Abstract
In breast and colon cancers, transforming growth factor (TGF)-β signaling initially has an antineoplastic effect, inhibiting tumor growth, but eventually exerts a proneoplastic effect, increasing motility and cancer spread. In prostate cancer, studies using human samples have correlated the loss of the TGF-β type II receptor (TβRII) with higher tumor grade. To determine the effect of an inhibited TGF-β pathway on prostate cancer, we bred transgenic mice expressing the tumorigenic SV40 large T antigen in the prostate with transgenic mice expressing a dominant negative TβRII mutant (DNIIR) in the prostate. Transgene( s) and TGF-β1 expression were identified in the prostate and decreased protein levels of plasminogen activator inhibitor type I, as a marker for TGF-β signaling, correlated with expression of the DNIIR. Although the sizes of the neoplastic prostates were not enlarged, increased amounts of metastasis were observed in mice expressing both transgenes compared to age-matched control mice expressing only the large T antigen transgene. Our study demonstrates for the first time that a disruption of TGF-β signaling in prostate cancer plays a causal role in promoting tumor metastasis.
Keywords: transforming growth factor-β, prostate cancer, metastasis, probasin, transgenic mice
Introduction
Prostate cancer is a heterogeneous disease that typically progresses from prostatic intraepithelial neoplasia (PIN) to locally invasive to potentially metastatic carcinoma. Surgical treatment of locally invasive cancer yields a 10-year progression-free survival probability of greater than 90%. In contrast, the development of metastatic cancer drops the survival probability to about 30% [1]. Metastasis occurs in late-stage prostate cancer wherein some cancers with the loss of p53, RB, and the TGF-β type II receptor (TβRII) have been identified [2–5]. The TβRII transduces signal for the ubiquitous, pleiotropic cytokine transforming growth factor (TGF)-β that is involved in many pathways including growth inhibition, apoptosis, and differentiation [6–8]. TGF-β signaling involves the binding of ligand to the extracellular domain of TβRII, the transphosphorylation of the TGF-β type I receptor by the cytoplasmic domain of TβRII, and the subsequent phosphorylation of Smad signaling molecules that can activate the transcription of downstream targets such as plasminogen activator inhibitor type I (PAI-I) [9,10]. Although the loss of the TβRII has been associated with prostate cancer, its effect on tumor progression has not been determined. We have studied transgenic mouse models of prostate cancer to examine the effect of blocking the TGF-β signaling pathway on tumor development and progression.
The large probasin (LPB) promoter directed the prostatic epithelial cell expression of the SV40 large T antigen (Tag) [11], with a deletion in the early region to remove the small t antigen. The oncogenic Tag has been reported to induce neoplasia by sequestering and inactivating two tumor suppressor genes, p53 and RB, that have been associated with prostate cancer [2,3]. Two LPB-Tag lines, 12T-7f and 12T-10, were used to study different aspects of prostate tumor progression. The 12T-7f line developed prostate tumors of lethal size by 23 weeks of age that contained high-grade prostatic intraepithelial neoplasia (HGPIN) and occasional foci of invasive carcinoma (IC) [12]. We chose this line to test whether blocking TGF-β signaling would promote progression from cancer precursors to more locally invasive and/or metastatic cancer. However, the 12T-10 line developed small prostate lesions containing HGPIN and invasive neuroendocrine carcinoma that became metastatic after 6 months of age [13]. This line was selected to determine whether inhibiting the TGF-β pathway would modify the metastatic phenotype.
Although multiple lines were made with the probasin promoter to target the dominant negative TβRII mutant (DNIIR) in the prostate of transgenic mice, none of the lines expressed the DNIIR transgene (unpublished observation). The zinc-inducible metallothionein (MT) promoter was well suited to express the transgene in the prostate because the prostate contains naturally high levels of zinc and expresses several MTs in the epithelium [14]. The DNIIR construct had a Flag epitope and contained the extracellular, transmembrane, and juxtamembrane domains (aa 1–191) of the human TβRII, but lacked the cytoplasmic kinase domain for downstream signaling [15]. Overexpressing the DNIIR transgene has been shown to create a dominant negative mutation in transgenic mice by competing with the naturally low levels of endogenous TβRII for TGF-β ligand and by binding to endogenous TGF-β receptor type I [16]. By breeding the MT-DNIIR mice with 12T-7f and 12T-10 mice, we hypothesized that expression of the DNIIR transgene in the Tag tumors would inhibit TGF-β signaling and alter the neoplastic phenotype.
Materials and Methods
Generating and Genotyping Transgenic Mice
LPB-Tag transgenic mice lines were generated with the 5′-flanking region of the rat LPB gene (-10,834 to +28 bp) linked to the Tag gene deletion mutant (d1 2005), which removed expression of the small t antigen [12]. Both 12T-7f and 12T-10 lines were maintained in the CD1 mouse strain. MT-DNIIR transgenic mice (MT-DNIIR-27) were generated using a truncated human TβRII with a Flag epitope as the DNIIR transgene under the control of the MT promoter [15]. The MT-DNIIR line used in this study was maintained in the B6D2 mouse strain. 12T-7fTTag+/- females were mated with MT-DNIIRDNIIR+/+ males to generate 12T-7f/MT-DNIIR and MT-DNIIR mice in the similar mouse background. The 12T-7f/MT-DNIIR and MT-DNIIR mice were supplemented with 25 mM zinc sulfate in the drinking water to enhance transgene expression. 12T-7fTag+/- females were mated with B6D2F1 males to generate control 12T-7f and nontransgenic (NT) mice in the same mouse background. Some of the control 12T-7f and NT mice were supplemented with 25 mM zinc sulfate in the drinking water. 12T-10Tag+/+ females were mated with MT-DNIIRDNIIR+/+ males to generate 12T-10/MT-DNIIR mice. To control for genetic background, 12T-10Tag+/+ females were mated with B6D2F1 males to generate 12T-10 mice. None of the 12T-10/MT-DNIIR or 12T-10 mice were supplemented with 25 mM zinc sulfate in the drinking water. Mice were genotyped for the DNIIR transgene by polymerase chain reaction (PCR) analysis of genomic DNA isolated from mouse tails using proteinase K digestion and ethanol extraction. The forward TβRII primer (5′-TCCCACCGCACGTTCAGAAG-3′) and reverse Flag primer (5′-ATCGTCATCGTCTTTGTAGTC-3′) produced an amplimer of 506 bp [15]. Mice were genotyped for the Tag transgene by PCR analysis using the forward LPB primer (5′-TAGCATCTTGTTCTTAGTCTT-3′) and reverse Tag primer (5′-CTCCTTTCAAGACCTAGAAGGTCCA-3′) to produce an amplimer of 430 bp [12]. Exon 7 of the endogenous mouse casein gene served as an internal control for the Tag PCR reaction using the forward casein primer (5′;-GATGTGCTCCAGGCTAAAGTT-3′) and the reverse casein primer (5′;-AGAAACGGAATGTTGTGGAGT-3′) to generate a 540-bp amplimer [12].
