Abstract
We report that human acetyl-CoA synthetase 2 (AceCS2) is a mitochondrial matrix protein. AceCS2 is reversibly acetylated at Lys-642 in the active site of the enzyme. The mitochondrial sirtuin SIRT3 interacts with AceCS2 and deacetylates Lys-642 both in vitro and in vivo. Deacetylation of AceCS2 by SIRT3 activates the acetyl-CoA synthetase activity of AceCS2. This report identifies the first acetylated substrate protein of SIRT3. Our findings show that a mammalian sirtuin directly controls the activity of a metabolic enzyme by means of reversible lysine acetylation. Because the activity of a bacterial ortholog of AceCS2, called ACS, is controlled via deacetylation by a bacterial sirtuin protein, our observation highlights the conservation of a metabolic regulatory pathway from bacteria to humans.
Keywords: sir2, SIRT3, SIRT5, sirtuin
Reversible lysine acetylation is a highly regulated posttranslational protein modification, which is controlled by protein deacetylases and acetyltransferases (1, 2). Although the importance of reversible lysine acetylation of nuclear nonhistone and histone proteins is well established, the role of protein modification by reversible lysine acetylation in mitochondria is unknown.
The nicotinamide (NAM) adenine dinucleotide (NAD+)-dependent deacetylase silent information regulator 2 (sir2) is an important mediator of longevity in response to caloric restriction signals in Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster (3–9). Seven mammalian Sir2 homologs (SIRT1–7) are known (10–13). The recent discovery that SIRT3, SIRT4, and SIRT5 are found in mitochondria (14–17) suggests the existence of mitochondrial sirtuin substrate proteins.
Results and Discussion
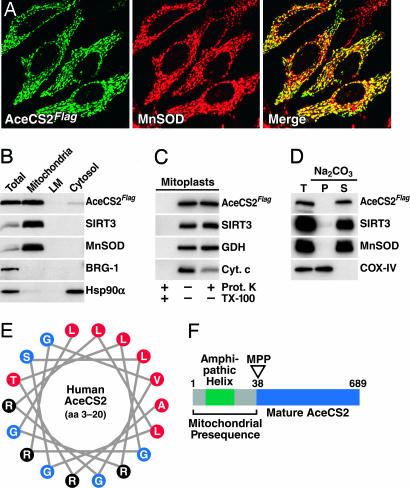
As a strategy to identify lysine-acetylated mitochondrial proteins that could be targeted by SIRT3, -4, or -5, we searched for human proteins with sequence similarity to the region surrounding the acetylated lysine residue found in acetyl-CoA synthetase (ACS) from Salmonella enterica. We chose the acetyl-lysine-containing region of S. enterica ACS, because it is a known substrate for the sirtuin CobB (18, 19). In a second step, human proteins showing high similarity were analyzed for their mitochondrial localization probability by using mitoprot ii and predotar Ver. 1.03 subcellular prediction software (20, 21). Our search yielded a human acetyl-CoA synthetase protein with a high probability of being localized to mitochondria (mitoprot ii, 0.997; predotar, 0.94; maximum score = 1), which displayed high sequence similarity to the murine acetyl-CoA synthetase 2, AceCS2 (22). To test whether the human AceCS2 enzyme is a mitochondrial protein, we cloned the ORF of human AceCS2 and expressed it in HeLa cells as a Flag-tagged protein. Confocal laser scanning microscopy showed a mitochondrial staining pattern for AceCS2Flag that overlapped with the staining pattern observed for the endogenous mitochondrial matrix protein manganese superoxide dismutase (MnSOD; Fig. 1A).
Fig. 1.
Human AceCS2 is a soluble mitochondrial matrix protein. (A) Mitochondrial localization of human AceCS2Flag in HeLa cells. (B) Equal amounts (30 μg) of total cell extract (Total), mitochondria, light membranes (LM), and cytosolic proteins were analyzed by immunoblotting. (C) Submitochondrial localization of AceCS2Flag. (D) AceCS2Flag is a soluble protein. Mitochondria were isolated and either directly solubilized in SDS sample buffer (T, total) or extracted with sodium carbonate (Na2CO3, pH 11.5). The alkaline extract was separated into a membrane fraction (P, pellet) and a fraction containing sodium carbonate soluble proteins (S, supernatant) by ultracentrifugation. (E) The N terminus of human AceCS2 contains an amphipathic α-helix. A region of AceCS2 (amino acids 3–20) predicted by computational secondary structure analysis to form an α-helix was plotted as a helical wheel. Black, basic residues; red, hydrophobic residues; blue, neutral residues. (F) Illustration of the mitochondrial presequence and cleavage site of human AceCS2 as determined by N-terminal protein sequencing and liquid chromatography-tandem MS analysis. For details, see text. MPP, mitochondrial matrix processing peptidase.
