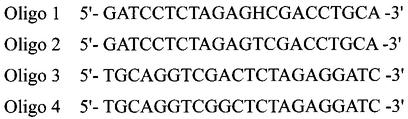Abstract
The oxidation and deamination of 5-methylcytosine (5mC) in DNA generates a base-pair between 5-hydroxymethyluracil (5hmU) and guanine. 5hmU normally forms a base-pair with adenine. Therefore, the conversion of 5mC to 5hmU is a potential pathway for the generation of 5mC to T transitions. Mammalian cells have high levels of activity of 5hmU-DNA glycosylase, which excises 5hmU from DNA. However, glycosylases that similarly excise 5hmU have not been observed in yeast or Escherichia coli. Recently, we found that E.coli MutM, Nei and Nth have DNA glycosylase activity for 5-formyluracil, which is another type of oxidation product of the thymine methyl group. In this study, we examined whether or not E.coli MutM, Nei and Nth have also DNA glycosylase activity that acts on 5hmU in vitro. When incubated with synthetic duplex oligonucleotides containing 5hmU:G or 5hmU:A, purified MutM, Nei and Nth cleaved the 5hmU:G oligonucleotide 58, 5 and 37 times, respectively, more efficiently than the 5hmU:A oligonucleotide. In E.coli, the 5hmU-DNA glycosylase activities of MutM, Nei and Nth may play critical roles in the repair of 5hmU:G mispairs to avoid 5mC to T transitions.
INTRODUCTION
In aerobic organisms, cellular DNA is continuously exposed to reactive oxygen species produced during normal metabolism and by exogenous agents such as ionizing radiation. Reactive oxygen species generate various types of DNA damage (1–3), which may contribute to the loss of genetic stability, altered cellular regulation associated with aging and many diseases, including cancer (3–6). Therefore, the damaged residues must be excised from DNA. Oxidative base modifications are primarily handled by base excision repair in mammalian cells, yeast and Escherichia coli (1,7–9).
When thymine in DNA suffers oxidative damage, several peroxides are formed (2,10). 5-hydroxyperoxymethyluracil spontaneously decomposes to generate two kinds of methyl group-oxidized thymine, 5-formyluracil (5-foU) and 5-hydroxymethyluracil (5hmU) (2,10). Recent studies revealed that 5-foU is potentially mutagenic (11–13), and so several enzymes recognize and remove the damage from DNA: AlkA, MutM, Nei and Nth in E.coli and hNTH1 in human cells (14–17). On the other hand, despite its identification as a major radiation product (18,19), the biological effects of 5hmU remain uncertain. 5hmU in the template directs the incorporation of only A during DNA replication (20,21). 5hmU:A oligonucleotides show the same melting behavior as normal T:A oligonucleotides (21). Furthermore, the substitution of T by 5hmU does not affect digestion by HincII or SalI, whereas 5-foU-containing oligonucleotides are resistant to cleavage by these restriction enzymes (21,22). These facts indicate that the oxidation of the thymine methyl group generates a stable base-pair between 5hmU and A, suggesting that there is no obvious need for the removal of 5hmU from 5hmU:A in DNA. However, using synthetic oligonucleotides containing 5hmU:A, 5hmU-DNA glycosylase activities have been detected in mammalian cells (1,7,19,23,24). However, corresponding enzymatic activities have not been detected in yeast or E.coli (7,25,26).
Recently, Rusmintratip and Sowers (27) found a 5hmU-DNA glycosylase activity that removes 5hmU mispaired with G in human cell extracts. The 5hmU:G excision activity was 60 times stronger than the corresponding 5hmU:A activity (27). The observation of an unexpectedly high 5hmU:G glycosylase activity suggests that human cells may encounter 5hmU:G much more frequently than previously expected. This 5hmU:G would arise from the damage of normal 5mC:G pairs in DNA (27,28). Therefore, it is likely that the oxidation and deamination of 5mC occurs much more frequently in DNA than expected. The findings and suggestions of Cannon-Carlson et al. (28) and Rusmintratip and Sowers (27) led us to examine whether or not E.coli cells have 5hmU-DNA glycosylase activity that removes 5hmU from 5hmU:G in DNA.