Tissue Preparation and Histopathologic Analysis
Mice were sacrificed by cervical dislocation after the inhalation of an anesthetic agent according to the policy of the Vanderbilt University Animal Care Committee. The prostates were generally dissected into four different lobes (ventral, lateral, dorsal, and anterior lobes) under a dissecting microscope. When it was not possible to separate the lateral and dorsal lobes, the tissues were taken together as the dorsolateral lobe. Seminal vesicle, vas deferens, testis, periurethral gland, bladder, bulbourethral gland, para-aortic lymph nodes (LNs), neck LNs, lumbar spine, liver, lung, kidney, spleen, brain, adrenal, parotid gland, and submandibular gland were also harvested for histologic examination. Tissues were either frozen on dry ice and stored at -80°C, or fixed in 10% formalin and processed and embedded in paraffin using standard techniques. Paraffinembedded tissues were cut at 5 Am and sections were used for H&E staining, immunohistochemistry, and in situ hybridization. Histology was classified in a blinded manner by two pathologists (S.B.S. and R.L.R.) based on histopathologic definitions from the Prostate Pathology Committee of the National Cancer Institute Mouse Models of Human Cancer Consortium [17].
Immunohistochemistry
Immunostaining was performed on 5-µm-thick paraffin sections, which were deparaffinized and rehydrated using standard techniques [13]. The following primary antibodies were used (with the indicated dilutions in PBS): (a) SV40 Tag, Ab-2 (1:100; Oncogene Research Products, Boston, ME); (b) CG, bovine SP-1 (1:1000; DiaSorin, Stillwater, MN); and (c) TGF-β1, sc-146 (1:200; Santa Cruz Biotechnology, Santa Cruz, CA). For Tag-immunostained sections, the same concentration of control mouse ascites fluid (Sigma, St. Louis, MO) was used as a negative control. For CG-immunostained sections, the same concentration of normal rabbit immunoglobulin, X903 (Dako, Carpinteria, CA), was used as a negative control. For TGF-β1-immunostained sections, control peptide (sc-146P) was added at 10 times the primary antibody concentration along with the primary antibody as a negative control, or the same concentration of normal rabbit immunoglobulin was used as negative control. Color development was performed with 3,3′-diaminobenzidine tetrahydrochloride (Dako). Slides were counterstained with hematoxylin, dehydrated, and coverslipped. Some of the immunostaining was quantitated using the MetaMorph image analysis program.
In Situ Hybridization for DNIIR
In situ hybridization was performed on 5-µm-thick paraffin sections, which were deparaffinized and rehydrated using standard techniques [15]. Sections were hybridized to 35S-labeled sense and antisense riboprobes. The MT-DNIIR plasmid was linearized with EcoRI, and the antisense probe was made with T7 polymerase. The sense probe was made from XbaI-linearized plasmid and T3 polymerase. Slides were exposed to photographic emulsion at 4°C for 1 month and then developed with D19 developer, fixed in 1% acetic acid, and cleared in 30% sodium thiosulfate. Sections were counterstained with 0.2% toluidine blue. Kodak Ektachrome film (Eastman Kodak, Rochester, NY) was used to take photographs under phase contrast, bright field, and dark field illumination using a microscope (Zeiss, Thornwood, NY).
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
RNA was extracted from frozen samples using the RNeasy Midi Kit 75144 including treatment with DNase 79254 (Qiagen, Valencia, CA). RNA concentration was determined based on absorbance on a spectrophotometer, and RNA quality was assessed by agarose gel electrophoresis. DNIIR copy numbers were determined using a Lightcycler fluorescence temperature rapid air cycler (Roche Molecular Biochemicals, Indianapolis, IN) with cDNA standard curves and the double-stranded DNA-binding fluorescent probe SYBR Green. The cDNA template was generated from the MT-DNIIR plasmid. The forward DNIIR primer (5′;-AGAAGAATATAACACCAGCAATCC-3′) and reverse DNIIR primer (5′;-ATCCAACGCGGTAGCAGTAGAAGA-3′) generated a 128-bp amplimer. The specificity of the amplimer in each reaction was confirmed by the melting curve analysis. Copy numbers of mRNA were calculated, using the Lightcycler software (version 3.5), from serially diluted (1:10) standard curves (109-103 copies). The serially diluted standards were simultaneously amplified with the unknown samples to generate a linear standard curve using the fit points method of analysis with seven points. Additionally, real-time RT-PCR was developed for β-actin as a loading control using a cDNA template, the forward β-actin primer (5′;-ACGGCCAGGTCATCACTATTG-3′) and the reverse h-actin primer (5′;-ATGCTACTCAGGCCGGGGA-3′).
Western Blot and Band Quantification
Protein was extracted by sonication and centrifugation of frozen tissues in RIPA buffer (PBS, pH 7.4, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1 mM PMSF and Complete Mini protease inhibitor cocktail, 1836153 (Roche, Mannheim, Germany). Protein concentration was determined by the Lowry method using the Bio-Rad Protein Assay 500-0006 (Bio-Rad Laboratories, Hercules, CA). Protein lysates were treated with β-mercaptoethanol and heated at 70°C for 10 minutes then electrophoresed in NuPage 7% Tris-acetate gels EA035A (Invitrogen, Carlsbad, CA). Proteins were transferred overnight at 30 V to Invitrolon PVDF membranes LC2005 (Invitrogen). Membranes were blocked with blocking buffer containing 5% skim milk in TBS with 0.1% Tween. For PAII detection, anti-PAI-I polyclonal antibody TP223 (Torrey Pines Biolaboratories, Houston TX) at 1:1000 dilution in blocking buffer was used, followed by horseradish peroxidase-linked antirabbit Ig NA9340 (Amersham Biosciences, Buckinghamshire, England, UK) at 1:2000 dilution in blocking buffer. Bands were visualized using ECL plus RPN2132 (Amersham Biosciences) and Kodak BioMax MR Film 870-1302 (Eastman Kodak).