To further verify the mitochondrial localization of human AceCS2, subcellular fractions were prepared from human embryonic kidney 293 (HEK293) cells stably expressing AceCS2Flag. We detected AceCS2Flag in the mitochondrial fraction along with SIRT3 and MnSOD, as expected (Fig. 1B). Immunoblotting of each fraction for the nuclear marker protein BRG-1 and the cytosolic marker protein Hsp90α confirmed the purity of the fractions (Fig. 1B).
To further define the submitochondrial localization of AceCS2, mitoplasts were prepared from mitochondria containing AceCS2Flag. Mitoplast preparation ruptures the outer mitochondrial membrane and makes the intermembrane space accessible to proteinase K. Treatment of mitoplasts with proteinase K led to a loss of the intermembrane space protein cytochrome c, whereas it did not affect the mitochondrial matrix proteins glutamate dehydrogenase (GDH) and SIRT3 (Fig. 1C). AceCS2Flag was also not affected by proteinase K treatment of mitoplasts, suggesting that it was localized in the mitochondrial matrix, like GDH and SIRT3 (Fig. 1C). All proteins were completely digested by proteinase K when the nonionic detergent TX-100 was added during the incubation of the mitoplasts with proteinase K (Fig. 1C, left lane).
To determine whether AceCS2 is integrally attached to the inner side of the inner mitochondrial membrane or soluble in the mitochondrial matrix, we extracted mitochondria with sodium carbonate (pH 11.5). This treatment releases soluble proteins but not integral membrane proteins. AceCS2 was completely released by sodium carbonate treatment and exhibited a distribution pattern like the soluble matrix proteins SIRT3 and MnSOD (Fig. 1D). Under the conditions used, the integral membrane protein COX-IV stayed associated with the membranes (Fig. 1D).
Nuclear-encoded proteins destined for the mitochondrial matrix often carry an N-terminal presequence, which is recognized by the mitochondrial translocase of the outer membrane complex and is cleaved off after import into the matrix (23). This N-terminal presequence often contains an α-helix and several basic residues. These characteristics are found in the N terminus of human AceCS2, including several arginine residues (Fig. 1E). N-terminal sequencing of immunoprecipitated AceCS2Flag by Edman degradation revealed that the first 37 amino acids of the ORF were missing from the protein, consistent with N-terminal processing of AceCS2 in the mitochondrial matrix. The identification of alanine 38 as the N-terminal amino acid of immunoprecipitated AceCS2Flag by liquid chromatography-tandem MS confirmed the protein sequencing results (data not shown). The existence of a mitochondrial matrix processing peptidase (MPP) R-2 motif immediately upstream of the N terminus of the mature AceCS2 protein suggests that AceCS2 is processed by MPP in the mitochondrial matrix (ref. 24; see Fig. 1F). These findings confirm the existence of a human mitochondrial acetyl-CoA synthetase located in the mitochondrial matrix.
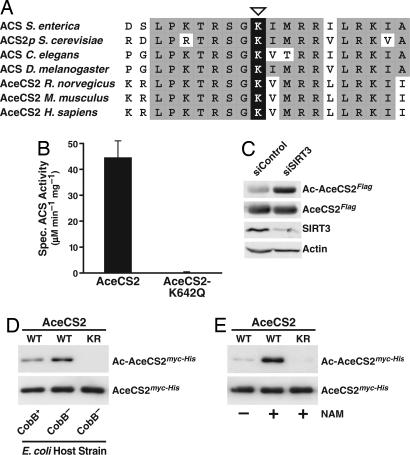
Based on the conservation of the active-site region of S. enterica ACS with human and murine AceCS2 and acetyl-CoA synthetases from other species (Fig. 2A), we tested whether mutation of Lys-642 would affect the acetyl-CoA synthetase activity of overexpressed human AceCS2 immunoprecipitated from HEK293 cells. Replacement of Lys-642 with glutamine, which mimics a constitutively acetylated lysine residue, yielded a completely inactive AceCS2 protein (Fig. 2B), indicating that Lys-642 is of critical importance for AceCS2 function. Both wild-type AceCS2 and mutant AceCS2-K642Q were expressed at similar levels (Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site).
Fig. 2.