Recently, we found that E.coli MutM, Nei and Nth recognize and excise 5-foU from DNA in vitro and in vivo (15,17). Both 5-foU and 5hmU are formed from 5-hydroxyperoxymethyluracil and have a common structure containing an oxidized methyl group (2,10,21). Hence, we first examined with synthetic duplex oligonucleotides containing 5hmU at a defined site whether or not the MutM, Nei and Nth recognize and remove 5hmU from DNA. Here we report that E.coli MutM, Nei and Nth have DNA glycosylase/AP lyase activities that remove 5hmU preferentially from 5hmU:G mispairs in DNA.
MATERIALS AND METHODS
Chemicals and enzymes
Tetracycline hydrochloride, kanamycin, chloramphenicol, phenylmethylsulfonyl fluoride and NaBH4 were purchased from Wako Pure Chemicals (Osaka, Japan). Ampicillin was obtained from Meiji Seika (Tokyo, Japan). Plasmids pGEX-4T-1 and pKK223-3, glutathione–Sepharose 4B, thrombin and prepacked columns for FPLC were purchased from Amersham Pharmacia Biotech. (Uppsala, Sweden). T4 polynucleotide kinase, Taq DNA polymerase and restriction enzymes were obtained from Takara Shuzo (Kyoto, Japan) and TOYOBO (Osaka, Japan). [γ-32P]ATP (>148 TBq/mmol) was obtained from ICN Biomedicals Inc. (Costa Mesa, CA).
Synthesis of 5hmU-containing substrates
Oligonucleotides containing 5hmU at a defined site were synthesized as described previously (21,29). Briefly, 22mer oligonucleotides containing 5-foU (21,29) were incubated with NaBH4 for 10 min at room temperature to convert 5-foU to 5hmU. The resulting oligonucleotides containing 5hmU were purified by HPLC. The purified solution was centrifuged at 30°C to remove CH3CN by evaporation, and the solution was lyophilized to remove the H2O containing ammonium formate by evaporation. The 5hmU-containing oligonucleotide was dissolved in H2O. The presence of 5hmU in the oligonucleotides was verified by HPLC and HPLC/mass spectrometry analyses (29). The nucleotide sequences of oligonucleotides used in this study are shown in Figure 1.
Figure 1.
Nucleotide sequences of oligonucleotides. H represents 5-hydroxymethyluracil.
Oligo 1 and Oligo 2 were labeled at the 5′-end with [γ-32P]ATP by incubation with T4 polynucleotide kinase at 37°C for 30 min and annealed with the phosphorylated Oligo 3 or Oligo 4 strand.
Expression and purification of MutM, Nth and Nei
Escherichia coli MutM and Nei were overproduced and purified from E.coli KSR7 (mutM nth nei) carrying pKK-MutM and pKK-Nei, respectively, as previously described (15,30). Nth was purified from E.coli BL21 carrying pGEX-Nth, as previously described (15,16,30).
DNA trapping assay with purified MutM, Nei and Nth
32P-labeled duplex oligonucleotides (50 fmol) were incubated with various amounts of purified MutM, Nei and Nth in a reaction mixture (15 µl) containing 20 mM HEPES–KOH (pH 7.5), 10 mM dithiothreitol, 1.5 mM MgCl2, 5 mM KCl, 2 mM EDTA, 1% glycerol and 0.5 µg of bovine serum albumin in the presence of 100 mM NaBH4. After incubation at 37°C for 30 min, the reaction was terminated by the addition of 2× sample buffer, and the samples were heated at 95°C for 5 min. Trapped protein–oligonucleotide complexes were separated by 15% SDS–polyacrylamide gel electrophoresis (SDS– PAGE). The gels were dried and autoradiographed using Fuji RX films at –80°C.
DNA cleavage assay with purified MutM, Nei and Nth
32P-labeled duplex oligonucleotides (20 fmol) were incubated with various amounts of purified MutM, Nei and Nth in a reaction mixture (10 µl) containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 50 mM NaCl. The reaction was performed at 37°C for 15 min. After incubation, the reaction was terminated by the addition of stop solution (95% formamide, 0.1% bromophenol blue, 0.1% xylene cyanol and 20 mM EDTA). The samples were then heated at 95°C for 5 min and immediately cooled on ice, and loaded onto 20% polyacrylamide gels in 90 mM Tris–borate (pH 8.3) containing 7 M urea and 2 mM EDTA. After electrophoresis at 1300 V, the gels were dried and autoradiographed using Fuji RX films at –80°C. The intensity of each band was determined by image analysis using Scion Image software.
