Abstract
The incorporation of noncanonical amino acids into recombinant proteins in Escherichia coli can be facilitated by the introduction of new aminoacyl-tRNA synthetase activity into the expression host. We describe here a screening procedure for the identification of new aminoacyl-tRNA synthetase activity based on the cell surface display of noncanonical amino acids. Screening of a saturation mutagenesis library of the E. coli methionyl-tRNA synthetase (MetRS) led to the discovery of three MetRS mutants capable of incorporating the long-chain amino acid azidonorleucine into recombinant proteins with modest efficiency. The Leu-13 → Gly (L13G) mutation is found in each of the three MetRS mutants, and the MetRS variant containing this single mutation is highly efficient in producing recombinant proteins that contain azidonorleucine.
Keywords: azide–alkyne ligation, azidohomoalanine, azidonorleucine, click chemistry
The aminoacyl-tRNA synthetases (aaRS) ensure the fidelity of protein synthesis in cells through specific ligation of each amino acid to its cognate tRNA(s). Despite the specificity of these enzymes for their natural substrates, protein engineers have exploited the promiscuity of aaRS to incorporate a wide array of noncanonical amino acids into recombinant proteins (1, 2). Elevation of the aaRS activity of the expression host can be critical for efficient in vivo production of proteins in which one of the natural amino acids is globally replaced by a noncanonical amino acid (3–6). Alternatively, mutant aaRS activity may be introduced into an expression host to permit incorporation of noncanonical amino acids that are inert with respect to the wild-type (WT) protein synthesis machinery (7–14). The discovery of new mutant aaRS activities for global replacement of natural amino acids by noncanonical counterparts can be greatly accelerated by the efficient screening of libraries of mutant aaRS. Such an approach has proved fruitful in generating novel aaRS activity for the related problem of site-specific incorporation of noncanonical amino acids (15, 16). Here we describe a rapid, flow-cytometry-based screening protocol to examine libraries of mutant aaRS for their ability to enable incorporation of reactive amino acids into proteins, and we demonstrate its application to the Escherichia coli methionyl-tRNA synthetase (MetRS).
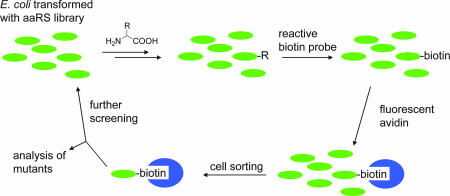
Our screening protocol (Fig. 1) relies on our previous observation that introduction of a noncanonical amino acid into the E. coli cell surface protein OmpC (outer membrane protein C) and the subsequent covalent tagging of this amino acid provides a sensitive mode of detection of the translational activity of the noncanonical amino acid (17, 18). E. coli cells displaying recombinant OmpC expressed in medium supplemented with azidohomoalanine (AHA; 1) (Fig. 2) were covalently biotinylated by means of Cu-catalyzed azide–alkyne ligation (19) and subsequently stained with fluorescent avidin. These cells were readily differentiable from unlabeled cells in flow-cytometric analyses. This observation suggests that cell-surface display should enable rapid screening for aaRS variants that activate noncanonical amino acids with high efficiency.
Fig. 1.
Protocol for screening libraries of mutant aaRS. Cells transformed with an aaRS library are induced to express OmpC in medium supplemented with a noncanonical amino acid bearing a reactive side chain. Cells that successfully incorporate the noncanonical amino acid display the reactive side chain on their surfaces. Labeled cells are covalently tagged by a biotin probe and subsequently stained with fluorescent avidin. Fluorescence-activated cell sorting isolates the tagged cells from the remainder of the library. Sorted cells may be subjected to additional rounds of screening or analyzed immediately.
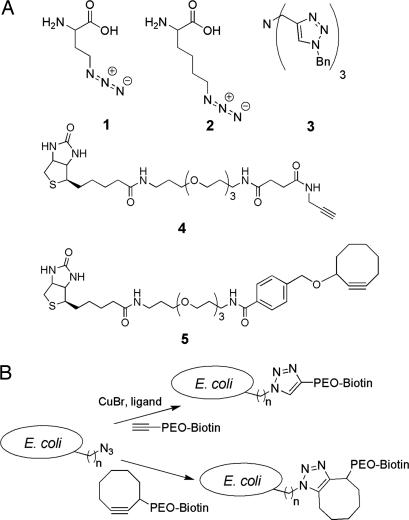
Fig. 2.