The conserved active-site lysine of human AceCS2 is subject to reversible acetylation. (A) Multiple sequence alignment of the active-site region of acetyl-CoA synthetases from different species and human AceCS2. The active-site lysine is marked by an open triangle. (B) Lys-642 is critical for AceCS2 function. AceCS2HA or AceCS2-K642QHA were expressed in HEK293 cells and immunoprecipitated, and the specific acetyl-CoA synthetase (ACS) activity was determined. Data are means + SD from three independent acetyl-CoA synthetase activity assays. (C) Reduction of endogenous SIRT3 levels increases the acetylation level of AceCS2Flag. Double-stranded siRNAs directed against human SIRT3 (siSIRT3) or firefly luciferase GL3 (siControl) were transfected into HEK293 cells stably expressing AceCS2Flag. Acetylation of immunoprecipitated AceCS2Flag was analyzed by immunoblotting with antibodies to acetylated lysine (Ac-AceCS2Flag). Membranes were stripped and reprobed for total AceCS2 amounts (AceCS2Flag). Immunoblotting of total cell extracts with α-SIRT3 and α-actin antibodies confirmed the efficiency and specificity of the siRNA treatment. (D) Removal or inhibition of the sirtuin CobB results in hyperacetylation of the active-site lysine of human AceCS2 in bacteria. Equal amounts of purified human myc-His-tagged AceCS2 (WT) or AceCS2-K642R (KR) from CobB− or CobB+ E. coli were analyzed by immunoblotting with an acetyl-lysine-specific antibody (Ac-AceCS2myc-His). Blots were stripped and reprobed with α-myc antibodies to show total levels of AceCS2 (AceCS2myc-His). (E) Inhibition of bacterial sirtuins by NAM during expression of recombinant human AceCS2 protein causes hyperacetylation of human AceCS2 at Lys-642. Equal amounts of the purified recombinant proteins were analyzed as in D.
To determine whether human AceCS2 is acetylated in vivo, AceCS2Flag was immunoprecipitated from cells after small interfering RNA (siRNA)-mediated knockdown of SIRT3 and prepared for MS (Fig. 7, which is published as supporting information on the PNAS web site). Liquid chromatography-tandem MS analysis of the tryptic digests indicated the presence of an acetylated lysine residue (acK) in position 642 of AceCS2 (Fig. 8, which is published as supporting information on the PNAS web site).
To further address whether SIRT3 has a role in deacetylating AceCS2 in cells, we treated AceCS2Flag-expressing HEK293 cells with siRNAs against SIRT3 or control siRNAs and examined the acetylation status of immunoprecipitated AceCS2Flag by immunoblotting with antibodies to acetylated lysine (Fig. 2C). Acetylation of AceCS2Flag was increased after knockdown of endogenous SIRT3, suggesting that SIRT3 affects AceCS2Flag acetylation in vivo (Fig. 2C).
Like S. enterica, Escherichia coli contains the sirtuin CobB. Given a potentially conserved role for sirtuin-mediated deacetylation in the control of acetyl-CoA synthetase proteins throughout evolution, we tested whether expression of human AceCS2 in an E. coli K-12 strain lacking the sirtuin CobB would induce hyperacetylation of AceCS2. Human AceCS2 purified from a CobB-knockout strain reacted more strongly with an acetyl-lysine-specific antibody than AceCS2 purified from the parental CobB wild-type strain (Fig. 2D). This acetylation was specific to AceCS2 Lys-642, because a mutation of Lys-642 to arginine, which cannot be acetylated, abrogated the reactivity with the α-acetyl-lysine antibody (Fig. 2D, right lane). The specificity of the acetyl-lysine-specific antibody was further confirmed by immunoblot analysis of synthetic peptides coupled to a carrier protein. The acetyl-lysine-specific antibody reacted only with the peptide containing an acetylated K642 residue and showed no reactivity with the nonacetylated K642-containing peptide (Fig. 9, which is published as supporting information on the PNAS web site).
We further showed that treatment of a wild-type E. coli strain (DH5α; Invitrogen) with the sirtuin inhibitor NAM resulted in the hyperacetylation of recombinant human AceCS2 (Fig. 2E). This increase in acetylation was specific to Lys-642 of AceCS2, because NAM treatment did not induce acetylation of a mutant AceCS2 protein carrying an arginine residue in position 642 (Fig. 2E, right lane). Analysis of purified recombinant AceCS2 from NAM-treated bacteria by liquid chromatography-tandem MS verified acetylation of a single lysine in the active site of AceCS2, Lys-642 (Fig. 10, which is published as supporting information on the PNAS web site).