RESULTS
Formation of trapped complexes between purified MutM, Nei and Nth and duplex oligonucleotides containing 5hmU:A or 5hmU:G
MutM, Nei and Nth were purified from E.coli KSR7 carrying pKK-MutM and pKK-Nei and E.coli BL21 carrying pGEX-Nth, respectively, as described previously (15,16,30). These enzymes catalyze both the cleavage of the glycosylic bond to remove damaged bases and the cleavage of the phosphodiester backbone at the resulting AP site via β- or β- and δ-elimination reactions (1,8,9,30,31). It was of interest to examine whether or not these enzymes have a DNA glycosylase activity for 5hmU in DNA. In this study, we used a trapping assay to identify 5hmU DNA glycosylase/AP lyase activities of MutM, Nth and Nei by using oligonucleotides containing 5hmU at specific sites. The trapping assay is based on the fact that DNA glycosylases with associated AP lyase activity form a Schiff base intermediate that can be trapped by NaBH4 to generate a stable covalent enzyme–DNA complex (8,9,31–33).
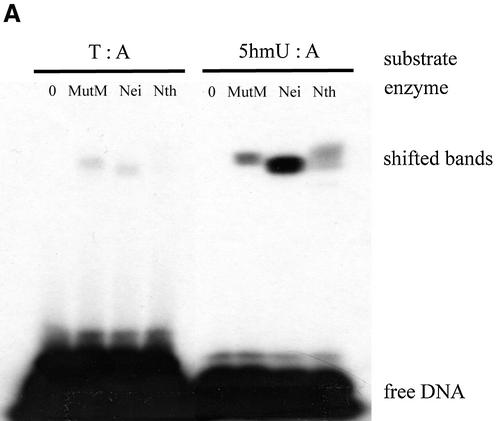
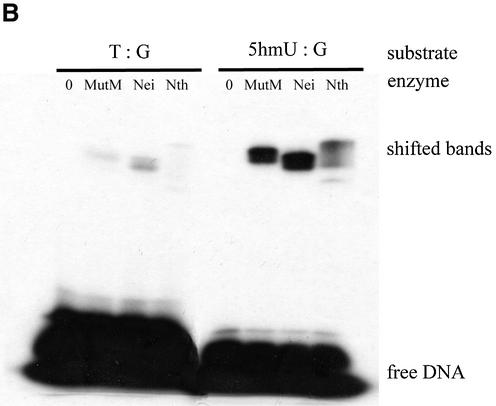
First, purified MutM, Nei and Nth were incubated with duplex oligonucleotides containing a single 5hmU:A, 5hmU:G, T:A or T:G in the presence of 100 mM NaBH4. Trapped DNA–protein complexes were analyzed by SDS–PAGE. Figure 2A and B show that purified MutM, Nei and Nth formed trapped complexes with the 5hmU-containing oligonucleotides.
Figure 2.
Trapping assay for 5hmU-DNA glycosylase activities of purified MutM, Nei and Nth with duplex oligonucleotides. The duplex oligonucleotides (T:A, Oligo 2/Oligo 3; T:G, Oligo 2/Oligo 4; 5hmU:A, Oligo 1/Oligo 3; 5hmU:G, Oligo 1/Oligo 4) (50 fmol) were incubated at 37°C for 30 min without or with purified proteins. The products were separated by electrophoresis on 15% polyacrylamide gels. MutM, 11.7 pmol; Nei, 10.1 pmol; Nth, 9.17 pmol.
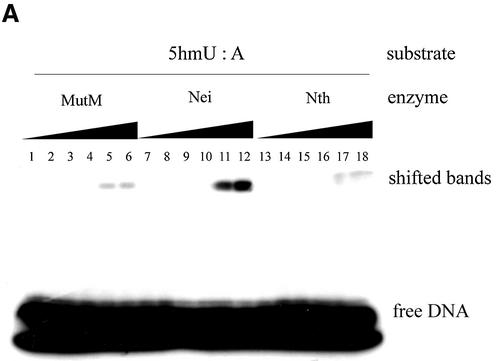
Next, 5hmU-containing oligonucleotides were incubated with various amounts of MutM, Nei and Nth. The results are shown in Figure 3A and B. The formation of trapped complexes increased with the amount of protein. Nei was trapped with 5hmU:A-containing oligonucleotide more efficiently than MutM and Nth. On the other hand, MutM and Nth formed the trapped complexes with 5hmU:G-oligonucleotide to a much greater extent than with 5hmU:A-oligonucleotide (Figs 2A and B, 3A and B).
Figure 3.