Noncanonical amino acids and tagging reagents used in this study. (A) 1, AHA; 2, ANL; 3, Tris(triazolyl)amine ligand for copper-catalyzed azide–alkyne ligation; 4, biotin–PEO–propargylamide; 5, biotin–PEO–cyclooctyne. (B) Scheme for biotin tagging of azide-functionalized E. coli cell surfaces. 1 or 2 is incorporated into OmpC to display the azide on the cell surface. Cell surface azides react either by Cu-catalyzed azide–alkyne ligation (top route) or by strain-promoted azide–alkyne ligation (bottom route).
As a first test of the screening protocol, we designed and built a saturation mutagenesis library of MetRS variants. High-resolution crystal structures of E. coli MetRS are available both without (20) and with (21) Met bound. Additional structures are available for MetRS bound to several analogs of Met and methionyl-AMP (22). This wealth of structural data allows informed selection of residues for saturation mutagenesis in the binding pocket of MetRS. Furthermore, MetRS lacks the “sieve-type” editing activity found in related aaRS such as the valyl-(23), isoleucyl- (24), and leucyl-tRNA synthetases (25), so engineering only of the synthetic site of the synthetase needs to be considered. Screening of the MetRS library led to the discovery of three different mutants that enable incorporation of the long-chain amino acid azidonorleucine (ANL) (2) into recombinant proteins with modest protein yields. Furthermore, a single amino acid mutation, Leu-13 → Gly (L13G), which occurs in each of the three active mutants, is sufficient to engender activity toward ANL. In fact, the L13G MetRS mutant enables incorporation of ANL into recombinant proteins more efficiently than any of the mutants obtained in the screen.
Results and Discussion
Cell-Surface Labeling with Biotin–Polyethyleneoxide (PEO)–Cyclooctyne.
We have previously labeled azide-functionalized E. coli cell surfaces by means of copper-catalyzed azide–alkyne ligation with biotin–PEO–propargylamide 4 as outlined in Fig. 2. As noted, however, the requisite copper catalyst is toxic to E. coli cells that overexpress OmpC (17, 18). Although screening experiments have been performed successfully on cells of compromised viability (26), the use of live cells can greatly simplify such screening experiments. Therefore, we investigated the efficiency of cell-surface labeling with biotin–PEO–cyclooctyne 5, which requires no copper catalyst (27). Cells displaying OmpC containing AHA (OmpC–AHA) were covalently biotinylated by treatment either with 100 μM CuBr, 200 μM 3, and 50 μM 4 for 16 h at 4°C, or with 100 μM 5 for 16 h at 37°C. After staining with fluorescent avidin, the cells were subjected to flow cytometry. The median fluorescence of cells tagged with 5 is ≈35% of the median fluorescence of cells tagged by means of the copper-catalyzed reaction (Fig. 3) but is nevertheless ≈25-fold higher than the background fluorescence of unlabeled cells. Although cells subjected to Cu-catalyzed azide–alkyne ligation under the conditions described here are unable to divide, cells that have been tagged with 5 and subjected to cell sorting are readily regrown in selective rich medium.
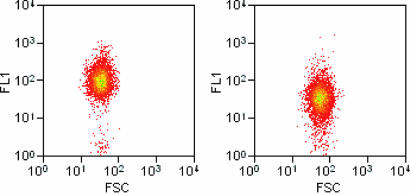
Fig. 3.
Comparison of the extent of cell-surface labeling by Cu-catalyzed azide–alkyne ligation and strain-promoted azide–alkyne ligation. Cells displaying OmpC containing AHA were biotinylated either by means of Cu-catalyzed azide–alkyne ligation (50 μM 4, 200 μM 3, and 100 μM CuBr at 4°C) (Left) or by strain-promoted azide–alkyne ligation (100 μM 5 at 37°C) (Right). Data are plotted as forward scatter (FSC; x axis), which is an indication of cell size, vs. fluorescence (FL1; y axis). The median fluorescence of cells labeled by means of Cu-catalyzed azide–alkyne ligation is 3-fold larger than that of cells labeled by means of the strain-based reaction.
MetRS Library Design and Construction.
