Abstract
Up to 15% of acute myeloid leukemias (AMLs) are characterized by the abnormal expression of the eight-twenty-one (ETO) transcriptional corepressor within an AML1-ETO fusion protein. The t(8;21) chromosomal translocation serves not only to disrupt WT AML1 function but also to introduce ETO activity during hematopoiesis. AML1-ETO was recently shown to inhibit E protein transactivation by physically displacing WT coactivator proteins in an interaction mediated by ETO. Here, we present the 3D solution structure of the human ETO TAFH (eTAFH) domain implicated in AML1-ETO:E protein interactions and report an unexpected fold similarity to paired amphipathic helix domains from the transcriptional corepressor Sin3. We identify and characterize a conserved surface on eTAFH that is essential for ETO:E protein recognition and show that the mutation of key conserved residues at this site alleviates ETO-based silencing of E protein transactivation. Our results address uncharacterized aspects of the corepression mechanism of ETO and suggest that eTAFH may serve to recruit ETO (or AML1-ETO) to DNA-bound transcription factors. Together, these findings imply that a cofactor exchange mechanism, analogous to that described for E protein inhibition, may represent a common mode of action for ETO.
Keywords: corepressor, NMR, protein structure, sin3
Core binding factor (CBF) is an essential hematopoietic transcriptional regulator that is frequently mutated in acute myeloid leukemias (AMLs) (1, 2). In up to 15% of AML cases, a translocation between chromosomes 8 and 21 replaces a portion of the gene encoding the CBF subunit AML1 with a gene specifying an unrelated corepressor protein, eight-twenty-one (ETO) (3, 4). The subsequent fusion protein, AML1-ETO, contains the DNA-binding domain from AML1 fused to most of ETO. Expression of this chimeric protein and the consequent disruption of WT AML1 function are key events in the production of an AML phenotype (5). In WT cells CBF regulates chromatin remodeling of target genes by recruiting coactivators and histone acetyl transferases (6). In t(8;21) cells, the AML1-ETO fusion still binds CBF target DNA sequences through the AML1 DNA-binding domain; however, the ETO fusion partner functionally dominates by recruiting proteins involved in chromatin repression (e.g., corepressors and histone deacetylases) (1, 7). The formation of a corepressor complex involving AML1-ETO serves to shut down gene expression of genes normally activated by CBF.
As an alternative or complementary mechanism involving distinct target genes, recent studies have shown that AML1-ETO can silence E protein transactivation (8). E proteins comprise a family of widely expressed transcription factors involved in regulating cell growth, differentiation, and apoptosis (9–11). DNA-bound E proteins interact with the histone acetyl transferase p300/CREB binding protein, leading to histone modifications that facilitate transcription initiation (8, 12). However, AML1-ETO very strongly interacts with E proteins through an N-terminal activation domain (AD1), which not only prevents p300/CREB binding protein-mediated transactivation but leads to active transcription repression through formation of a histone deacetylase-containing complex (8). In this capacity ETO acts independently of CBF and functionally resembles corepressors like nuclear receptor corepressor (N-CoR), Sin3, and Silencing-Mediator for Retinoid/Thyroid hormone receptors (SMRT). ETO contains four domains that exhibit homology to the Drosophila protein NERVY (3). The domain of ETO responsible for the interaction with E proteins has been classified as a member of a TATA box-binding protein-associated factor homology (TAFH) family (also called NERVY homology region 1). As only limited structural characterization of ETO has been reported, significant questions regarding the structure–function relationship of this AML-implicated corepressor remain unanswered.
Here, we report the 3D structure of the ETO TAFH (eTAFH) domain of human ETO elucidated by solution NMR spectroscopy. The 3D structure shows the conformation of eTAFH when interacting with a 17-residue peptide derived from the N-terminal AD of the human E protein HeLa E-box-binding protein (HEB) (AD112–28). We establish by a thorough mutational analysis that eTAFH is the critical domain involved in E protein silencing by ETO and further demonstrate that point mutations of key conserved residues abrogates this interaction.
Results
Solution Structure of eTAFH.