Trapping assay of various amounts of purified MutM, Nei and Nth duplex oligonucleotides containing 5hmU:A or 5hm:G mispairs. The 5hmU:A or 5hmU:G base-pair (Oligo 1/Oligo 3 and Oligo 1/Oligo 4, respectively)-containing duplex oligonucleotides (50 fmol) were incubated at 37°C for 30 min without (lanes 1, 7 and 13) or with purified proteins (MutM, lanes 2–6; Nei, lanes 8–12; Nth, lanes 14–18). The products were separated by electrophoresis on 15% polyacrylamide gels. MutM (lane 1, 0 pmol; lane 2, 0.44 pmol; lane 3, 0.88 pmol; lane 4, 1.8 pmol; lane 5, 3.5 pmol; lane 6, 7.0 pmol); Nei (lane 7, 0 pmol; lane 8, 0.42 pmol; lane 9, 0.84 pmol; lane 10, 1.7 pmol; lane 11, 3.4 pmol, lane 12, 6.7 pmol); Nth (lane 13, 0 pmol; lane 14, 0.38 pmol; lane 15, 0.76 pmol; lane 16, 1.5 pmol; lane 17, 3.1 pmol; lane 18, 6.1 pmol).
Cleavage of 5hmU-containing oligonucleotide by purified MutM, Nei and Nth
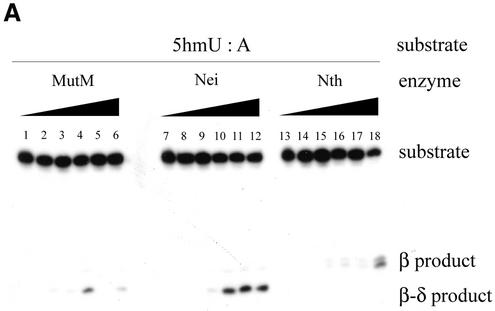
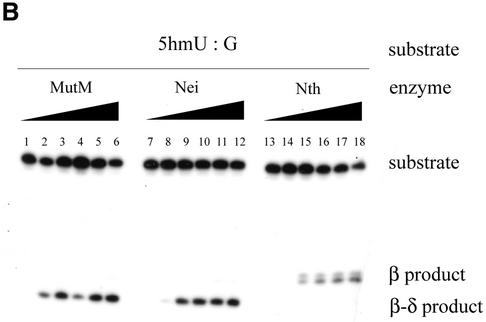
To further verify that the MutM, Nei and Nth are DNA glycosylases with associated AP lyase that excises 5hmU from DNA, we performed DNA cleavage assays with purified MutM, Nei and Nth. Duplex oligonucleotides containing 5hmU:A (Oligo 1/Oligo 3) or 5hmU:G (Oligo 1/Oligo 4) were incubated with the enzymes at 37°C, and then the products were analyzed by polyacrylamide gel electrophoresis. The results are shown in Figure 4A and B. It was observed that all of the enzymes cleaved the oligonucleotides at the site of 5hmU. The products of the cleavage reaction increased with the amount of the enzymes (Fig. 4A and B). Comparison of the mobilities of the products on the gels with those of molecular size markers revealed that MutM and Nei cleaved the oligonucleotides by β- and δ-elimination reactions, whereas Nth cleaved them by a β-elimination reaction (Fig. 4A and B).
Figure 4.
Cleavage of 22mer oligonucleotides containing 5hmU by purified MutM, Nei and Nth. The 5hmU:A and 5hmU:G (Oligo 1/Oligo 3 and Oligo 1/Oligo 4, respectively)-containing duplex oligonucleotides (20 fmol) were incubated at 37°C for 15 min without (lanes 1, 7 and 13) or with purified proteins (MutM, lanes 2–6; Nei, lanes 8–12; Nth, lanes 14–18). The products were separated by electrophoresis on denaturing 20% polyacrylamide gels containing 7 M urea. MutM (lane 1, 0 pmol; lane 2, 0.18 pmol; lane 3, 0.53 pmol; lane 4, 1.8 pmol; lane 5, 5.3 pmol, lane 6, 11 pmol); Nei (lane 7, 0 pmol; lane 8, 0.17 pmol; lane 9, 0.51 pmol; lane 10, 1.7 pmol; lane 11, 5.1 pmol; lane 12, 10 pmol); Nth (lane 13, 0 pmol; lane 14, 0.15 pmol; lane 15, 0.46 pmol; lane 16, 1.53 pmol; lane 17, 4.6 pmol; lane 18, 9.2 pmol).