For ′-actin detection, the PAI-I western membranes were stripped by incubation in stripping buffer (2% SDS, 62.5 mM Tris-HCl, pH 7.4, 100 mM β-mercaptoethanol) for 30 minutes at 70°C. Then the membranes were blocked and probed with monoclonal anti-β-actin antibody A5441 (Sigma) at 1:5000 in blocking buffer, followed by horseradish peroxidase-linked antimouse Ig NA931V (Amersham Biosciences) at 1:2000 dilution in blocking buffer. Bands were visualized using ECL plus RPN2132 and Kodak BioMax MR Film 870-1302.
For band quantitation, the exposed film was scanned to obtain TIFF images in Adobe Photoshop. With each membrane image, the band intensities in a constant area were measured using the Scion Image analysis program (Scion, Frederick, MD). The band intensity values were transferred to the Microsoft Excel program for statistical analysis.
Statistical Analysis
Heteroskedastic t-test was performed to compare the Western blot data between two groups. Fisher's exact test as well as the Generalized Linear Mode (Logistic Regression Model) were used to compare the metastasis data between two groups. P value < .05 was considered statistically significant. The analyses were performed in collaboration with the Vanderbilt Cancer Center Biostatistics Shared Resource.
Results
Generation and Characterization of Transgenic Mice
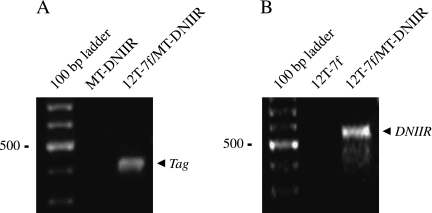
The 12T-7f line was heterozygous for the Tag transgene, whereas the 12T-10 line was homozygous for the Tag transgene. The MT-DNIIR line was homozygous for the DNIIR transgene. Therefore, 12T-7f mice were bred with MT-DNIIR mice to generate 12T-7f/MT-DNIIR offspring and MT-DNIIR control offspring. 12T-7f mice were bred with B6D2F1 mice to generate 12T-7f control offspring and NT offspring. 12T-10 mice were bred with MT-DNIIR mice to generate 12T-10/MT-DNIIR mice. 12T-10 mice were bred with B6D2F1 mice to generate 12T-10 control offspring. Offspring were genotyped by PCR using genomic DNA isolated from mouse tail (arrow, Figure 1, A and B).
Figure 1.
Detection of transgene in transgenic mice. (A) An example of PCR genotyping using primer sets for the Tag transgene. The expected size of the Tag-amplified product is 430 bp. Transgene was detected in the 12T-7f/MT-DNIIR sample but not in the MT-DNIIR sample. (B) An example of PCR genotyping using primer sets for the DNIIR transgene. The expected size of the DNIIR-amplified product is 506 bp. Transgene was detected in the 12T-7f/MT-DNIIR sample but not in the 12T-7f sample.
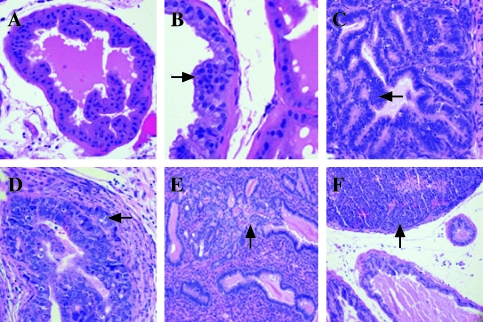
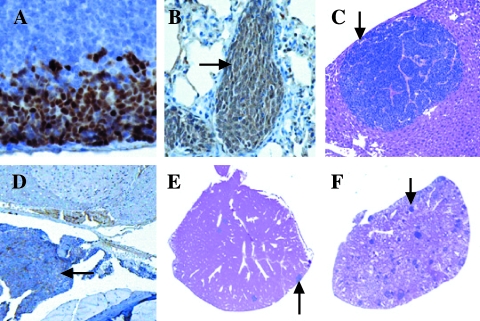
Histologic examination of the prostates showed progressive tumor development in the mice expressing the T antigen, but minimal or no lesions in the other mice. The prostates were classified in a blinded manner by two pathologists (S.B.S. and R.L.R.) as containing no histologic abnormality (NHA), low-grade prostatic intraepithelial neoplasia (LGPIN), HGPIN, microinvasive carcinoma (MI), IC, or undifferentiated carcinoma (UC) [13,17]. Compared to normal areas, LGPIN and HGPIN were characterized by stratification and crowding of the epithelial cells within basement membrane-bound preexisting glands along with cytologic abnormalities such as nuclear enlargement (arrow, Figure 2, A–C). HGPIN was distinguished from LGPIN based on the accentuation of these features including marked nuclear atypia, more hyperchromatic nuclei, and higher mitotic and apoptotic rates (arrow, Figure 2, B and C). MI was recognized as foci of single cells or groups of cells breaking through the basement membrane of HGPIN-containing glands into the surrounding stroma (arrow, Figure 2D). Lesions were designated as IC when they were more extensive than those described above as MI, with foci of glands demonstrating invasion (arrow, Figure 2E). UC was characterized by invasive lesions that frequently showed destructive overgrowth of normal prostate architecture and lacked glandular differentiation but had cytologic and histologic features typical of neuroendocrine differentiation (arrow, Figure 2F). The MT-DNIIR and NT mice showed predominately NHA or LGPIN and rarely HGPIN but not cancer in the prostate. Given that carcinoma was identified only in the mice expressing the T antigen and predominantly in the dorsolateral lobes, we focused on the dorsolateral prostate (DLP) in these mice for further experiments (Table 1).
Figure 2.
Illustrations of histopathology of transgenic animals (H&E). (A) NHA in a DLP of a 12-week-old NT mouse. (B) LGPIN (arrow) in a DLP of a 23-week-old MT-DNIIR mouse. (C) HGPIN (arrow) in a DLP of an 18-week-old 12T-7 mouse. (D) MI (arrow) in a DLP of a 9-month-old 12T-10 mouse. (E) IC (arrow) in a DLP of a 20-week-old 12T-7f/MT-DNIIR mouse. (F) UC (arrow) in a VP of a 10-month-old 12T-10/MT-DNIIR mouse.
Table 1.
