriptomeres (12q13, 52–63 Mbp; 17q21, 38–50 Mbp) appear to overlap with the corresponding p53-regulated transcriptional islands. Several others cancer type-specific transcriptomeres demonstrated similar overlapping positional patterns with the p53-regulated genes (Tables 1S-8S, Supplement), suggesting a mechanism of consistent recurrent transcriptional targeting in multiple human cancers of p53-regulated chromosomal regions.
A recently generated human transcriptome map revealed an apparent clustering of highly expressed genes in 12 normal and pathological tissue types to specific chromosomal domains called regions of increased gene expression, RIDGES [4]. As described here, clustering of cancer-associated genes to the discrete regions of chromosomes may be related to the specific RIDGES, implying that selected chromosomal domains of increased gene expression are preferential targets for transcriptional activation in human cancer cells (Figure 4).
Figure 4.
Cancer-associated transcriptomeres located on chromosome 11 correspond well to the region of increased gene expression identified on human chromosome 11 [4]. The experimental protocols are described in the legends to Figures 1–3 and in the Materials and Methods section. The distribution of regions of increased gene expression and gene density along human chromosome 11 are shown in the box and originally described in Ref. [4].
The stated goal of a systematic analysis of chromosomal positions of cancer-associated genes was achieved by performing such analysis for the transcripts that were previously defined as being cancer-associated in published peerreviewed papers [5–8] as well as for a set of 165 upregulated transcripts in xenograft-derived human prostate cancer cell lines (this study). Our results imply that at least some of the transcripts defined previously as tumor-associated may in fact be the bystanders of the enhanced transcriptional readouts reflecting the increased proportion of cycling cells in tumors and/or activation of the p53 response pathway. Our analysis argues that without follow-up experiments, the distinction between so-called cancer-associated and proliferative transcripts is ambiguous at least for some genes (particularly those that are located in the chromosomal regions targeted for transcriptional activation during the cell cycle) and may indeed reflect the relative enrichment of clinical tumor samples with actively proliferating cells. Alternatively, these regions may have been targeted for recurrent transcriptional activation because they harbor important cell cycle control and/or survival genes.
We do not intend to imply that the chromosomal regions are more important than the genes that may be associated with malignancy. We believe that specific chromosomal regions were targeted for transcriptional activation precisely because they harbor the important genes. However, the transcriptional readout from the particular chromosomal region is, in our opinion, less reliable and is a more variable endpoint that could be influenced by many variables such as transcript stability, assay sensitivity, experimental conditions, sample handlings, etc. Identification of different overexpressed transcripts derived from the same chromosomal region in multiple pathological and experimental conditions may indicate that cells maintain the accessibility of the region for direct transcriptional regulation, thus implying its potential significance. Therefore, gene-specific induction can be easily achieved when growth and/or survival requirements are in place. Identification of common chromosomal regions of transcriptional activation would facilitate a detailed and precise gene-by-gene analysis of these regions by employing the most sophisticated state-of-the-art approaches such as high-resolution array-based CGH, Q-PCR-based analysis, promoter methylation survey, and direct sequencing.
Our data do not necessarily imply that the mechanism of transcriptional activation within identified chromosomal regions is exclusively epigenetic. In fact, some of these regions are within the boundaries of well-established cancer-associated amplicons (e.g., 17q21 and 17q23 for breast cancer), suggesting that at least in some cancer cell lines and/or subset of tumors, activation of the transcription in these regions could be associated with DNA amplification. However, during the cell cycle progression of normal cells and in response to the p53 overexpression, the mechanisms of transcriptional activation are most likely epigenetic. One of our main conclusions is that these regions are commonly targeted for transcriptional activation under a wide range of pathological and experimental conditions suggesting their potential relevance. Most likely, transcription activation effect can be achieved by engaging either epigenetic mechanisms (normal cells and some cancer cells) or DNA amplification (cancer cells).