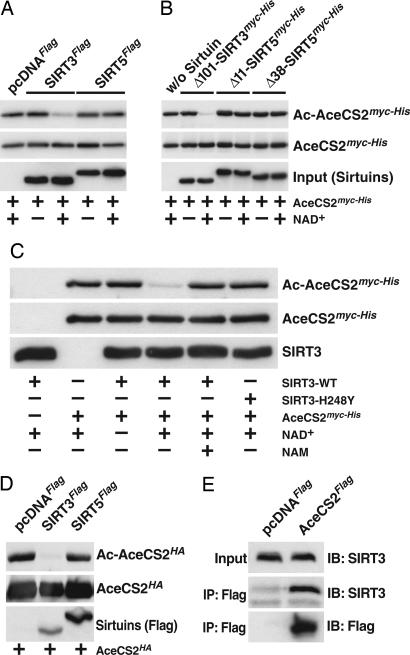
Acetylation of the active-site lysine in S. enterica ACS inactivates the enzyme (19). S. enterica CobB, which deacetylates ACS, is a class III sirtuin that is most closely related to human SIRT5. SIRT5 localizes to mitochondria and is subject to N-terminal processing (15). To test whether SIRT5 could deacetylate AceCS2, in vitro deacetylation assays were performed. We prepared immunoprecipitated Flag-tagged sirtuins from HEK293 cells as described by North et al. (25). Although immunoprecipitated Flag-tagged SIRT5 has low but detectable activity on a chemically acetylated H4 peptide (25), it failed to deacetylate AceCS2 (Fig. 3A). In contrast, immunoprecipitated Flag-tagged SIRT3 deacetylated AceCS2 in a NAD+-dependent manner (Fig. 3A). Flag-tagged SIRT4, another mitochondrial sirtuin with no reported deacetylase activity (25), did not deacetylate AceCS2 (data not shown).
Fig. 3.
SIRT3 but not SIRT5 deacetylates AceCS2. (A) Recombinant human AceCS2myc-His purified from NAM-treated E. coli was incubated with Flag-tagged SIRT3 or SIRT5 immunoprecipitated from HEK293 cells. As a control, α-Flag immunoprecipitates from pcDNAFlag-expressing HEK293 cells were used in the reactions (left lane). Deacetylation reactions were incubated in the presence or absence of NAD+. All reactions were carried out in the presence of trichostatin A. Reactions were stopped by the addition of SDS sample buffer, boiled, and analyzed by immunoblotting. Acetylated AceCS2myc-His was detected with acetyl-lysine-specific antibodies (Ac-AceCS2myc-His). Blots were stripped and probed for total levels of AceCS2 with α-myc antibodies (AceCS2myc-His). The presence of Flag-tagged SIRT3 and SIRT5 was verified by immunoblotting with α-Flag antibodies {bottom blot [Input (Sirtuins)]}. (B) Recombinant human AceCS2 purified from NAM-treated E. coli was incubated with purified recombinant sirtuins in the presence or absence of NAD+ and in the presence of trichostatin A and analyzed as described in A. Recombinant SIRT3 or SIRT5 proteins were detected by probing with α-myc-antibodies {bottom blot [Input (Sirtuins)]}. (C) SIRT3, but not a catalytically inactive mutant of SIRT3, deacetylates AceCS2 in vitro. Deacetylation assays were performed and analyzed as described above. Where indicated, NAM was included during the incubation. (D) Overexpression of SIRT3 decreases the acetylation of ectopically expressed AceCS2. COS-1 cells were cotransfected with AceCS2HA and pcDNAFlag, SIRT3Flag, or SIRT5Flag. Acetylation of immunoprecipitated AceCS2HA was analyzed by immunoblotting with antibodies to acetylated lysine (Ac-AceCS2HA). Membranes were stripped and reprobed for total AceCS2 amounts (AceCS2HA) by probing with an α-hemagglutinin antibody. The expression of SIRT3Flag and SIRT5Flag in the total cell lysate was verified by immunoblotting with α-Flag antibodies [Sirtuins (Flag)]. (E) AceCS2 and SIRT3 coimmunoprecipitate from cells. α-Flag immune complexes from HEK293 cell lines stably expressing AceCS2Flag or an empty Flag-control vector (pcDNAFlag) were analyzed for the presence of endogenous SIRT3.
To confirm our findings regarding SIRT3 and SIRT5, we expressed and purified recombinant sirtuins. Based on the finding that SIRT5 is N-terminally truncated (15) and on the presence of two putative mitochondrial matrix processing peptidase R-2 motifs in its N terminus, two different recombinant SIRT5 proteins were used, lacking either the first 11 or 38 amino acids, respectively. Again, although recombinant SIRT3 effectively deacetylated AceCS2, both recombinant SIRT5 proteins failed to do so (Fig. 3B). As expected, deacetylation of AceCS2 by SIRT3 strictly depended on the presence of NAD+ (Fig. 3B). Also, the sirtuin inhibitor NAM completely prevented the deacetylation of AceCS2 by SIRT3, and the catalytically inactive SIRT3-H248Y mutant (17) did not deacetylate AceCS2, despite the presence of NAD+ (Fig. 3C).
To determine whether SIRT5 deacetylates AceCS2 in cells, we coexpressed AceCS2HA and SIRT3Flag or SIRT5Flag in COS-1 cells, immunoprecipitated AceCS2HA, and analyzed the immune complexes with antibodies to acetylated lysine (Fig. 3D). Overexpression of SIRT3 decreased the acetylation levels of ectopically expressed AceCS2, whereas SIRT5 had no effect, despite much higher levels of expression (Fig. 3D).