Histopathology of Mice Expressing the Tag Transgene.
| Line | Mouse number | Age | AP | DLP | VP | Metastatic site |
| 12T-7f/MT-DNIIR | 9237 | 12 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f/MT-DNIIR | 9240 | 12 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f/MT-DNIIR | 9241 | 12 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f/MT-DNIIR | A3474 | 12 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3476 | 12 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f/MT-DNIIR | A3477 | 12 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f/MT-DNIIR | A3478 | 12 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3231 | 12 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3242 | 12 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3246 | 12 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3479 | 12 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3483 | 12 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3484 | 12 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f/MT-DNIIR | 9201 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9207 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9208 | 16 weeks | HGPIN | HGPIN, IC | HGPIN | Lung |
| 12T-7f/MT-DNIIR | A3187 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3190 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3199 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3203 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3204 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3207 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3208 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3209 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3499 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3505 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3117 | 16 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f | A3120 | 16 weeks | HGPIN | HGPIN | HGPIN | Lung |
| 12T-7f | A3218 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3221 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3228 | 16 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3232 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3233 | 16 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3234 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3235 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3239 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3243 | 16 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3244 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3247 | 16 weeks | HGPIN | HGPIN | LGPIN | Lung |
| 12T-7f | A3249 | 16 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3251 | 16 weeks | HGPIN | HGPIN | NHA | None |
| 12T-7f | A3252 | 16 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3276 | 16 weeks | HGPIN | HGPIN, IC, UC | HGPIN, IC, UC | LN |
| 12T-7f/MT-DNIIR | 9215 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9218 | 18 weeks | HGPIN | HGPIN, IC | HGPIN | Lung, liver, LN, spine |
| 12T-7f/MT-DNIIR | 9239 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3028 | 18 weeks | HGPIN | HGPIN | HGPIN | Lung |
| 12T-7f/MT-DNIIR | A3078 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3194 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3195 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3490 | 18 weeks | HGPIN | HGPIN | HGPIN | Lung |
| 12T-7f/MT-DNIIR | A3491 | 18 weeks | HGPIN | HGPIN | HGPIN | Lung, liver |
| 12T-7f | A3104 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3108 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3283 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3285 | 18 weeks | HGPIN, IC | HGPIN | LGPIN | None |
| 12T-7f | A3286 | 18 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9212 | 20 weeks | HGPIN, IC | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9219 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9223 | 20 weeks | HGPIN, IC | HGPIN, IC | HGPIN | None |
| 12T-7f/MT-DNIIR | A3087 | 20 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f/MT-DNIIR | A3210 | 20 weeks | HGPIN | HGPIN, IC | LGPIN | Lung |
| 12T-7f | 9194 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | 9195 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | 9197 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3271 | 20 weeks | HGPIN | HGPIN | HGPIN | Lung, liver, spine |
| 12T-7f | A3274 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3384 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3387 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3391 | 20 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9220 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | 9227 | 23 weeks | HGPIN | HGPIN, IC | NHA | Lung |
| 12T-7f/MT-DNIIR | A3018 | 23 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f/MT-DNIIR | A3030 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f/MT-DNIIR | A3081 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | 9245 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | 9246 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-7f | A3258 | 23 weeks | HGPIN | HGPIN, IC | HGPIN | None |
| 12T-7f | A3265 | 23 weeks | HGPIN | HGPIN | LGPIN | None |
| 12T-7f | A3278 | 23 weeks | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3130 | 6 months | HGPIN | HGPIN | HGPIN, MI | None |
| 12T-10/MT-DNIIR | A3131 | 6 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10/MT-DNIIR | A3132 | 6 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10/MT-DNIIR | A3133 | 6 months | HGPIN | HGPIN | HGPIN | Lung |
| 12T-10/MT-DNIIR | A3134 | 6 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3158 | 6 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3159 | 6 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3160 | 6 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3161 | 6 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3164 | 6 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3142 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3143 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3144 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3145 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3146 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3162 | 7 months | HGPIN | HGPIN, MI | HGPIN | Lung |
| 12T-10 | A3163 | 7 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3169 | 7 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3170 | 7 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3171 | 7 months | HGPIN | HGPIN, MI | HGPIN | Lung |
| 12T-10/MT-DNIIR | A3150 | 8 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3151 | 8 months | HGPIN | HGPIN, IC | HGPIN | None |
| 12T-10/MT-DNIIR | A3152 | 8 months | HGPIN | HGPIN, MI, IC | HGPIN | None |
| 12T-10/MT-DNIIR | A3407 | 8 months | HGPIN | HGPIN, MI | HGPIN, MI | Lung, liver |
| 12T-10 | A3175 | 8 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3176 | 8 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3177 | 8 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10 | A3178 | 8 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3179 | 8 months | HGPIN | HGPIN | HGPIN | None |
| 12T-10/MT-DNIIR | A3403 | 9 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10/MT-DNIIR | A3404 | 9 months | HGPIN | HGPIN, MI | HGPIN, MI | Lung, liver, LN |
| 12T-10/MT-DNIIR | A3405 | 9 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10/MT-DNIIR | A3406 | 9 months | HGPIN | HGPIN, MI | HGPIN, MI | Lung, liver |
| 12T-10/MT-DNIIR | A3408 | 9 months | HGPIN | HGPIN, MI | HGPIN | Lung, liver |
| 12T-10 | A3394 | 9 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3395 | 9 months | HGPIN | HGPIN, MI | HGPIN | Lung |
| 12T-10 | A3396 | 9 months | HGPIN, MI | HGPIN, MI, IC | HGPIN | Lung, liver |
| 12T-10 | A3397 | 9 months | HGPIN | HGPIN, MI | HGPIN | Lung |
| 12T-10/MT-DNIIR | A3135 | 10 months | HGPIN, MI | HGPIN, MI | HGPIN, IC, UC | Lung, liver, LN |
| 12T-10/MT-DNIIR | A3137 | 10 months | HGPIN | HGPIN, IC | HGPIN, IC, UC | Lung, LN |
| 12T-10/MT-DNIIR | A3138 | 10 months | HGPIN, IC | HGPIN, MI | HGPIN | Lung |
| 12T-10 | A3168 | 10 months | HGPIN | HGPIN, IC | HGPIN, MI | None |
| 12T-10 | A3180 | 10 months | HGPIN | HGPIN, MI | HGPIN | Lung |
| 12T-10 | A3184 | 10 months | HGPIN | HGPIN, MI | HGPIN | None |
| 12T-10 | A3185 | 10 months | HGPIN | HGPIN | HGPIN, MI | Lung, liver |
AP = anterior prostate; VP = ventral prostate.