To select residues in the binding pocket of MetRS for saturation mutagenesis, the crystal structure of Met-bound MetRS (21) was examined by using swiss pdb viewer software (www.expasy.ch/spdbv). Five residues (Leu-13, Trp-253, Pro-257, Tyr-260, and His-301) were found within a radius of 4 Å of either the sulfur atom or the methyl group of bound Met. Alignment of MetRS sequences from different organisms (see Fig. 7, which is published as supporting information on the PNAS web site) reveals that W253, Y260, and H301 are universally conserved, whereas L13 and P257 are highly conserved. These residues have been suggested to be critical to Met binding; the backbone amide of L13 and the hydroxyl group of Y260 form hydrogen bonds with the sulfur atom of bound Met (21). H301 also is believed to form a hydrogen bond to the sulfur atom, whereas W253 forms hydrophobic contacts with the Met side chain (22). Four of these five residues, excluding W253, were selected for mutagenesis to all other possible amino acids; their orientations with respect to bound Met are shown in Fig. 4. W253 was excluded from mutagenesis because of its importance in forming the wall of the Met-binding pocket. The library was built by means of a modified PCR gene assembly process (see Materials and Methods) using primers containing degenerate NNK (N is all bases; K is G,T) codons at the mutagenesis sites. The use of NNK codons permits complete coverage of the possible amino acid space while eliminating two of the nonsense codons and decreasing the theoretical size of the library required for complete sequence coverage. The library was subsequently ligated into the plasmid pAJL-20, which encodes a variant of OmpC containing six additional surface-exposed Met residues under inducible control, and the resulting plasmids were introduced to the Met auxotroph M15MA.
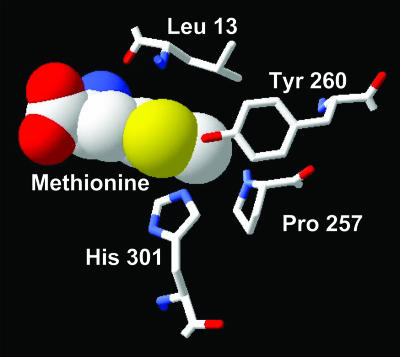
Fig. 4.
Residues in the Met-binding pocket of MetRS selected for saturation mutagenesis. Four residues (stick models) found within 4 Å of the sulfur atom or methyl group of Met (space-filling model) were mutated to all other possible amino acids. Figure was drawn from coordinates in ref. 21 (Protein Data Bank ID code 1F4L).
Screening and Identification of Active Mutants.
The MetRS library was screened for evidence of efficient protein synthesis in Met-depleted minimal medium supplemented with the long-chain amino acid ANL. We have reported very low levels of incorporation of ANL in cells characterized by elevated WT MetRS activity (17), but the amounts of protein produced in such experiments were vanishingly small. We therefore sought to identify a MetRS mutant that would allow efficient incorporation of ANL into recombinant proteins.
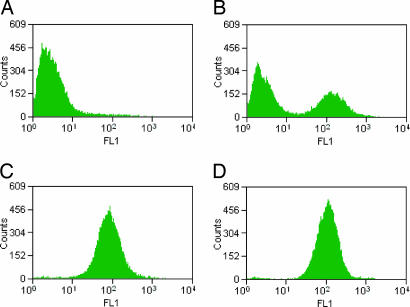
An aliquot of M15MA transformed with the MetRS library in pAJL-20 was used to inoculate minimal medium supplemented with all 20 canonical amino acids. Upon reaching midlog phase, the cells were washed and transferred to minimal medium lacking Met. This culture was supplemented with high levels (8 mM) of ANL, and OmpC expression was induced by addition of isopropyl-β-thiogalactoside (IPTG). The cells were treated sequentially with biotin–PEO–cyclooctyne 5 and fluorescent avidin to render fluorescent the surfaces of cells that had successfully incorporated ANL. The same cells also were biotinylated by means of Cu-catalyzed azide–alkyne ligation for comparison. Cells treated with 5 were analyzed by flow cytometry, and the top 1% of the cells in the fluorescence channel were sorted and regrown in selective liquid medium to ensure plasmid retention. A portion of the sorted cell population also was rescued by plating on selective agar plates, allowing analysis of single clones. The sorted pool of cells was subjected to another round of OmpC expression in ANL-supplemented medium and biotinylated by means of azide–alkyne ligation to determine the extent of enrichment of highly fluorescent clones. Although only 1% of the cells in the original library resided in the high-fluorescence regime (>80 arbitrary fluorescence units), 40% of the sorted cells fell in this range, demonstrating selective enrichment of highly fluorescent clones (Fig. 5A and B). Two individual clones from this population (clones 2.6.1 and 2.6.2) retained the high fluorescence characteristic of extensive azide functionalization of the cell surface (Fig. 5 C and D) and were analyzed further. In addition, the plasmid DNA from the pooled sorted cells was isolated, and the MetRS genes were moved into a variant of the plasmid pAJL-20 encoding an OmpC with only two additional Met sites in its exposed loops (as opposed to the six additional sites previously used). These plasmids were reintroduced to M15MA, and the screening protocol was carried out as described above, except that ANL was added at a concentration of 3.2 mM. This screen yielded one additional highly fluorescent mutant, clone 3.2.7.