Interestingly, endogenous SIRT3 coimmunoprecipitated with AceCS2Flag from HEK293 cells (Fig. 3E). Together with the findings described above, this suggests that SIRT3 is the bona fide deacetylase of mitochondrial AceCS2.
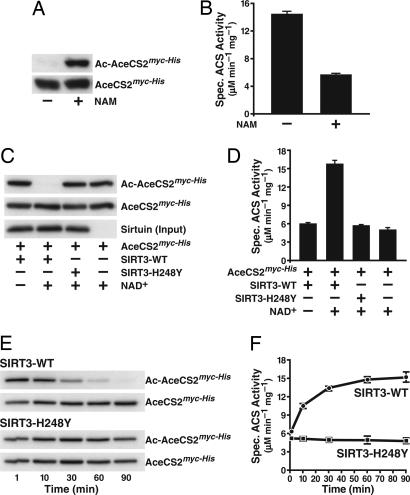
To address whether acetylation of AceCS2 at Lys-642 controls its acetyl-CoA synthetase activity, we took advantage of our finding that NAM treatment of E. coli during expression of human AceCS2 significantly increased its levels of acetylation (Fig. 4A). Measurement of the specific activity of AceCS2 purified from bacteria, treated or not with NAM, indicated that hyperacetylated AceCS2 had a significantly lower specific activity (Fig. 4B). These observations suggested that acetylation of AceCS2 inhibited its enzymatic activity, consistent with what has been shown for ACS in bacteria (19).
Fig. 4.
Acetylation of Lys-642 controls the acetyl-CoA synthetase activity of human AceCS2. (A) Recombinant human AceCS2 was expressed in bacteria in the presence or absence of NAM. Acetylation levels and total amounts of AceCS2 were analyzed by immunoblotting with acetyl-lysine-specific antibodies or α-myc antibodies. (B) Specific activity of purified AceCS2 expressed in the presence or absence of NAM. The data are means + SD from three independent experiments. (C) NAD+-dependent deacetylation of AceCS2 by SIRT3 but not by SIRT3-H248Y. The bottom blot indicates that equal amounts of SIRT3 or SIRT3-H248Y were added to the deacetylation reactions. (D) Deacetylation of AceCS2 by SIRT3 activates its acetyl-CoA synthetase activity. Equal amounts of AceCS2 were incubated in the absence or presence of SIRT3 or a catalytically inactive SIRT3 mutant (SIRT3-H248Y). Where indicated, NAD+ was added during the incubation. The deacetylation reaction was stopped by the addition of NAM, and the specific activity of AceCS2 was determined. Data are means + SD from three independent activity determinations. (E) Time-dependent activation of AceCS2 by SIRT3. Recombinant human AceCS2 purified from NAM-treated bacteria was incubated with recombinant SIRT3 or SIRT3-H248Y. Samples were brought to 32°C, and NAD+ was added. At the indicated time points, aliquots of the reaction were removed, mixed with NAM, and incubated on ice. The immunoblots show the progressive deacetylation of AceCS2 by SIRT3 but not by SIRT3-H248Y. (F) The specific activity of AceCS2 after incubation with recombinant SIRT3 or SIRT3-H248Y for the indicated time was determined. Means ± SD from three independent activity assays are shown.
To further test this hypothesis, we incubated hyperacetylated AceCS2 purified from NAM-treated E. coli with SIRT3 in the presence of NAD+. This treatment led to a complete deacetylation of AceCS2 (Fig. 4C). No deacetylation of AceCS2 was observed when NAD+ was omitted from the reaction, or when a catalytically inactive SIRT3 mutant (H248Y) was used (Fig. 4C). Measurement of the enzymatic activity of AceCS2 with different levels of acetylation, as described above, showed that deacetylation by SIRT3 was associated with a significant increase in enzymatic activity (Fig. 4D).
In an additional experiment, we found that incubation of AceCS2 with SIRT3 and NAD+ caused a progressive deacetylation of AceCS2 over time, which could not be observed with the inactive SIRT3-H248Y mutant (Fig. 4E). Again, deacetylation of AceCS2 was associated with a progressive increase in enzymatic activity (Fig. 4F). We conclude that the acetylation status of Lys-642 controls the activity of human mitochondrial AceCS2.