In summary, accumulation of cancer-associated transcripts in the mRNA abundance space seems to occur from the discrete continuous chromosomal regions comprising a set of transcriptional islands of coregulated physically adjacent genes (the transcriptomeres). Most of the cancerassociated transcriptomeres appear to exhibit a cancer type-specific pattern of chromosomal distribution. However, several of the MARTAs exhibited a recurrent overlapping pattern of chromosomal distribution in human prostate, breast, ovarian, and colon cancers, suggesting a mechanism of preferential targeting for transcriptional activation in multiple types of human cancer of the selected chromosomal regions.
Acknowledgements
We thank M. McClelland and J. Welsh for helpful discussions. Supplemental Information accompanies the paper on Neoplasia's website (www.neoplasia.com). Original gene expression profiling data sets are presented elsewhere (Glinsky GV, et al., submitted).
Footnotes
This work was supported, in part, by a grant from the National Cancer Institute (1RO1 CA89827-01) to G.V.G. AKH is a postdoctoral fellow in D. Mercola lab at the Sidney Kimmel Cancer Center and was supported by the grant RO1 CA76173 to D. Mercola and the fellowship from the Deutscher Akademischer Austaushdienst. The results of the RT-PCR analysis (Figure 1) were obtained in collaboration with Dr. D. Mercola lab at the Sidney Kimmel Cancer Center and presented at the 93rd Annual Meeting of the American Association for Cancer Research, April 6 – 10, San Francisco, CA [22].
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Welsh J, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA-copy number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 3.Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi O-P. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- 4.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Stuis P, Hermus M-C, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portrait of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 8.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacologic targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 9.Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- 10.Bae VL, Jackson-Cook CK, Brothman AR, Maygarden SJ, Ware J. Tumorigenicity of SV40 T antigen immortalized human prostate epithelial cells: association with decreased epidermal growth factor receptor (EGFR) expression. Int J Cancer. 1994;58:721–729. doi: 10.1002/ijc.2910580517. [DOI] [PubMed] [Google Scholar]
- 11.Jackson-Cook C, Bae V, Edelman W, Brothman A, Ware J. Cytogenetic characterization of the human prostate cancer cell line P69SV40T and its novel tumorigenic sublines M2182 and M15. Cancer Genet Cytogenet. 1996;87:14–23. doi: 10.1016/0165-4608(95)00232-4. [DOI] [PubMed] [Google Scholar]
- 12.Bae VL, Jackson-Cook CK, Maygarden SJ, Plymate SR, Chen J, Ware JL. Metastatic subline of an SV40 large T antigen immortalized human prostate epithelial cell line. Prostate. 1998;34:275–282. doi: 10.1002/(sici)1097-0045(19980301)34:4<275::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.Glinsky GV, Glinsky VV. Apoptosis and metastasis: a superior resistance of metastatic cancer cells to programmed cell death. Cancer Lett. 1996;101:43–51. doi: 10.1016/0304-3835(96)04112-2. [DOI] [PubMed] [Google Scholar]
- 14.Glinsky GV, Price JE, Glinsky VV, Mossine VV, Kiriakova G, Metcalf JB. Inhibition of human breast cancer metastasis in nude mice by synthetic glycoamines. Cancer Res. 1996;56:5319–5324. [PubMed] [Google Scholar]
- 15.Glinsky GV, Glinsky VV, Ivanova AB, Hueser CJ. Apoptosis and metastasis: increased apoptosis resistance of metastatic cancer cells is associated with the profound deficiency of apoptosis execution mechanisms. Cancer Lett. 1997;115:185–193. doi: 10.1016/s0304-3835(97)04738-1. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays [see comments] Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 17.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 18.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic function as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 20.Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated signaling. J Biol Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 21.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack D, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 22.Glinsky GV, Glinskii AB, McClelland M, Krones-Herzig A, Mercola D, Welsh J. Microarray gene expression analysis of tumor progression in the nude mouse model of human prostate cancer; In Proceedings of the 93rd Annual Meeting of the American Association for Cancer Research, April 6 – 10, San Francisco, CA; 2002. p. 462. Abstract#4480. [Google Scholar]