Fig. 5.
Fluorescence histograms of cells induced to produce OmpC in medium supplemented with ANL. All cells were biotinylated by means of Cu-catalyzed azide–alkyne ligation before flow cytometry. (A) Cells harboring naïve MetRS library. (B) Cells enriched from the top 1% of cells in A. (C) Cells harboring clone 2.6.1 MetRS. (D) Cells harboring clone 2.6.2 MetRS.
The MetRS mutations found in these three clones are presented in Table 1. The net effect of these mutations is an expansion of the binding pocket to create room for the bulky substrate ANL. The character of the binding pocket remains hydrophobic. Most striking is the recurrence of the L13G mutation, which is present in all three mutants analyzed. The L13G MetRS single mutant therefore was constructed and tested for its ability to incorporate ANL as described below.
Table 1.
Mutations found in MetRS clones that enable incorporation of ANL into recombinant proteins
| MetRS | Leu-13 | Pro-257 | Tyr-260 | His-301 |
|---|---|---|---|---|
| 2.6.1 | G | L | T | A |
| 2.6.2 | G | S | T | L |
| 3.2.7 | G | L | L | V |
Recombinant Protein Production with MetRS Mutants.
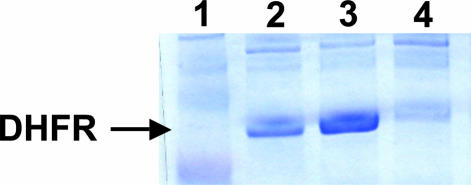
Each of the four MetRS mutants (clones 2.6.1, 2.6.2, and 3.2.7 and L13G) was transferred to the plasmid pAJL-61, which encodes a 6xHis-tagged dihydrofolate reductase (DHFR) under inducible control as well as the repressor protein lacIq. The resulting plasmids were transformed into the expression host M15MA, which also contains a plasmid-borne copy of lacIq. The high levels of lacIq were introduced to minimize leaky expression of DHFR, because any DHFR produced before induction will not contain ANL. Midlog phase cultures of M15MA[pAJL-61] were shifted to minimal medium lacking Met but supplemented with 8 mM ANL as described in Materials and Methods. After 3 h, the cells were lysed, and the DHFR was purified by using Ni–nitrilotriacetic acid (NTA) chromatography. The yield of protein produced per liter of culture was calculated by measuring the A280 value of a solution of the purified protein, and the extent of incorporation of ANL was estimated by N-terminal sequencing (Table 2). The L13G mutant enables the most efficient incorporation of ANL; ≈95% of the N-terminal Met is replaced by ANL in cells that express this mutant. Met was the only other amino acid detected above background in N-terminal sequencing analyses. SDS/PAGE analysis of whole-cell lysates from M15MA[pAJL-20] cells harboring the L13G mutant demonstrates that comparable amounts of DHFR are produced when the cells are supplemented with either Met or ANL (Fig. 6). The yields of purified protein obtained from experiments using the L13G mutant are within a factor of 2 of the protein yield of native DHFR produced with WT MetRS (27) and are sufficient for demanding applications such as biomaterials synthesis.
Table 2.
Purified protein yield and extent of incorporation of ANL into DHFR produced in M15MA[pAJL-61] harboring different MetRS variants
| MetRS | DHFR yield, mg/liter culture | Extent of incorporation, % |
|---|---|---|
| WT | —* | —† |
| Clone 2.6.1 | 4.5 | 55 |
| Clone 2.6.2 | 3.2 | 39 |
| Clone 3.2.7 | 1.5 | —† |
| L13G | 18.1 | 95 |
*No protein was detected.