In S. enterica, acetylation of ACS by the protein acetyltransferase Pat inactivates the enzyme, whereas deacetylation by the sirtuin CobB reactivates it (19, 26). The acetyl-CoA synthetase reaction, which results in the formation of acetyl-CoA from acetate, ATP, and CoA, proceeds in two steps. In a first step, acetate is activated to acetyl-adenosine monophosphate (acetyl-AMP). In a second step, acetyl-AMP is converted to acetyl-CoA by the thioester bond-forming activity of ACS, and acetyl-CoA and AMP are released sequentially (reviewed in ref. 27). It has been shown that lysine acetylation of the active-site lysine (Lys-609) of bacterial ACS specifically inhibits the ATP-dependent adenylation of acetate to acetyl-AMP, whereas the second step of the ACS reaction remains unaffected (19). As illustrated in Fig. 2A, Lys-642 of human AceCS2 corresponds to the active-site lysine Lys-609 of ACS from S. enterica. Given the conservation of the active-site region between bacterial ACS and human AceCS2, we propose that acetylation of Lys-642 of AceCS2 interferes with the first reaction step involving the activation of acetate to acetyl-AMP.
AceCS2 is the first example of a mitochondrial protein being controlled by reversible lysine acetylation and the first target protein for a mitochondrial sirtuin deacetylase. A schematic overview of the regulation of AceCS2 by reversible lysine acetylation is given in Fig. 5. Our data support the model that activation of AceCS2 by SIRT3 is an evolutionarily conserved metabolic control mechanism. The interconversion of acetyl-CoA (in former times known as “activated acetate”) and acetate, referred to as the “acetate switch,” exists in archaea, eubacteria, and eukaryotes and provides the cell with opportunities to recover NAD+, produce ATP, and replenish CoA stores (reviewed in ref. 28). The acetate switch is controlled by the acetate-scavenging enzyme acetyl-CoA synthetase. Metabolic pathways such as energy production in the tricarboxylic acid cycle and cholesterol and fatty acid biosynthesis require acetyl-CoA as an intermediate.
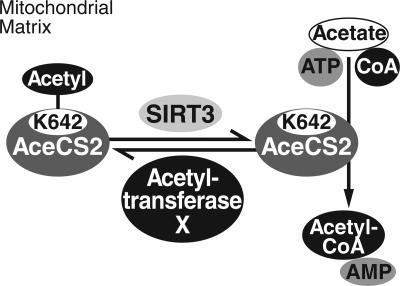
Fig. 5.
Schematic illustration of AceCS2 regulation by reversible lysine acetylation of Lys-642. Deacetylation of AceCS2 by SIRT3 activates its acetyl-CoA synthetase activity. The identity of the protein acetyltransferase of AceCS2 is unknown and is therefore labeled Acetyltransferase X.
Yeast critically require acetyl-CoA synthetases for growth, but this is not the case for mammalian cells. The majority of acetyl-CoA in mammalian cells is produced in pathways that do not depend on acetyl-CoA synthetase activity. These are the conversion of pyruvate to acetyl-CoA by pyruvate dehydrogenase and β-oxidation, which results in the formation of acetyl-CoA as an end product. However, in mammals under some circumstances, large amounts of acetate are produced that need to be activated by acetyl-CoA synthetase. For example, under ketogenic conditions such as prolonged fasting or diabetes, the liver releases substantial amounts of acetate into the bloodstream (29–31). In addition, the hepatic acetyl-CoA hydrolase, which produces acetate, is activated under ketogenic conditions (32). Utilization of the released acetate in extrahepatic tissues requires the action of acetyl-CoA synthetases. The findings that murine AceCS2 is abundant in heart and skeletal muscle but absent from the liver and induced under ketogenic conditions suggest that AceCS2 plays an important role in acetate conversion for energy production under ketogenic conditions (22). Based on the finding that a bacterial sirtuin controls the activity of acetyl-CoA synthetase in S. enterica and on the presence of sirtuins in all three kingdoms, a universal connection between central metabolism and sirtuins has been proposed (18, 19). Our findings showing that AceCS2 can be inactivated by acetylation of its active-site lysine and reactivated by a mitochondrial sirtuin support these claims and demonstrate the conservation of these pathways from bacteria to mammalian mitochondria.
Materials and Methods
Cell Culture and Plasmid Construction.
HEK293, COS-1, and HeLa cells were cultured in DMEM supplemented with 10% FCS. All expression constructs were generated by using PCR-based standard cloning strategies, and all expression constructs were verified by DNA sequencing. The human AceCS2 coding sequence was PCR-amplified from human full-length Mammalian Gene Collection cDNA (GenBank accession no. BC039261; obtained through Open Biosystems, www.openbiosystems.com) and cloned into the pcDNA3.1+ (Invitrogen)-derived vectors pcDNAFlag or pcDNAHA to yield AceCS2 with a C-terminal Flag- or hemagglutinin-tag. Based on N-terminal protein-sequencing results, the ORF corresponding to mature AceCS2 (amino acids 38–689) was cloned into pTrcHis2C (Invitrogen). Recombinant expression vectors encoding mature human SIRT3 (amino acids 102–399; ref. 17) or SIRT5 (amino acids 12–310 or 39–310) were constructed by PCR amplification and cloning into pTrcHis2C. The templates used for PCR amplification of the SIRT3 and SIRT5 coding sequences were as described (17, 25).