Table 1 represents the specific activities of cleavage of the 5hmU-containing oligonucleotides by the enzymes. MutM, Nei and Nth cleaved the 5hmU:G oligonucleotide 58, 5 and 37 times more efficiently, respectively, than the 5hmU:A oligonucleotide. The relative activities of cleavage of the 5hmU:G oligonucleotide by E.coli MutM and Nth were consistent with those observed in human cell extracts (27).
Table 1. Cleavage of oligonucleotides containing 5hmU:A and 5hmU:G mispairs by MutM, Nei and Nth proteins.
| Enzyme | Cleavage activity (nM oligonucleotide cleaved/pg protein/min) | b/a | |
|---|---|---|---|
| 5hmU:Aa | 5hmU:Gb | ||
| MutM | 2 | 115 | 58 |
| Nei | 21 | 102 | 4.8 |
| Nth | 39 | 147 | 37 |
DISCUSSION
5hmU is one of the major types of oxidative base damage produced in DNA (2,10,21). It is formed by ionizing radiation with a yield comparable with thymine glycol, 5-foU and 8-oxoguanine (18,19). However, the biological consequences of the formation of 5hmU in DNA remain uncertain. In the genome of Bacillus subtilis bacteriophage SPO1, thymine is completely replaced by 5hmU (34). When 5hmU arises in DNA via the oxidation of thymine, it makes a normal base-pair with A (20,21,35). Furthermore, 5hmU in the template does not disturb the functions of DNA or RNA polymerases (21,35,36), and only dAMP is incorporated opposite the damage during DNA synthesis in vitro (20,21,35). These facts lead us to conclude that 5hmU produced by the oxidation of thymine is not a serious kind of damage in DNA and does not cause substantial mutations.
Previously, Cannon-Carlson et al. (28) suggested another pathway of 5hmU formation: oxidation and deamination of 5mC. 5mC occurs naturally in DNA as a product of cytosine methylation (27,28). Therefore, a normal base-pair between 5mC and G generates a mismatched base-pair between 5hmU and G. If left unrepaired, 5hmU would cause 5mC:G to T:A transition mutations. Cannon-Carlson et al. (28) reported that the major pathway of 5hmU formation is the modification of 5mC. Recently, Rusmintratip and Sowers (27) found a 5hmU-DNA glycosylase activity that removes 5hmU mispaired with G in human cell extracts. This activity excised 5hmU from 5hmU:G mispairs with 60 times higher efficiency than from 5hmU:A base-pairs (27).
Cannon-Carlson et al. (28) suggested that the principal function of 5hmU-glycosylase activities is the maintenance in DNA of methylated cytosine, which plays a critical role in gene regulation in mammals. They mentioned that bacteria do not have 5hmU-DNA glycosylase activities because 5mC residues are not thought to be an essential component of gene regulation in bacteria (28). However, the genome of E.coli includes the dcm gene, whose product methylates the second cytosine residues in the sequence -CC(A/T)GG- (37,38). The 5mC residues are a hot spot for C to T transitions, since the deamination of 5mC leads to the conversion to thymine (39). The formation of T:G mismatches in DNA is corrected by the very short patch (VSP) repair pathway, which removes thymine from T:G mismatches (40,41). The existence of the VSP repair pathway suggests that 5mC residues also play some important biological role in cellular regulation. The modification of 5mC to 5hmU would be harmful, and so bacterial cells might have enzymatic activity that excises the 5hmU from DNA.
The excision activities for 5hmU from 5hmU:G of E.coli MutM, Nei and Nth were much greater than those for 5hmU from 5hmU:A (Fig. 4A and B). There is no doubt that 5hmU is harmful and so E.coli needs to possess a strong ability to excise 5hmU from 5hmU:G mispairs. In E.coli, MutM and Nth excised 5hmU mispaired with G ∼40–50 times more efficiently than 5hmU paired with A, whereas the relative efficiency for Nei was only 5-fold (Table 1). Based on these results, we hypothesized that the DNA glycosylases share roles in repairing 5hmU in DNA to prevent mutations: the 5hmU-DNA glycosylase activity of Nei probably removes 5hmU from 5hmU:A base pairs, while MutM and Nth function cooperatively to remove 5hmU mispairs with G that are generated in DNA. 5hmU:G in DNA would be a serious mispair. Therefore, MutM, Nei and Nth activities would act respectively as a backup activities for each other.