Both Transgenes and TGF-β1 Were Expressed in Prostate Lesions
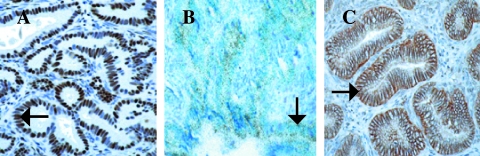
Because human prostate carcinomas arise from the glandular epithelium, it was important to demonstrate the presence of both transgenes in the epithelium. Both Tag and DNIIR transgenes were expressed in luminal epithelial cells of 12T-7f/MT-DNIIR and 12T-10/MT-DNIIR mice according to immunohistochemical analyses for Tag (arrow, Figure 3A) and in situ hybridization analyses for DNIIR (arrow, Figure 3B). We found stable levels of DNIIR expression throughout tumor progression as measured by real-time RT-PCR (Table 2). Additionally, TGF-β1 was identified in the prostate and localized to the epithelium according to immunohistochemical analyses (arrow, Figure 3C). The staining intensities of TGF-β1 were similar based on Meta-Morph analysis. The presence of TGF-β justified the use of a dominant negative TβRII to block TGF-β signaling. Thus, expression of both transgenes and TGF-β1 was seen in the prostatic epithelial lesions that developed in the 12T-7f/MT-DNIIR and 12T-10/MT-DNIIR.
Figure 3.
Detection of transgene expression in transgenic mice. (A) An example of immunohistochemical analysis shows expression of Tag in an HGPIN lesion (arrow) in a DLP of a 12T-7f/MT-DNIIR mouse. (B) A light field view of in situ hybridization shows expression of DNIIR in an HGPIN lesion (arrow) in a DLP of a 12T-7f/MT-DNIIR mouse. (C) An example of immunohistochemical analysis shows localization of TGF-β1 in an HGPIN lesion (arrow) in a DLP of a 12T-7f mouse.
Table 2.
Quantitation of DNIIR Transgene Expression by Real-Time RT-PCR.
| Line | Mouse number | Age | Tissue | DNIIR level | β-Actin | level Ratio of DNIIR/β-Actin |
| 12T-7f/MT-DNIIR | 9237 | 12 weeks | DLP | 3.65E + 04 | 1.00E + 10 | 3.7E - 06 |
| 12T-7f/MT-DNIIR | 9241 | 12 weeks | DLP | 1.85E + 05 | 4.24E + 10 | 4.4E - 06 |
| 12T-7f/MT-DNIIR | 9201 | 16 weeks | DLP | 7.65E + 04 | 1.19E + 10 | 6.4E - 06 |
| 12T-7f/MT-DNIIR | 9208 | 16 weeks | DLP | 1.06E + 05 | 1.03E + 10 | 1.0E - 05 |
| 12T-7f/MT-DNIIR | 9218 | 18 weeks | DLP | 1.80E + 04 | 5.53E + 09 | 3.3E - 06 |
| 12T-7f/MT-DNIIR | A3028 | 18 weeks | DLP | 6.02E + 04 | 2.60E + 10 | 2.3E - 06 |
| 12T-7f/MT-DNIIR | 9212 | 20 weeks | DLP | 4.98E + 04 | 4.35E + 10 | 1.1E - 06 |
| 12T-7f/MT-DNIIR | 9223 | 20 weeks | DLP | 3.10E + 04 | 4.07E + 10 | 7.6E - 07 |
| 12T-7f/MT-DNIIR | 9227 | 23 weeks | DLP | 2.40E + 04 | 1.07E + 10 | 2.2E - 06 |
| 12T-7f/MT-DNIIR | A3030 | 23 weeks | DLP | 5.76E + 04 | 1.08E + 10 | 5.3E - 06 |
| 12T-7f/MT-DNIIR | A3407 | 8 months | Liver metastasis | 1.69E + 07 | 5.89E + 10 | 2.9E - 04 |
| 12T-7f/MT-DNIIR | A3404 | 9 months | Liver metastasis | 1.79E + 03 | 5.67E + 10 | 3.2E - 08 |
| 12T-7f/MT-DNIIR | A3135 | 10 months | Liver metastasis | 1.30E + 03 | 6.02E + 10 | 2.2E - 08 |
Levels are listed as copy numbers per 200 ng of total RNA.
Expression of the DNIIR Transgene Decreased Levels of a TGF-β Downstream Target, PAI-I
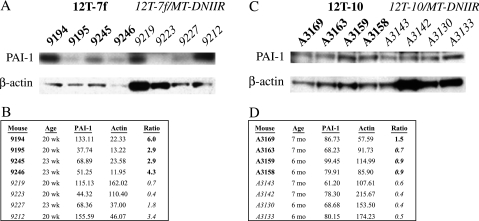
PAI-I expression is positively regulated by TGF-β [9,10]. Using PAI-I as a marker for TGF-β signaling, the presence of the DNIIR transgene was shown to disrupt TGF-β signaling. Western blot analyses were performed to determine the PAI-I and β-actin levels in tissues from age-matched mice. The PAI-I levels were quantitated using the Scion Image analysis program and normalized for protein loading by the β-actin levels. The average normalized level of PAI-I in 12T-7f/MTDNIIR mice was significantly lower than the average normalized level of PAI-I in 12T-7f mice (P < .05) (Figure 4, A and B). Similarly, the average normalized level of PAI-I in 12T-10/MT-DNIIR mice was significantly lower than the average normalized level of PAI-I in 12T-10 mice (P < .05) (Figure 4, C and D). Therefore, the DNIIR transgene actively inhibited TGF-β signaling in the prostate.
Figure 4.
Quantitation and normalization of PAI-I levels in transgenic mice. (A) Western blots for PAI-I and for β-actin as a loading control in 12T-7f and 12T-7f/MTDNIIR mice. (B) Summary of band intensities from Western blots in (A) along with ratios of PAI-I to β-actin. The average ratio is significantly lower in 12T-7f/MT-DNIIR mice than in 12T-7f mice (P < .05). (C) Western blots for PAI-I and for β-actin as a loading control in 12T-10 and 12T-10/MT-DNIIR mice. (B) Summary of band intensities from Western blots in (C) along with ratios of PAI-I to β-actin. The average ratio is significantly lower in 12T-10/MT-DNIIR mice than in 12T-10 mice (P < .05).