Expression and Purification of Recombinant Proteins.
Transformed E. coli DH5α bacteria (Invitrogen) were grown to an A600 nm = 0.4, and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 16 h. 6×His-tagged proteins were purified under native conditions at 4°C by using Ni-NTA agarose (Qiagen, Valencia, CA). Purified proteins were dialyzed, adjusted to 0.5 g per liter, and stored frozen at −80°C. To induce acetylation of recombinant AceCS2, NAM (50 mM) was added during protein expression (16 h, 25°C). Parental E. coli K-12 BW25113 and a CobB-deficient single-gene knockout (KO) mutant (JW1106) of the same strain (33) were obtained from the Systematic Knock Out Strains of E. coli K-12 Collection of GenoBase (http://ecoli.aist-nara.ac.jp/gb5/Resources/deletion/deletion.html). Technical details regarding the generation of the single-gene KO mutants can be found at http://ecoli.aist-nara.ac.jp/gb5/Resource_download.html.
Immunoblotting.
Antibodies used were α-mtHsp70 (Affinity Bioreagents, Neshanic Station, NJ), α-Hsp90α, and α-manganese superoxide dismutase (Stressgen Biotechnologies, Victoria, Canada), α-cytochrome c oxidase subunit IV (Molecular Probes), α-Flag M2 or rabbit polyclonal α-Flag (Sigma), α-HA (12CA5 and 3F10; Roche Diagnostics), acetylated-lysine polyclonal antibody (Cell Signaling Technology, Beverly, MA), α-actin C4 (ICN), α-cytochrome c (Pharmingen), and α-BRG-1 and α-c-myc (Santa Cruz Biotechnology). SIRT3 antiserum was raised as described (17). Immunoblots were developed with enhanced chemiluminescence (Amersham Pharmacia Biosciences) or West SuperSignal reagent (Pierce).
Immunoprecipitation.
Cells were lysed in ice-cold NP1 buffer (1% Nonidet P-40/150 mM NaCl/0.5 mM EDTA/50 mM Tris·HCl, pH 7.4) containing protease inhibitor mixture (Roche). Flag-tagged proteins were immunoprecipitated and washed four times in NP1 buffer. In coimmunoprecipitation experiments, NP1 buffer containing 300 mM NaCl was used. Immunoprecipitated Flag-tagged sirtuins to be used in deacetylation assays were washed three times in NP1 buffer containing 500 mM NaCl and twice in sirtuin deacetylase buffer (SDAC) [50 mM Tris·HCl (pH 9.0)/4 mM MgCl2/50 mM NaCl/0.5 mM DTT].
Confocal Microscopy.
HeLa cells were fixed and permeabilized as described (17). Cells were costained with monoclonal α-Flag M2 (1:500) and polyclonal α-MnSOD (1:300) antibodies, followed by incubation with α-mouse-Cy2 antibodies and α-rabbit-Cy5 antibodies suitable for multilabeling experiments (Jackson ImmunoResearch).
Subcellular Fractionation and Submitochondrial Localization Experiments.
Subcellular fractionation was performed as described (17, 34). Mitoplast formation and protease accessibility experiments were performed according to published protocols (17, 35). Carbonate extraction of mitochondria was performed as described (17, 36).
N-Terminal Protein Sequencing by Edman Degradation.
Immunoprecipitated AceCS2Flag was subjected to SDS/PAGE, transferred to poly(vinylidene difluoride) membrane, visualized by Ponceau S staining (Sigma), cut, and submitted to the Stanford PAN Facility (Stanford, CA) for N-terminal sequencing by Edman degradation according to standard protocols.
In Vitro Deacetylation Assays.
Equimolar amounts of purified recombinant AceCS2 and purified recombinant sirtuins were incubated in SDAC buffer in the presence or absence of NAD+ (1 mM), in the presence or absence of NAM (10 mM), in the presence of trichostatin A (500 nM) for 3 h at 32°C. For time-course deacetylation experiments, aliquots of the deacetylation reaction were removed at the indicated time points, mixed with 10 mM NAM, and incubated on ice until further analysis.
Acetyl-CoA Synthetase Activity Assays.
The activity of purified AceCS2 was measured as described (37, 38). Each reaction contained 100 mM hydroxylamine (preneutralized with KOH), 50 mM Tris·HCl (pH 8.0), 20 mM potassium acetate, 10 mM MgCl2, 10 mM ATP, 2 mM DTT, and 1 mM CoA. Reactions were preincubated at 35°C for 5 min before the addition of purified AceCS2. Samples without AceCS2 served as a blank, and formation of acetylhydroxamate by acetyl-phosphate (Sigma) served as a standard (38). No acetyl-CoA synthetase activity was detectable in the absence of CoA.