Base excision repair systems are highly conserved from bacteria to mammalian cells (1,42,43). Many homologs of E.coli base excision repair enzymes have been identified: hNth1, the human structural and functional homolog of E.coli Nth is a good example. hNTH1 acts on 5-foU, thymine glycol and 5,6-dihydrouracil in DNA via the same mechanisms as E.coli Nth (1,15,44,45). It would be of interest to know whether hOGG1 and hNEI1, homologs of E.coli MutM and Nei, respectively, are also concerned with the excision of 5hmU from DNA. Cell extracts of E.coli KSR7 transformed with the ntg2 gene, which encodes a functional Nth homolog of Saccharomyces cerevisiae, or with the hNTH1 gene formed a trapped complex with duplex oligonucleotides containing 5hmU:G mispairs (our unpublished results).
The transition from 5mC to T is the most frequent base substitution found in human cancer (46). In the p53 gene, about half of the observed base substitutions are C:G to T:A changes at methylated CpG sequences (47,48). These base substitutions may arise from the conversion of 5mC to T. Therefore, it is important to clarify the mechanisms of repair for 5hmU in DNA. The 5hmU:G glycosylase activity found by Rusmintratip and Sowers (27) may thus be important in preventing transforming mutations in human cells. Recently, Baker et al. (49) reported that the substrate specificity of the partially purified 5hmU-DNA glycosylase of human HeLa cells is distinct from that of previously reported DNA glycosylases, i.e., uracil DNA glycosylase, a DNA glycosylase that recognizes mismatched uracil (MUG), thymine DNA glycosylase and single-stranded mono-functional uracil DNA N-glycosylase (SMUG1) (49,50). However, the biochemical properties of mammalian homologs of these proteins and their roles in the repair of 5hmU in DNA are not fully understood. The results of this study provide some insights into the mechanisms and roles of 5hmU DNA glycosylases in preventing the development of cancer in mammalian cells.
Acknowledgments
ACKNOWLEDGEMENT
This study was partly supported by Grants-in-Aid for Scientific Research (A) and (B) and for Scientific Research on Priority Areas (C) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Wallace S.S. (1997) Oxidative damage to DNA and its repair. In Scandalios,J.G. (ed.), Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 49–90.
- 2.Cadet J., Delatour,T., Douki,T., Gasparutto,D., Pouget,J.-P., Ravanat,J.-L. and Sauvaigo,S. (1999) Hydroxyl radicals and DNA base damage. Mutat. Res., 424, 9–21. [DOI] [PubMed] [Google Scholar]
- 3.Marnett L.J. (2000) Oxyradicals and DNA damage. Carcinogenesis, 21, 361–370. [DOI] [PubMed] [Google Scholar]
- 4.Ames B.N., Shigenaga,M. and Hagen,T.M. (1993) Oxidants, antioxidants, diseases and the degenerative of aging. Proc. Natl Acad. Sci. USA, 90, 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner H.R. (1994) Superoxide dismutase, aging and degenerative disease. Free Radic. Biol. Med., 17, 249–258. [DOI] [PubMed] [Google Scholar]
- 6.Beckman K.B. and Ames,B.N. (1998) The free radical theory of aging matures. Physiol. Rev., 78, 547–581. [DOI] [PubMed] [Google Scholar]
- 7.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 8.David S.S. and Williams,S.D. (1998) Chemistry of glycosylase and endonuclease involved in base-excision repair. Chem. Rev., 98, 1221–1261. [DOI] [PubMed] [Google Scholar]
- 9.Mol C.D., Parikh,S.S., Putnam,C.D., Lo,T.P. and Tainer,J.A. (1999) DNA repair mechanisms for the recognition and removal of damaged bases. Annu. Rev. Biophys. Biomol. Struct., 28, 101–128. [DOI] [PubMed] [Google Scholar]