Biophysical characterization of the hydrodynamic properties of human eTAFH (residues 90–192; GenBank accession no. AAP88873.1) revealed the unliganded domain has a tendency for oligomerization at elevated concentrations. The process of resonance assignment of apo-eTAFH was complicated by low signal-to-noise values and poor transfer of magnetization in 3D NMR experiments. With a 2H,13C,15N-labeled sample of apo-eTAFH it was possible to establish backbone (HN, N, Cα, and Cβ) resonance assignments for 93 of 98 expected NH correlations (see Fig. 5, which is published as supporting information on the PNAS web site). Empirical prediction of Φ/Ψ angles with talos (13) based on backbone chemical-shift values shows apo-eTAFH to contain four α-helical regions (see Fig. 6A, which is published as supporting information on the PNAS web site). The quality of additional 3D data was insufficient to permit a more thorough structural evaluation of apo-eTAFH.
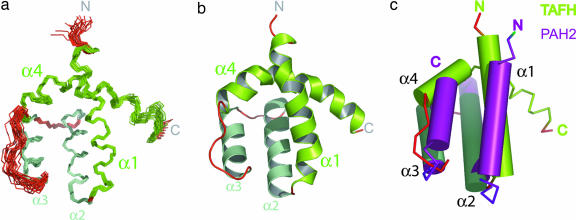
A substantial improvement in linewidth and magnetization transfer was observed for spectra recorded of eTAFH in complex with unlabeled AD112–28 from the human E protein HEB (see Fig. 7, which is published as supporting information on the PNAS web site). An eTAFH:AD112–28 complex was observed to elute as a single species by analytical size exclusion chromatography (data not shown). Titration of 15N-labeled eTAFH with unlabeled AD112–28 showed considerable perturbation of NH resonances (compare Figs. 5, 6D, and 7). The change in chemical shift exhibited a dependence on AD112–28 concentration up to 1:1 stoichiometry, indicative of an exchange regime on the fast-to-intermediate NMR time scale and a midmicromolar dissociation constant. Backbone (98%) and side-chain (95% nonlabile 1H; 93% aliphatic carbon) resonance assignments were duly collected by using standard protocols. The 3D structure of eTAFH was elucidated by using a combination of manual and automatic nuclear Overhauser effect (NOE) assignment procedures (Table 1, which is published as supporting information on the PNAS web site). The calculated 3D structure of eTAFH shows the conformation when interacting with HEB AD112–28. The ensemble of the 20 lowest-energy NMR structures shows four well defined α-helical regions (α1–α4) arranged in an unusual left-handed, up-and-down four-helix bundle topology (Fig. 1a and b). eTAFH contains a network of apolar amino acids (L100, V118, L121, F135, L139, L155, and L162) that form a well resolved hydrophobic core with contributions from residues on all four α-helices (see Fig. 8a, which is published as supporting information on the PNAS web site). Within this core the residues located on the internal faces of α2 and α3 exhibit the greatest precision in the NMR ensemble (see Fig. 6C). Analysis of the local per-residue rms deviation values shows a decrease in the precision for the C-terminal residues beyond K174, suggesting that this region is unstructured and flexible. Given the flexibility of the C terminus, it appears that residues A90–K174 define a general TAFH fold.
Fig. 1.
3D structure of the AD1-bound state of the human ETO TAFH domain. (a) Ensemble of 20 lowest-energy NMR structures showing four α-helical regions (colored green). (b) Cartoon representation of the lowest-energy structure of eTAFH. (c) Superposition of the 3D structures of eTAFH (green) and the SID-bound state of mSin3A PAH2 (purple) (15) with α-helices represented by cylinders. All structural representations were drawn with pymol (www.pymol.org).
Residues located in the α1/α2 and α3/α4 loops displayed noteworthy spectral properties. Resonances from amino acids situated in these loops generally suffered from lower signal-to-noise than the helical regions, most likely due to broadening by chemical exchange. On binding AD112–28 there is an appreciable increase in signal-to-noise overall, which is pronounced for residues in α1/α2 and α3/α4 loops. A comparison of predicted Ψ angles suggests little change in overall secondary structure for eTAFH on binding AD112–28 (see Fig. 6 A and B). However, there are changes in the predicted backbone geometry of residues in the α1/α2 loop. The α3/α4 loop is well conserved across the TAFH family (see Fig. 8b) and contains a high content of proline residues (144NFPLRPFVIPF154). In the structure of eTAFH the local rms deviation values for the α3/α4 loop are considerably greater than the structure regions of the domain (see Fig. 6C). This reduction in precision is likely a reflection of the local decrease in signal-to-noise, which in turn reduced the number of observable NOEs and therefore structural constraints. These observations suggest some flexibility of the orientations of the front (α1 and α4) and back (α2 and α3) helical elements with respect to each other with α1/α2 and α3/α4 loops acting as hinge regions.
eTAFH Is Structurally Related to Sin3 Paired Amphiphatic Helix (PAH) Domains.