12T-7f/MT-DNIIR Mice Tended to Develop More IC But Not Larger Prostates Compared to 12T-7f Mice
The histologic progression of prostatic neoplasia in 12T-7f/MT-DNIIR (n = 39) mice was similar to that of 12T-7f mice (n = 41) from 12 to 23 weeks old. Both lines predominantly developed HGPIN with glandular proliferation and hypercellular stroma in the prostate. To determine whether the loss of TGF-β inhibition in the tumors stimulated growth, we measured the wet prostatic weights and normalized for differences in animal size by calculating the percentage of body weight. Comparing the prostate weights, as percentage of body weight, of 12T-7f/MT-DNIIR and 12T-7f mice revealed no increase in tumor size (data not shown). Invasive periurethral and bulbourethral tumors were rarely observed in 12T-7f/MT-DNIIR mice (n = 2) and 12T-7f mice (n = 1). Focal lesions of locally invasive carcinoma were observed more frequently in routine sections of the prostates of 12T-7f/MT-DNIIR mice (n = 6) than in 12T-7f mice (n = 3) (Table 1). None of the control MT-DNIIR mice (n = 50) and NT mice (NT, n = 56) from 12 weeks to 10 months old developed IC.
12T-7f/MT-DNIIR Mice Tended to Develop Metastasis More Frequently and Extensively Than 12T-7f Mice
Both 12T-7f/MT-DNIIR and 12T-7f mice developed microscopic neuroendocrine metastases to the para-aortic LNs (arrow, Figure 5A), lungs (arrow, Figure 5B), liver (Figure 5C), and rarely bones (12T-7f/MT-DNIIR, n = 1; 12T-7f, n = 1) (arrow, Figure 5D). Metastases were identified histologically and showed typical features of neuroendocrine differentiation. The metastatic lesions were immunopositive for Tag (arrow, Figure 5A), TGF-β1 (arrow, Figure 5B), and the neuroendocrine marker chromogranin A (CG), confirming their transgene expression and neuroendocrine differentiation (arrow, Figure 5D). More 12T-7f/MT-DNIIR mice (n = 7) developed metastasis than age-matched 12T-7f mice (n = 4) (Table 1). Lung metastasis occurred most commonly whereas liver metastasis occurred in a subset of those animals, seemingly indicating more widespread metastasis. Comparing liver metastasis in the 12T-7f line (n = 1) (arrow, Figure 5E) and the 12T-7f/MT-DNIIR line (n = 2) (arrow, Figure 5F) showed more extensive metastasis in the 12T-7f/MT-DNIIR mice. These results in the 12T-7f mouse model suggested that loss of the TGF-β pathway in prostate tumors promotes metastatic disease.
Figure 5.
Metastatic lesions along with immunohistochemical analyses in 12T-7f/MT-DNIIR mice and 12T-7f mice. (A) Tag-expressing metastatic lesion (arrow) in the para-aortic lymph node of a 12T-7f/MT-DNIIR mouse. (B) TGF-β1-expressing metastatic lesion (arrow) in the lung of a 12T-7 mouse. (C) Metastatic lesion (arrow) on H&E stain in the liver of a 12T-7 mouse. (D) CG-expressing metastatic lesion (arrow) in the lumbar spine of a 12T-7f mouse. (E) Micrometastases (arrow) in the liver of a 20-week-old 12T-7f mouse. (F) Extensive micrometastases (arrow) in the liver of an 18-week-old 12T-7f/MT-DNIIR mouse.
12T-10/MT-DNIIR Mice Tended to Develop More IC But Not Larger Prostates Compared to 12T-10 Mice
Characterizing the effects of DNIIR overexpression on the tumors in the 12T-10 line showed an even more pronounced effect on tumor progression. The histologic progression of the primary prostatic neoplasia in 12T-10/MT-DNIIR mice was similar to that of 12T-10 mice. The 12T-10/MT-DNIIR mice (n = 22) and 12T-10 mice (n = 23) from 6 to 10 months old progressively developed HGPIN, locally invasive neuroendocrine carcinoma in the prostate, and neuroendocrine metastasis. Comparing the prostate weights, as percent of body weight, of 12T-10/MT-DNIIR and 12-T10 mice showed no increase in tumor size (data not shown). Invasive periurethral and bulbourethral tumors were rarely observed in 12T-10/MT-DNIIR (n = 3) and 12T-10 (n = 1) mice. The incidence of IC was higher in the 12T-10/MT-DNIIR mice (n = 5) than 12T-10 mice (n = 2) (Table 1).
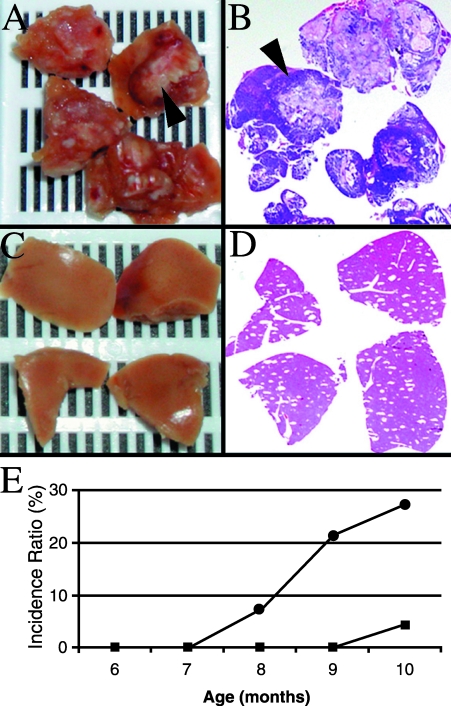
12T-10/MT-DNIIR Mice Developed Significantly More Extensive Metastasis Than 12T-10 Mice
The 12T-10/MT-DNIIR and 12T-10 mice developed LN, lung, and liver metastases similar to, but more frequently than, 12T-7f/MT-DNIIR and 12T-7f mice. According to immunohistochemical and real-time RT-PCR analyses, the metastatic lesions expressed Tag, TGF-β1, CG, and DNIIR (Table 2). Based on histologic examination, the cumulative incidence of metastasis was not different between the 12T-10/MT-DNIIR (n = 8) and 12T-10 (n = 7) mice (Table 1). However, the extent of metastatic disease was prominently increased in the 12T-10/MT-DNIIR mice compared to 12T-10 mice. Grossly evident metastatic nodules in the liver and lungs from 12T-10/MT-DNIIR mice (arrow, Figure 6, A and B) were noted but no grossly evident metastases were observed in those organs from age-matched 12T-10 mice (Figure 6, C and D). Comparing the cumulative incidence of gross metastasis over time between 12T-10/MT-DNIIR mice (●, n = 6/22) and 12T-10 (■, n = 1/23) mice showed a statistically significant (P < .05) increase in the 12T-10/MT-DNIIR mice (Figure 6E). Furthermore, comparing only mice with metastasis showed a statistically significant (P < .05) increase in gross metastasis in 12T-10/MT-DNIIR mice (n = 6/8) compared with 12T-10 mice (n = 1/7). These results in the 12T-10 mouse model demonstrated that a loss of TGF-β signaling in metastatic prostate cancer increases the amount of metastatic lesions.