RNA Interference Experiments.
Double-stranded siRNAs (100 nM; Dharmacon Research, Lafayette, CO) directed against human SIRT3 or firefly luciferase GL3 control siRNAs were transfected into HEK293 by using oligofectamine (Invitrogen) according to the manufacturer’s recommendations. Five days after transfection, cells were lysed in NP1 buffer containing protease inhibitors, 10 μM trichostatin A, and 10 mM NAM.
Supplementary Material
Acknowledgments
We thank B. J. North, R. Vries, S. Kauder, T. Mahmoudi, M. Parra, N. Darani, and R. Streeper for helpful comments on the manuscript and J. Carroll for help with graphics. J.B. is supported by a grant from the Carlsberg Foundation. The Center for Experimental Bioinformatics is supported by a grant from the Danish National Research Foundation. E.V. is a senior scholar of the Ellison Medical Foundation. This work was supported by funds from the Sandler Foundation Program in Basic Sciences (to E.V.).
Abbreviations
- ACS
acetyl-CoA synthetase
- AceCS2
acetyl-CoA synthetase 2
- acetyl-AMP
acetyl-adenosine monophosphate
- NAM
nicotinamide
- NAD+
NAM adenine dinucleotide
- HEK293
human embryonic kidney 293
- SIRTn
silent information regulator homolog n
- siRNA
small interfering RNA
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Han K. K., Martinage A. Int. J. Biochem. 1992;24:19–28. doi: 10.1016/0020-711x(92)90225-p. [DOI] [PubMed] [Google Scholar]
- 2.Yang X. J. BioEssays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M., McVey M., Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tissenbaum H. A., Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 5.Lin S. J., Defossez P. A., Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 6.Rogina B., Helfand S. L. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S. J., Ford E., Haigis M., Liszt G., Guarente L. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S. J., Kaeberlein M., Andalis A. A., Sturtz L. A., Defossez P. A., Culotta V. C., Fink G. R., Guarente L. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 9.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frye R. A. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 11.Frye R. A. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 12.Blander G., Guarente L. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 13.North B. J., Verdin E. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T., Wang F., Stieren E., Tong Q. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 15.Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onyango P., Celic I., McCaffery J. M., Boeke J. D., Feinberg A. P. Proc. Natl. Acad. Sci. USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwer B., North B. J., Frye R. A., Ott M., Verdin E. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starai V. J., Takahashi H., Boeke J. D., Escalante-Semerena J. C. Genetics. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 20.Small I., Peeters N., Legeai F., Lurin C. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 21.Claros M. G., Vincens P. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 22.Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. J. Biol. Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 23.Rehling P., Brandner K., Pfanner N. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 24.Ito A. Biochem. Biophys. Res. Commun. 1999;265:611–616. doi: 10.1006/bbrc.1999.1703. [DOI] [PubMed] [Google Scholar]
- 25.North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 26.Starai V. J., Escalante-Semerena J. C. J. Mol. Biol. 2004;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Starai V. J., Escalante-Semerena J. C. Cell Mol. Life Sci. 2004;61:2020–2030. doi: 10.1007/s00018-004-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe A. J. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckley B. M., Williamson D. H. Biochem. J. 1977;166:539–545. doi: 10.1042/bj1660539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seufert C. D., Graf M., Janson G., Kuhn A., Soling H. D. Biochem. Biophys. Res. Commun. 1974;57:901–909. doi: 10.1016/0006-291x(74)90631-7. [DOI] [PubMed] [Google Scholar]
- 31.Yamashita H., Kaneyuki T., Tagawa K. Biochim. Biophys. Acta. 2001;1532:79–87. doi: 10.1016/s1388-1981(01)00117-2. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga T., Isohashi F., Nakanishi Y., Sakamoto Y. Eur. J. Biochem. 1985;152:331–336. doi: 10.1111/j.1432-1033.1985.tb09202.x. [DOI] [PubMed] [Google Scholar]
- 33.Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Liu X., Bhalla K., Kim C. N., Ibrado A. M., Cai J., Peng T. I., Jones D. P., Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 35.Ryan M. T., Voos W., Pfanner N. In: Mitochondria. Pon L. A., Schon E. A., editors. Vol. 65. New York: Academic; 2001. pp. 190–213. [Google Scholar]
- 36.Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones M. E., Lipmann F. Methods in Enzymology. Vol. 1. New York: Academic; 1955. pp. 585–591. [Google Scholar]
- 38.Barak R., Prasad K., Shainskaya A., Wolfe A. J., Eisenbach M. J. Mol. Biol. 2004;342:383–401. doi: 10.1016/j.jmb.2004.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.