Submission of the structural coordinates of eTAFH to the dali search engine (14) revealed significant similarities between the fold of eTAFH and the PAH domains found in mammalian Sin3 (mSin3) corepressors (dali score = 3.5) (15–17). The PAH family comprises small independently folding domains that adopt a left-handed, up-and-down four-helix bundle fold comparable to that described here for eTAFH. This similarity between eTAFH and PAH2 was unexpected given the lack of apparent sequence homology across the two families. Superposition of the lowest-energy structures of eTAFH and mSin3A PAH2 in complex with a peptide derived from the Sin3 interaction domain (SID) of Mad1 (15) yielded a rms deviation between structured backbone atoms of 3.3 Å (Fig. 1C). The agreement is particularly acute for α2 and α3 (rms deviation = 1.5 Å; see Fig. 8c), which form the mainstay of the hydrophobic core of eTAFH. Two notable differences between the fold families involve the extension in eTAFH of the loop connecting α3 and α4 and the trajectory of α4. In eTAFH α4 curves over the top of the fold, whereas in all currently known PAH structures α4 packs against α1 and α3, producing a more traditional and compact four-helix topology (15–18). The contrasting positions of α4 raises an intriguing question over the location of the E protein interaction site on eTAFH. A PAH-like α1/α2 binding surface is not feasible in eTAFH because of the presence of α4, suggesting differences between these two domain families at the level of target recognition.
eTAFH Binds AD1 via an Interface Involving α1/α4.
The location of the AD1-binding site of eTAFH was investigated by using NMR chemical-shift mapping. The backbone NH groups of residues K95, L96, T102, L103, R148, I152, F150, and F154 all registered larger than average chemical-shift changes on titration with unlabeled AD112–28 (see Fig. 8d). These residues map clearly to an extended channel formed by α1, α4, and the proline-rich loop connecting α3 and α4 (Figs. 2A and 6D). The apolar nature of this channel (including residues F99, L100, L103, P149, F150, and F154) is consistent with the leucine-rich hydrophobic AD1 peptide and other examples of domains interacting with LxxLL peptide motifs (19–21). A striking correlation is readily apparent between sites perturbed upon AD1 binding and the location of residues conserved across the TAFH family (Fig. 2). F99, which undergoes the largest change in chemical shift on AD1 binding, and L103 from α1 are perfectly conserved across the domain family (see Fig. 8b). In the α3/α4 loop, F154, which appears to form an aromatic patch with F99 (Figs. 2 and 8d), is well conserved and is absent only from NERVY TAFH (see Fig. 8b). The α2/α3 face is generally less well conserved compared with the AD1-binding surface. However, a hotspot of conservation is present on the α2/α3 face and comprises two glutamate residues from the conserved tripeptide 133EEF135 and a conserved lysine (K156). The magnitude of chemical-shift changes recorded for residues in this patch is considerably lower than for residues located in the α1/α4 channel. The evolutionary motivations behind the conservation of these solvent-exposed residues are currently unclear.
Fig. 2.
Analysis of the 3D structure of eTAFH. (a) An annotated molecular surface representation of eTAFH showing the location of residues undergoing chemical-shift perturbation on interaction with AD112–28 (same orientation as Fig. 1 a and b). The scale indicates the magnitude of the compound [1H, 15N] chemical-shift change (average = 0.11) as described by Mulder et al. (36). (b) An annotated molecular surface representation of eTAFH (same orientation as a) showing distribution of conserved residues. The scale bar indicates the degree of residue conservation from 0.0 to 0.9 taken from Fig. 8b.
Characterization of the AD1 Binding Site on eTAFH.