†Not determined.
Fig. 6.
SDS/PAGE analysis of whole-cell lysates of M15MA[pAJL-61] encoding the L13G MetRS mutant. (Lane 1) Molecular weight marker. Cells were grown to midlog phase in medium containing all 20 canonical amino acids and then shifted to 19-aa medium (no Met) supplemented with 40 mg/liter Met (lane 2), 172 mg/liter ANL (lane 3), or no amino acid (lane 4). Expression of DHFR is comparable in the cultures supplemented either with Met or ANL.
In Vitro Activation Kinetics of L13G MetRS.
Previous work has shown that the rates of activation of Met analogs correlate strongly with the efficiencies with which those analogs are incorporated into recombinant proteins (27, 28). The kinetic parameters for activation of both Met and ANL were determined by using the purified L13G mutant of MetRS (Table 3). Met is still activated by the mutant synthetase, although the specificity constant kcat/Km is nearly 300-fold smaller than that characteristic of the WT enzyme. Nevertheless, the activation rate of ANL by the L13G mutant (1.56 × 10−3 μM−1·s−1) compares favorably with the rates previously determined for the activation of the excellent Met surrogates AHA and homopropargylglycine by the WT MetRS (28). We have reported that activation of ANL by WT MetRS is undetectable in similar assays (18).
Table 3.
Kinetic parameters for activation of Met, 1 and 2 by WT and L13G MetRS
Conclusions
We have presented a rapid, high-throughput screening method for the identification of new aaRS activities in vivo. By using this screening protocol, we have identified three mutants of the E. coli MetRS that enable incorporation of the long-chain amino acid ANL into recombinant proteins in response to Met codons. The L13G mutation occurs in each of these three mutants, and the MetRS variant harboring this single amino acid mutation proves to be the most efficient synthetase for incorporation of ANL into recombinant proteins. The expanded binding pocket of L13G MetRS may permit incorporation of other long-chain amino acids into recombinant proteins.
A relatively small library of 106 members was screened in the current study, but the ultra-high-throughput nature of the flow cytometric screen should permit analysis of libraries several orders of magnitude larger, up to the limits of bacterial transformation efficiency. Protocols of the kind described here need not be limited to azide-bearing amino acids and may be readily adapted to other reactive amino acid side chains.
Materials and Methods
Amino Acids and Tagging Reagents.
AHA (1) (29), ANL (2) (18), biotin–PEO–propargylamide 4 (17), biotin–PEO–cyclooctyne 5 (30), and the Tris(triazolyl)amine ligand 3 (19, 31) were prepared as described. CuBr (99.999% purity) was purchased from Aldrich.
MetRS Library Construction.
Restriction enzymes were from New England Biolabs or Roche. T4 DNA ligase was purchased from Invitrogen or New England Biolabs. The plasmid pAJL-20 was used as a template for construction of the MetRS library. pAJL-20 has been described (18) and encodes a variant of OmpC under control of the isopropyl-β-thiogalactoside (IPTG)-inducible T5 promoter. In addition, pAJL-20 contains a cassette encoding the E. coli MetRS under control of its natural promoter. Oligonucleotides encoding degenerate NNK (N is A,T,G,C; K is G,T) codons at the sites corresponding to Leu-13, Pro-257, Tyr-260, and His-301 in E. coli MetRS were obtained from Qiagen (Valencia, CA). Four separate PCRs were performed with PicoMaxx polymerase (Stratagene) by using pAJL-20 as a template and the following pairs of primers: Nhe lib forward and L13 reverse, L13 forward and P257 reverse, P257 forward and H301 reverse, H301 forward and lib reverse. The P257 forward and P257 reverse primers span both the Pro-257 and the Tyr-260 codons. Sequences for all primers may be found in Supporting Text, which is published as supporting information on the PNAS web site. The DNA fragments obtained from these PCRs were electrophoresed and purified by using Zymo-spin columns (Zymo Research, Orange, CA). Equimolar quantities (≈10 fmol) of the fragments were mixed and subjected to 10 rounds of PCR (95°C for 1 min, 55°C for 1 min, 72°C for 2 min) by using PicoMaxx as the polymerase. The primers Nhe lib forward and lib reverse were subsequently added, and the reaction mixture was subjected to 30 more rounds of PCR using the same parameters described above. The resulting 1.4-kb PCR product was digested with NotI and BsrGI and ligated into pAJL-20 digested with the same enzymes. The ligation mixture was transformed into chemically competent XL-1 Blue cells (Stratagene), yielding 106 independent transformants for a maximum of ≈62% coverage of the library. The plasmid DNA from the pooled transformants was isolated with Maxiprep (Qiagen), diluted 10-fold with water, and used to transform the Met auxotroph M15MA through electroporation, yielding 5 × 106 independent clones. Aliquots of cultures containing the pooled transformants (500 μl) were mixed with 500 μl of frozen stock solution (65% glycerol/25 mM Tris/100 mM MgSO4) and stored at −80°C until needed.