- 10.Tofigh S. and Frenkel,K. (1989) Effect of metals on nucleoside hydroperoxide, a product of ionizing radiation in DNA. Free Radic. Biol. Med., 7, 131–143. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q.-M., Sugiyama,H., Miyabe,I., Matsuda,S., Saito,I. and Yonei,S. (1997) Replication of DNA templates containing 5-formyluracil, a major oxidative lesion of thymine in DNA. Nucleic Acids Res., 25, 3969–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyabe I., Zhang,Q.-M., Sugiyama,H., Kino,K. and Yonei,S. (2001) Mutagenic effects of 5-formyluracil on a plasmid vector during replication in Escherichia coli. Int. J. Radiat. Biol., 77, 53–58. [DOI] [PubMed] [Google Scholar]
- 13.Klungland A., Paulsen,R., Rolseth,V., Yamada,Y., Ueno,Y., Wiik,P., Matsuda,A., Seeberg,E. and Bjelland,S. (2001) 5-formyluracil and its nucleoside derivatives confer toxicity and mutagenicity to mammalian cells by interfering with normal RNA and DNA metabolism. Toxicol. Lett., 119, 71–78. [DOI] [PubMed] [Google Scholar]
- 14.Bjelland S., Birkeland,N.K., Benneche,T., Volden,G. and Seeberg,E. (1994) DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J. Biol. Chem., 269, 30489–30495. [PubMed] [Google Scholar]
- 15.Zhang Q.-M., Miyabe,I., Matsumoto,Y., Kino,K., Sugiyama.H. and Yonei,S. (2000) Identification of repair enzymes for 5-formyluracil in DNA. Nth, Nei, and MutM proteins of Escherichia coli. J. Biol. Chem., 275, 35471–35477. [DOI] [PubMed] [Google Scholar]
- 16.Miyabe I., Zhang,Q.-M., Kino,K., Sugiyama,H., Takao,M., Yasui,A. and Yonei,S. (2002) Identification of 5-formyluracil DNA glycosylase activity of human hNTH1 protein. Nucleic Acids Res., 30, 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q.-M. (2001) Role of the Escherichia coli and human DNA glycosylases that remove 5-formyluracil from DNA in the prevention of mutations. J. Radiat. Res., 42, 11–19. [DOI] [PubMed] [Google Scholar]
- 18.Frenkel K., Cummings,A., Solomon,J., Cadet,J. and Teebor,G.W. (1985) Quantitative determination of the 5-(hydroxymethyl)uracil moiety in the DNA of gamma-irradiated cells. Biochemistry, 24, 4527–4533. [DOI] [PubMed] [Google Scholar]
- 19.Teebor G.W., Boorstein,R.J. and Cadet,J. (1988) The repairability ofoxidative free radical mediated damage to DNA: a review. Int. J. Radiat. Biol., 54, 131–150. [DOI] [PubMed] [Google Scholar]
- 20.Levy D.D. and Teebor,G.W. (1991) Site directed substitution of 5-hydroxymethyluracil for thymine in replicating phi X-174am3 DNA via synthesis of 5-hydroxymethyl-2′-deoxyuridine-5′-triphosphate. Nucleic Acids Res., 19, 3337–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q.-M., Sugiyama,H., Miyabe,I., Matsuda,S., Kino,K., Saito,I. and Yonei,S. (1999) Replication in vitro and cleavage by restriction endonuclease of 5-formyluracil- and 5-hydroxymethyluracil-containing oligonucleotides. Int. J. Radiat. Biol., 75, 59–65. [DOI] [PubMed] [Google Scholar]
- 22.Vilpo J.A. and Vilpo,L.M. (1995) Restriction, methylation and ligation of 5-hydroxymethyluracil-containing DNA. Mutat. Res., 316, 123–131. [DOI] [PubMed] [Google Scholar]
- 23.Hollstein M.C., Brooks,P., Linn,S. and Ames,B.N. (1984) Hydroxymethyluracil DNA glycosylase in mammalian cells. Proc. Natl Acad. Sci. USA, 81, 4003–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boorstein R.J., Chiu,L.-N. and Teebor,G.W. (1992) A mammalian cell line deficient in activity of the DNA repair enzyme 5- hydroxymethyluracil-DNA glycosylase is resistant to the toxic effects of the thymidine analog 5-hydroxymethyl-2′-deoxyuridine. Mol. Cell. Biol., 12, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedberg E.C., Ganesan,A.K. and Minton,K. (1975) N-glycosidase activity in extracts of Bacillus subtilis and its inhibition after infection with bacteriophage PBS2. J. Virol., 16, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boorstein R.J., Levy,D.D. and Teebor,G.W. (1987) 5-hydroxymethyl-uracil-DNA glycosylase activity is a differentiated function of mammalian cells. Mutat. Res., 183, 257–263. [DOI] [PubMed] [Google Scholar]