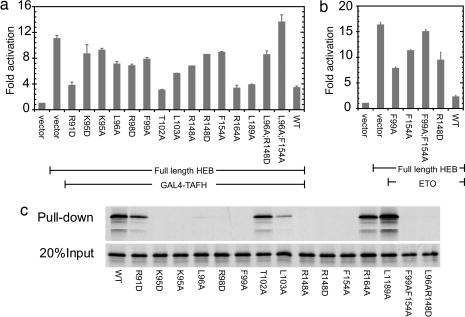
Residues with solvent-exposed side chains from α1 and α4 that are conserved or exhibited large chemical-shift perturbations were subjected to mutagenesis (see Fig. 8d). The effect of each mutation on the transcriptional corepression activity of the isolated eTAFH domain was investigated in a HEB-based activation assay (Fig. 3a). Mutations of two control residues located distal to the putative interaction site (R164A and L189A) had little effect on repression levels (Fig. 3a). Several mutations on α1 also produced negligible effects on repression (e.g., R91D and T102A). However, mutations of many residues identified by chemical-shift mapping (K95, L96, F99, R148, and F154) reduced the ability of eTAFH to repress HEB-driven transactivation. Combined mutations at two positions (L96A;R148D and L96A;F154A) displayed an additive effect on transcriptional repression. Similar results were obtained with full-length ETO (Fig. 3b and C.G. and J.Z., unpublished observations), with the largest reduction in activity resulting from an F99/F154 two-site mutation (Fig. 3b). In pull-down assays using GST-HEB11–27 fusions, mutations in eTAFH that suppress E protein silencing by ETO also inhibit AD1 binding (Fig. 3c), demonstrating a direct link between the eTAFH:AD1 interaction and E protein repression. These results are in excellent agreement with the NMR-derived structure of eTAFH bound to AD1.
Fig. 3.
The interaction with HEB is prevented by targeted mutation of eTAFH. (a) Mutations targeting the putative interface in the isolated eTAFH domain prevent inhibition of HEB-based activation of a luciferase reporter gene. (b) Two-site mutations of F99 and F154 have an additive effect on ETO-based silencing of HEB-based activation. Error bars indicate ± 1 SE. (c) GST-AD111–27 was used to pull down 35S-labeled full-length ETO. (Upper) Bound fraction. (Lower) Twenty percent input of full-length ETO.
The above NMR and mutagenesis results suggest that eTAFH uses both hydrophobic and electrostatic interactions to distinguish target peptides. The proposed AD1-binding site of eTAFH has a largely hydrophobic central pocket composed by F99, L103, and F154 with a high peripheral concentration of positively charged residues (R91, K95, K97, R98, and, notably, R148; see Fig. 8d). AD112–28 contains a highly conserved commonly used signaling motif LxxLL (where L = leucine and X = any amino acid; see Fig. 9a, which is published as supporting information on the PNAS web site) (22, 23), which is predicted to form a short amphipatic α-helix (8, 24). Mutation of any of these core leucines abrogates the interaction between ETO and HEB (8) and WT AD1 function (8, 24). A number of acidic residues in proximity to the core LxxLL motif (see Fig. 9a), including D14, E16, D19, and D22, may further contribute to specificity by interacting with positively charged residues from eTAFH. Indeed, mutation of D19 and D22 has been shown to abolish the ETO:E protein interaction (8).
Modeling of the TAFH:AD112–28 Complex.
A model of the eTAFH:AD1 complex was generated by using the restraint-driven docking program haddock (25). A docking-based approach was used because exchange broadening of resonances from AD112–28 complexed with unlabeled eTAFH (see Fig. 9b) prevented elucidation of the 3D structure of the eTAFH:AD1 complex by experimental means. Although it was possible to obtain backbone resonance assignments for 16 of the 17 residues in AD112–28 (see Fig. 9b), poor transfer of magnetization prevented the elucidation of a 3D structure of the AD1 portion. Using a combination of CD analysis (data not shown), secondary structure prediction, and previously published structural studies of complexes involving LxxLL peptides (23), we generated a model for the AD1 peptide motif. An ensemble of structures from the lowest-energy haddock cluster (see Fig. 9c) shows a well defined complex with a consistent orientation of the AD1 helix (Figs. 4a and 9d). The model of the eTAFH:AD112–28 complex shows that the core leucine residues of AD1 (L17, L20, and L21) and F23, M26, and F27 make multiple hydrophobic contacts with eTAFH. The results presented here show that eTAFH uses a peptide-binding site formed by α1 and α4 (Fig. 4 a and c). The location of a binding interface at this position in eTAFH is a considerable divergence from the α1/α2 site used by PAH1 or PAH2 domains (Fig. 4 b and d) (15–17, 26). However, both sites are characterized by a central hydrophobic cavity encircled by polar residues and are thus well suited for interactions with small amphiphatic α-helical motifs.