Other Plasmids.
The XL-1 Blue strain of E. coli (Stratagene) was used for all recombinant DNA manipulations. The plasmid pAJL-61 is functionally equivalent to the previously described pQE-15 MRS (3) except that it encodes a copy of the lac repressor protein lacIq. pAJL-61 was constructed by digesting the 0.6-kb DHFR coding region from pQE-15 MRS with BamHI and HindIII. This fragment was ligated into similarly digested pQE-80L (Qiagen) to generate pAJL-60. The 2.5-kb cassette encoding MetRS was isolated from pQE-15 MRS upon digestion with NheI and was ligated into NheI-digested pAJL-60 to generate pAJL-61. The plasmid pAJL-80 encodes a 551-aa variant of MetRS bearing a C-terminal 6xHis-tag for affinity purification. A 1.7-kb DNA fragment was amplified from pAJL-20 by using the primers Cterm MRS forward and Cterm MRS reverse (see Supporting Text for sequences). This fragment was digested with BsaI and BglII and subsequently ligated to pQE-60 (Qiagen) digested with NcoI and BglII to generate pAJL-80. The plasmid pAJL-84 encodes the L13G mutant of MetRS and was generated by using QuikChange site-directed mutagenesis with the primers L13G forward and L13G reverse (see Supporting Text). The integrity of all constructs was confirmed by DNA sequencing.
OmpC Expression and Cell-Surface Labeling.
The expression of recombinant OmpC containing either AHA or ANL was carried out essentially as described (17). Briefly, 30 ml of M9 medium (M9 salts/0.2% glucose/1 mM MgSO4/25 mg/liter thiamine) supplemented with 40 mg/liter of each of the 20 canonical amino acids, 200 mg/liter ampicillin, and 35 mg/liter kanamycin was inoculated with either 400 μl of an overnight culture of M15MA[pAJL-20] or with a 1-ml aliquot of M15MA transformed with the MetRS library. When the OD600 of these cultures reached 0.9–1.0, the cells were pelleted by centrifugation at 5,000 × g for 5 min. The cells were resuspended in 30 ml of M9 medium supplemented with 19 aa (no Met) and incubated with shaking for 10 min at 37°C. The cells were pelleted again and resuspended in fresh M9 medium supplemented with 19 aa. The resulting cultures were divided into four 5-ml aliquots, which were supplemented with the following: Met (0.27 mM, 40 mg/liter), AHA (0.28 mM, 40 mg/liter), ANL (8 mM, 1.38 g/liter), or no analog. Expression of recombinant OmpC was induced at 37°C upon addition of IPTG to a final concentration of 1 mM. After 3 h, a 1-ml sample of each culture was pelleted and washed with 1 ml of sterile PBS (pH 7.4). The surfaces of these cells were tagged either by means of Cu-catalyzed azide–alkyne ligation or by treatment of the cells with biotin–PEO–cyclooctyne. For the Cu-catalyzed reaction, the cells were treated with 50 μM biotin–PEO–propargylamide, 200 μM triazole ligand, and 100 μM CuBr, which was delivered as an aqueous suspension as described (18). The Cu-catalyzed reaction proceeded for 16 h at 4°C with agitation. Alternatively, 1 ml of the washed cells was treated with 100 μM biotin–PEO–cyclooctyne for 16 h at 37°C with agitation. After the labeling reaction, the cells were washed twice more with 1 ml of PBS and treated with 2.5 μl of a 1 mg/ml solution of an avidin–Alexa Fluor 488 conjugate (Molecular Probes) for 2 h at 4°C with agitation. The cells were washed three more times with 1 ml of PBS to remove nonspecifically bound avidin.
Flow Cytometry and Cell Sorting.