Figure 6.
Comparison of metastasis in 12T-10/MT-DNIIR mice and 12T-10 mice. (A and B) Gross metastatic lesions (arrow) in the liver of a 12T-10/MT-DNIIR mouse and accompanying histology. (C and D) No gross lesions in the liver of an age-matched 12T-10 mouse and accompanying histology. (E) Comparison of incidence (%) of gross metastasis over time in 12T-10/MT-DNIIR (●, n = 22) mice versus 12T-10 (■, n = 23) mice. The incidence ratio of metastasis is significantly greater in 12T-10/MT-DNIIR mice than in 12T-10 mice (P < .05).
Discussion
In summary, this study has demonstrated that disruption of TGF-β signaling modulates progression in late-stage prostate cancer, specifically metastasis. Although more IC foci were noted in routine sections in the mice expressing the DNIIR and Tag transgenes, the increases were not statistically significant (Table 3). Therefore, the effect on metastasis was not simply due to the development of more IC. Instead, the effect must have been either on the development of metastasis and/or growth at the metastatic site. Because the MT promoter is not prostate-specific, more direct effects on the immune system or angiogenesis cannot be ruled out. Nevertheless, the DNIIR transgene had a direct effect of disrupting the TGF-β pathway in the prostate tumors as demonstrated by the significant reduction in levels of PAI-I. Decreased levels of PAI-I, an inhibitor of urokinase-type plasminogen activator (uPA) that mediates extracellular matrix proteolysis, may be a possible mechanism for the increased metastasis in our model because it has been demonstrated that PAI-I is present at low levels in several human prostate cancer cell lines including PC-3 cells and increasing PAI levels by stable transfection decreases metastasis of PC-3 cells in mice [18,19]. Other intracellular effectors of TGF-β might also be causally involved in induction of prostate cancer metastasis. It has been reported in breast and colon cancers that TGF-β signaling initially has a negative effect but eventually exerts a positive effect on tumor progression. For example, blocking TGF-β is important for tumor growth, but subsequent restoration of the TGF-β pathway contributes to tumor cell motility, epithelial-to-mesenchymal transition, and ultimately metastasis [20,21]. Those reports focused on nonprostatic cancer and used different systems such as injection of cell lines into mice for tumor and metastasis formation.
Table 3.
Summary of Histopathology.
| Line | Age | IC | Metastasis |
| 12T-7f/MT-DNIIR | 12–23 weeks | 6 of 39 | 7 of 39 (2 to liver) |
| 12T-7f | 12–23 weeks | 3 of 41 | 4 of 41 (1 to liver) |
| 12T-10/MT-DNIIR | 6–10 months | 5 of 22 | 8 of 22 (6 gross*) |
| 12T-10 | 6–10 months | 2 of 23 | 7 of 23 (1 gross*) |
| MT-DNIIR | 12 weeks–10 months | 0 of 50 | 0 of 50 |
| NT | 12 weeks–10 months | 0 of 56 | 0 of 56 |
Significant difference with P < .05.
In human prostate cancer, the loss of the TβRII occurs most commonly in advanced stage disease [4,5], suggesting that this pathway does not function as a positive influence on the spread of this disease. These studies correlated a dysfunctional TGF-β receptor pathway with tumor grade at the primary site but did not prove that the loss of the receptor was responsible for cancer progression. Our study in two transgenic animal models provides proof, rather than a pathologic correlation, that disruption of TGF-β regulation is important for increasing metastatic burden. This is especially critical in human prostate cancer that metastasizes in end-stage disease mainly to the bone, where high TGF-β levels are naturally found [22]. Metastatic initiation and growth will be important targets for future study and possibly therapy because limiting metastasis would allow for more successful treatment of prostate cancer [1]. The majority of human prostate cancer metastases are moderate to poorly differentiated adenocarcinomas, but these mouse models, similar to other SV40 T antigen models, develop invasive and metastatic poorly differentiated carcinomas with neuroendocrine differentiation [17]. Murine trangenic models to study human prostate cancer have some limitations because mice do not naturally develop prostate cancer and results in mice do not always translate directly to humans. Nevertheless, a recent report has identified the neuroendocrine phenotype and TGF-β pathway as important prognostic factors in human prostate cancer [23]. Although TGF-β loss has been associated with human prostate cancer progression, it has not been determined whether that loss was a cause or consequence of tumor progression. Our animal models of prostate cancer demonstrate genetic and histopathologic changes analogous to those in human prostate cancer. Our data in these models suggest for the first time that there is a clinically relevant selective advantage for loss of TGF-β regulation as a cause of increased metastatic burden. We hope to use these models to identify novel TGF-β targets that would be tested in human samples.
Acknowledgement
The authors thank Simon Hayward for his helpful advice and critical reading of this manuscript.
Abbreviations
- AP
anterior prostate
- DLP
dorsolateral prostate
- DNIIR
dominant negative TβRII mutant
- HGPIN
high-grade prostatic intraepithelial neoplasia
- IC
invasive carcinoma
- LGPIN
low-grade prostatic intraepithelial neoplasia
- LN
lymph node
- Met
metastasis
- MI
microinvasive carcinoma
- MT
etallothionein
- NHA
no histologic abnormality
- NT
non-transgenic
- PAI-I
plasminogen activator inhibitor type I
- PIN
prostatic intraepithelial neoplasia
- RT-PCR
reverse transcriptase polymerase chain reaction
- Tag
SV40 large T antigen
- TβRII
TGF-β type II receptor
- TGF
transforming growth factor
- UC
undifferentiated carcinoma
- VP
ventral prostate
- wk
weeks
Footnotes
This study was supported, in part, by the DOD grant PC992022, NCI grant R01-CA-76142, NIH Mouse Models of Human Cancer Consortium grant NCI U01-CA-98013, and Frances Williams Preston Laboratories of the T.J. Martell Foundation. Transgenic mice were bred by the Transgenic Core/ES Cell Shared Resource of the Vanderbilt-Ingram Cancer Center (NCI grant 2P30-CA-68485).