- 27.Rusmintratip V. and Sowers,L.C. (2000) An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts. Proc. Natl Acad. Sci. USA, 97, 14183–14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannon-Carlson S.V., Gokhale,H. and Teebor,G.W. (1989) Purification and characterization of 5-hydroxymethyluracil-DNA glycosylase from calf thymus. Its possible role in the maintenance of methylated cytosine residues. J. Biol. Chem., 264, 13306–13312. [PubMed] [Google Scholar]
- 29.Sugiyama H., Matsuda,S., Kino,K., Zhang,Q.-M., Yonei,S. and Saito,I. (1996) New synthetic method of 5-formyluracil-containing oligonucleotides and their melting behavior. Tetrahedron Lett., 37, 9067–9070. [Google Scholar]
- 30.Matsumoto Y., Zhang,Q.-M., Takao,M., Yasui,A. and Yonei,S. (2001) Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res., 29, 1975–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 32.Nash H.M., Bruner,S.D., Scharer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 33.Rabow L.E. and Kow,Y.W. (1997) Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry, 36, 5084–5096. [DOI] [PubMed] [Google Scholar]
- 34.Kallen R.G., Simon,M. and Marmur,J. (1962) The occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA: 5-hydroxymethyl uracil. J. Mol. Biol., 5, 248–250. [DOI] [PubMed] [Google Scholar]
- 35.Mellac S., Fazakerley,G.V. and Sowers,L.C. (1993) Structures of base pairs with 5-(hydroxymethyl)-2′-deoxyuridine in DNA determined by NMR spectroscopy. Biochemistry, 32, 7779–7786. [DOI] [PubMed] [Google Scholar]
- 36.Herrala A.M. and Vilpo,J.A. (1989) Template-primer activity of 5-(hydroxymethyl)uracil-containing DNA for prokaryotic and eukaryotic DNA and RNA polymerases. Biochemistry, 28, 8274–8277. [DOI] [PubMed] [Google Scholar]
- 37.Marinus M.G. (1973) Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol. Gen. Genet., 127, 47–55. [DOI] [PubMed] [Google Scholar]
- 38.Hanck T., Schmidt,S. and Fritz,H.J. (1993) Sequence-specific and mechanism-based crosslinking of Dcm DNA cytosine-C5 methyltransferase of E. coli K-12 to synthetic oligonucleotide containing 5-fluoro-2′-deoxycytidine. Nucleic Acids Res., 21, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coulondre C., Miller,J.H., Farabauge,P.J. and Gilbert,W. (1978) Molecular basis of base substitution hotspots in Escherichia coli. Nature, 274, 775–780. [DOI] [PubMed] [Google Scholar]
- 40.Lieb M. (1991) Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics, 128, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieb M. and Bhagwat,A.S. (1996) Very short patch repair: reducing the cost of cytosine methylation. Mol. Microbiol., 20, 467–473. [DOI] [PubMed] [Google Scholar]
- 42.Eisen J.A. and Hanawalt,P.C. (1999) A phylogenomic study of DNA repair genes, proteins and processes. Mutat. Res., 435, 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aravind L., Walker,D.R. and Koonin,E.V. (2000) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- 45.Dizdaroglu M., Bauche,C., Rodriguez,H. and Laval,J. (2000) Novel substrates of Escherichia coli Nth protein and its kinetics for excision of modified bases from DNA damaged by free radicals. Biochemistry, 39, 5586–5592. [DOI] [PubMed] [Google Scholar]
- 46.Spruck C.H. II, Rideout,W.M.,III and Jones,P.A. (1993) DNA methylation and cancer. In Jost,J.P. and Saluz,H.P. (eds), DNA Methylation: Molecular Biology and Biological Significance. Birkhauser Verlag, Basel, Switzerland, pp. 487–509.
- 47.Jones P.A., Rideout,W.M.,III, Shen,J.-C., Spruck,C.H. and Tsai,Y.C. (1992) Methylation, mutation and cancer. BioEssays, 14, 33–36. [DOI] [PubMed] [Google Scholar]
- 48.Laird P.W. and Jaenisch,R. (1994) DNA methylation and cancer. Hum. Mol. Genet., 3, 1487–1495. [DOI] [PubMed] [Google Scholar]
- 49.Baker D., Liu,P., Burdzy,A. and Sowers,L.C. (2002) Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem. Res. Toxicol., 15, 33–39. [DOI] [PubMed] [Google Scholar]
- 50.Boorstein R.J., Cummings,A.,Jr, Marenstein,D.R., Chan,M.K., Ma,Y., Nuebert,T.A., Brown,S.M. and Teebor,G.W. (2001) Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem., 276, 41991–41997. [DOI] [PubMed] [Google Scholar]