All flow-cytometric analyses were carried out on a DAKO MoFlo cell sorter equipped with an argon ion laser emitting at 488 nm. All sorting was performed in sort single mode. Sort gates were set, and data analysis was performed with summit software (DAKO). Highly fluorescent cells were sorted from libraries expressing OmpC containing ANL by setting a gate in the fluorescence channel corresponding to the top 1% of the cells. Additional gates were set in the forward- and side-scatter channels to exclude scatterers of unusually large size. Approximately 105 cells were collected in a typical experiment, and 2–3 × 103 of the sorted cells were reanalyzed to check the quality of the sort. The remaining sorted cells were rescued upon inoculation into 10 ml of 2xYT medium supplemented with 200 mg/liter ampicillin and 35 mg/liter kanamycin. The regrown pools of cells were mixed 1:1 with frozen stock buffer and stored at −80°C. Alternatively, the sorted cells were plated on 2xYT agar supplemented with the same antibiotics to facilitate analysis of individual clones.
Recombinant DHFR Expression, Purification, and Analysis.
DHFR containing ANL was produced from M15MA[pAJL-61] and its variants harboring selected MetRS mutants under the culture conditions described above. The concentration of ANL was decreased to 1 mM (172 mg/liter) for production of DHFR in cells harboring the L13G mutant of MetRS. After protein expression, 10 ml of culture was pelleted and resuspended in 8 M urea. DHFR was purified under denaturing conditions by using Ni-NTA spin columns (Qiagen) as directed by the manufacturer. N-terminal sequencing was performed by the Peptide and Protein Microanalysis Laboratory at the California Institute of Technology. Protein yields were determined by measuring the A280 of a solution of the purified protein, assuming an extinction coefficient of 32,693 M−1·cm−1. Because ANL does not exhibit appreciable absorbance at 280 nm, the extinction coefficient was assumed to remain constant across samples of DHFR with differing extents of ANL incorporation.
MetRS Expression, Purification, and Activation Assays.
Superbroth medium (2 liters) was inoculated with 10 ml of an overnight culture of E. coli strain XL-1 Blue[pAJL-84]. When the culture reached an OD600 of 1.0, IPTG was added to a final concentration of 1 mM, and protein expression was induced at 37°C for 5 h. The cells were harvested by centrifugation (6,000 × g for 15 min). His-tagged MetRS was isolated under native conditions on Ni-NTA agarose (Qiagen) according to the manufacturer’s recommendations. The column eluent was buffer-exchanged into storage buffer (50 mM Tris/1 mM DTT) by using Bio-Rad PD-10 columns. The eluent was added to an equal mass of glycerol, mixed thoroughly, and frozen at −80°C until needed. Activation assays were carried out as described (32). The Met concentrations tested ranged from 150 μM to 5 mM, whereas the ANL concentrations ranged from 625 μM to 20 mM. The L13G mutant of MetRS was added to all reactions at a concentration of 100 nM. Data were fit to the Michaelis–Menten model by using sigmaplot (SPSS, Chicago) software.
Supplementary Material
Acknowledgments
We thank H. Jakubowski (New Jersey Medical School, Newark, NJ) and Y. Mechulam (École Polytechnique, Paliseau, France) for the generous donation of plasmids encoding MetRS. This work was supported by National Institutes of Health Grants GM58867 (to C.R.B.) and GM62523 (to D.A.T.) and by the U.S. Army Research Office through Institute for Collaborative Biotechnologies Grant DAAD19-03-D-0004.