References
- 1.Ohori M, Goad JR, Wheeler TM, Eastham JA, Thompson TC, Scardino PT. Can radical prostatectomy alter the progression of poorly differentiated prostate cancer? J Urol. 1994;152:1843–1849. doi: 10.1016/s0022-5347(17)32398-4. [DOI] [PubMed] [Google Scholar]
- 2.Quinn DI, Henshall SM, Head DR, Golovsky D, Wilson JD, Brenner PC, Turner JJ, Delprado W, Finlayson JF, Stricker PD, Grygiel JJ, Sutherland RL. Prognostic significance of p53 nuclear accumulation in localized prostate cancer treated with radical prostatectomy. Cancer Res. 2000;60:1585–1594. [PubMed] [Google Scholar]
- 3.Bookstein R, Rio P, Madreperla SA, Hong F, Allred C, Grizzle WE, Lee WH. Promoter deletion and loss of retinoblastoma gene expression in human prostate carcinoma. Proc Natl Acad Sci USA. 1990;87:7762–7766. doi: 10.1073/pnas.87.19.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim IY, Ahn HJ, Lang S, Oefelein MG, Oyasu R, Kozlowski JM, Lee C. Loss of expression of transforming growth factor-beta receptors is associated with poor prognosis in prostate cancer patients. Clin Cancer Res. 1998;4:1625–1630. [PubMed] [Google Scholar]
- 5.Guo Y, Jacobs SC, Kyprianou N. Down-regulation of protein and mRNA expression for transforming growth factor-beta (TGF-beta1) type I and type II receptors in human prostate cancer. Int J Cancer. 1997;71:573–579. doi: 10.1002/(sici)1097-0215(19970516)71:4<573::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 6.Morton DM, Barrack ER. Modulation of transforming growth factor β1 effects on prostate cancer cell proliferation by growth factors and extracellular matrix. Cancer Res. 1997;55:2596–2602. [PubMed] [Google Scholar]
- 7.Brodin G, ten Dijke P, Funa K, Heldin CH, Landstrom M. Increased smad expression and activation are associated with apoptosis in normal and malignant prostate after castration. Cancer Res. 1999;59:2731–2738. [PubMed] [Google Scholar]
- 8.Danielpour D. Transdifferentiation of NRP-152 rat prostatic basal epithelial cells toward a luminal phenotype: regulation by glucocorticoid, insulin-like growth factor-I and transforming growth factorbeta. J Cell Sci. 1999;112 (Part 2):169–179. doi: 10.1242/jcs.112.2.169. [DOI] [PubMed] [Google Scholar]
- 9.Datta PK, Blake MC, Moses HL. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-β-induced physical and functional interactions between Smads and Sp1. J Biol Chem. 2000;275:40014–40019. doi: 10.1074/jbc.C000508200. [DOI] [PubMed] [Google Scholar]
- 10.Hua X, Miller ZA, Wu G, Yigong S, Lodish HF. Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor, µE3, and Smad proteins. Proc Natl Acad Sci USA. 1999;96:13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Sheppard PC, Kasper S, Lin L, Hoare S, Kapoor A, Dodd JG, Duckworth ML, Matusik RJ. A large fragment of the probasin promoter targets high levels of transgene expression to the prostate of transgenic mice. Prostate. 1997; 32:129–139. doi: 10.1002/(sici)1097-0045(19970701)32:2<129::aid-pros8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Kasper S, Sheppard PC, Yan Y, Pettigrew N, Borowsky AD, Prins GS, Dodd JG, Duckworth ML, Matusik RJ. Development, progression and androgen-dependence of prostate tumors in transgenic: a model for prostate cancer. Lab Invest. 1998;78:319–334. [PubMed] [Google Scholar]
- 13.Masumori N, Thomas TZ, Case T, Paul M, Kasper S, Chaurand P, Caprioli RM, Tsukamoto T, Shappell SB, Matusik RJ. A probasin-large T antigen transgenic mouse line develops prostate adeno and neuroendocrine carcinoma with metastatic potential. Cancer. 2001;Res 61:2239–2249. [PubMed] [Google Scholar]
- 14.Ghatak S, Oliveria P, Kaplan P, Ho SM. Expression and regulation of metallothionein mRNA levels in the prostates of noble rats: lack of expression in the ventral prostate and regulation by sex hormones in the dorsolateral prostate. Prostate. 1996;29:91–100. doi: 10.1002/(SICI)1097-0045(199608)29:2<91::AID-PROS4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 15.Serra R, Johnson M, Filvaroff EH, LaBorde J, Sheehan DM, Derynck R, Moses HL. Expression of a truncated, kinase-defective TGF-beta type II receptor in mouse skeletal tissue promotes terminal chondrocyte differentiation and osteoarthritis. J Cell Biol. 1997;139:541–552. doi: 10.1083/jcb.139.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XJ, Greenhalgh DA, Bickenbach JR, Jiang A, Bundman DS, Krieg T, Derynck R, Roop DR. Expression of a dominantnegative type II transforming growth factor beta (TGF-beta) receptor in the epidermis of transgenic mice blocks TGF-beta-mediated growth inhibition. Proc Natl Acad Sci USA. 1997;94:2386–2391. doi: 10.1073/pnas.94.6.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matusik R, Masumori N, Thomas T, Case T, Paul M, Kasper S, Shappell S. Transgenic mouse models of prostate cancer. In: Matzuk M, Brown C, Kumar T, editors. Contemporary Endocrinology: Transgenics in Endocrinology. Totowa, Canada: Humana Press Inc; 2001. pp. 401–425. [Google Scholar]
- 18.Lyon PB, See WA, Xu Y, Cohen MB. Diversity and modulation of plasminogen activator activity in human prostate carcinoma cell lines. Prostate. 1995;27:179–186. doi: 10.1002/pros.2990270402. [DOI] [PubMed] [Google Scholar]
- 19.Soff GA, Sanderowitz J, Gately S, Verrusio E, Weiss I, Brem S, Kwaan HC. Expression of plasminogen activator inhibitor type 1 by human prostate carcinoma cells inhibits primary tumor growth, tumor-associated angiogenesis, and metastasis to lung and liver in an athymic mouse model. J Clin Invest. 1995;96:2593–3600. doi: 10.1172/JCI118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases. J Clin Invest. 1995;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 22.Pfeilschifter J, Mundy GR. Modulation of type beta transforming growth factor activity in bone cultures by osteotropic hormones. Proc Natl Acad Sci USA. 1987;84:2024–2028. doi: 10.1073/pnas.84.7.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd CP, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, Kantoff TR, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cells. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]