Abbreviations
- MetRS
methionyl-tRNA synthetase
- aaRS
aminoacyl-tRNA synthetase
- ANL
azidonorleucine
- AHA
azidohomoalanine
- OmpC
outer membrane protein C
- PEO
polyethyleneoxide
- DHFR
dihydrofolate reductase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Budisa N. Angew. Chem. Int. Ed. 2004;43:6426–6463. doi: 10.1002/anie.200300646. [DOI] [PubMed] [Google Scholar]
- 2.Link A. J., Mock M. L., Tirrell D. A. Curr. Opin. Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Kiick K. L., van Hest J. C. M., Tirrell D. A. Angew. Chem. Int. Ed. 2000;39:2148–2152. [PubMed] [Google Scholar]
- 4.Kiick K. L., Weberskirch R., Tirrell D. A. FEBS Lett. 2001;502:25–30. doi: 10.1016/s0014-5793(01)02657-6. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y., Tirrell D. A. J. Am. Chem. Soc. 2001;123:11089–11090. doi: 10.1021/ja016652k. [DOI] [PubMed] [Google Scholar]
- 6.Wang P., Fichera A., Kumar K., Tirrell D. A. Angew. Chem. Int. Ed. 2004;43:3664–3666. doi: 10.1002/anie.200454036. [DOI] [PubMed] [Google Scholar]
- 7.Ibba M., Hennecke H. FEBS Lett. 1995;364:272–275. doi: 10.1016/0014-5793(95)00408-2. [DOI] [PubMed] [Google Scholar]
- 8.Sharma N., Furter R., Kast P., Tirrell D. A. FEBS Lett. 2000;467:37–40. doi: 10.1016/s0014-5793(00)01120-0. [DOI] [PubMed] [Google Scholar]
- 9.Bentin T., Hamzavi R., Salomonsson J., Roy H., Ibba M., Nielsen P. E. J. Biol. Chem. 2004;279:19839–19845. doi: 10.1074/jbc.M401278200. [DOI] [PubMed] [Google Scholar]
- 10.Kirshenbaum K., Carrico I. S., Tirrell D. A. Chembiochem. 2002;3:235–237. doi: 10.1002/1439-7633(20020301)3:2/3<235::AID-CBIC235>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Datta D., Wang P., Carrico I. S., Mayo S. L., Tirrell D. A. J. Am. Chem. Soc. 2002;124:5652–5653. doi: 10.1021/ja0177096. [DOI] [PubMed] [Google Scholar]
- 12.Kim W. Y., George A., Evans M., Conticello V. P. Chembiochem. 2004;5:928–936. doi: 10.1002/cbic.200400052. [DOI] [PubMed] [Google Scholar]
- 13.Tang Y., Tirrell D. A. Biochemistry. 2002;41:10635–10645. doi: 10.1021/bi026130x. [DOI] [PubMed] [Google Scholar]
- 14.Doring V., Mootz H. D., Nangle L. A., Hendrickson T. L., de Crecy-Lagard V., Schimmel P., Marliere P. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Brock A., Herberich B., Schultz P. G. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Schultz P. G. Angew. Chem. Int. Ed. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 17.Link A. J., Tirrell D. A. J. Am. Chem. Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 18.Link A. J., Vink M. K. S., Tirrell D. A. J. Am. Chem. Soc. 2004;126:10598–10602. doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Chan T. R., Hilgraf R., Fokin V. V., Sharpless K. B., Finn M. G. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 20.Mechulam Y., Schmitt E., Maveyraud L., Zelwer C., Nureki O., Yokoyama S., Konno M., Blanquet S. J. Mol. Biol. 1999;294:1287–1297. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 21.Serre L., Verdon G., Choinowski T., Hervouet N., Risler J. L., Zelwer C. J. Mol. Biol. 2001;306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 22.Crepin T., Schmitt E., Mechulam Y., Sampson P. B., Vaughan M. D., Honek J. F., Blanquet S. J. Mol. Biol. 2003;332:59–72. doi: 10.1016/s0022-2836(03)00917-3. [DOI] [PubMed] [Google Scholar]
- 23.Fersht A. R., Kaethner M. M. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 24.Fersht A. R. Biochemistry. 1977;16:1025–1030. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- 25.Englisch S., Englisch U., Vonderhaar F., Cramer F. Nucleic Acids Res. 1986;14:7529–7539. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey B. R., Georgiou G., Hayhurst A., Jeong K. J., Iverson B. L., Rogers G. K. Proc. Natl. Acad. Sci. USA. 2004;101:9193–9198. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiick K. L., Tirrell D. A. Tetrahedron. 2000;56:9487–9493. [Google Scholar]
- 28.Kiick K. L., Saxon E., Tirrell D. A., Bertozzi C. R. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangold J. B., Mischke M. R., Lavelle J. M. Mutat. Res. 1989;216:27–33. doi: 10.1016/0165-1161(89)90020-4. [DOI] [PubMed] [Google Scholar]
- 30.Agard N. J., Prescher J. A., Bertozzi C. R. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 31.Chan T. R., Hilgraf R., Sharpless K. B., Fokin V. V. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson T. L., Nomanbhoy T. K., Schimmel P. Biochemistry. 2000;39:8180–8186. doi: 10.1021/bi0004798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.