Fig. 4.
The HEB AD1 interaction site is located between α1 and α4 on eTAFH. (a) Cartoon representation of a model of the eTAFH:AD1 complex generated by haddock (25). (b) mSin3A PAH2 interacts with Mad1 SID via an interface comprising α1/α2 (15). (c and d) Schematic representations of eTAFH:AD1 (c) and mSin3A PAH2:SID (d) complexes highlighting the similarities in fold and differences in binding site location. The topology of eTAFH is drawn to emphasize the fold overlap between the two structures; for simplicity, the true location of α4 in eTAFH is represented by a two semicircles connected by a dotted green line. Full circles represent α-helices, and red lines symbolize loop regions. The color scheme is consistent with Figs. 1 and 2.
Discussion
Currently, E proteins are the only characterized interaction partners for eTAFH. However, because of different tissue distributions (8), E proteins are not considered natural targets of ETO in t(8;21) cells. That this interaction occurs is a direct result of the misexpression of ETO following t(8;21). However, this does not exclude the possibility that this interaction might occur when ETO is expressed at a high level, as occurs in neuronal cells. It also is possible that there are other eTAFH-binding sequences that may participate in WT ETO function in nonhematopoietic cells or other uncharacterized t(8;21)-related systems. The structural similarity observed between eTAFH and PAH structures allows us to speculate as to the nature of such WT eTAFH targets. PAH domains interact with numerous short amphipathic peptides that closely resemble the E protein AD1 motif. Such AD1-like motifs are used prevalently in transcriptional regulation (22, 23), particularly for interactions involving transcription factors and their coactivators or corepressors. PAH1 (17) and PAH2 (15, 16) domains can each interact with several amphiphatic motifs. This multitarget capability is also common to nuclear receptors (23) and the p300/CREB binding protein KIX domain (27), which also bind α-helical peptides containing similar motifs to AD1. Indeed the KIX domain has also been proposed to mediate the interaction between E proteins and p300/CREB binding protein by directly targeting AD1 (12). Both PAH2 (18) and KIX domains (28) have been shown to adopt less well folded conformations in their ligand-free states. This property of structural plasticity may facilitate the interaction with different binding partners and play a role in target selectivity. It is also noteworthy that LxxLL motif-containing peptides are frequently unstructured until interaction with a binding partner. It has been proposed that PAH domains function as folding templates for binding peptides (18), for which a degree of conformation flexibility is a prerequisite. The chemical exchange broadening observed for resonances from amino acids in α1/α2 and α3/α4 loops suggests eTAFH also exhibits some degree of structural flexibility in its free state (see Fig. 5). Accordingly, eTAFH may also be capable of interacting with multiple, AD1-like, amphiphatic motifs, although further studies will be required to confirm this idea and identify these targets.
The TAFH family encompasses domains from the three ETO family members (ETO/MTG8, MTGR1, and MTG16/ETO-2) (3), the Drosophila protein NERVY, and human and Drosophila isoforms of the general transcription factor TAF4 (29) (see Fig. 8b). The sequence identity between the TAFH domains in the ETO/MTG proteins is particularly pronounced, suggesting that other MTG TAFH domains adopt an equivalent structure to eTAFH and therefore bind similar recognition sequences also at the α1/α4 interface. ETO-2 is transiently associated with the TAL-1/SCL-based E protein-containing complex that is involved in regulating erythroid-specific gene expression (30). In this context the E protein E2A is thought to physically recruit ETO-2 (31), whose presence in turn regulates expression of TAL-1/SCL-activated genes. Consistent with the observation that D18A and D21A mutations within the E2A AD1 domain abolish the E2A-MTG16/ETO-2 interaction (30), our eTAFH structure coupled with sequence conservation (see Fig. 8b) predicts that MTG16/ETO-2 interacts with E proteins via its eTAFH domain. Furthermore, mutation of the residues homologous to those described here would likely inhibit recruitment of MTG16/ETO-2 to TAL1/SCL and therefore cause disregulation of the expansion of erythroid progenitors.
Across the wider TAFH family there is more modest sequence identity (15/85 structured residues for eTAFH; see Fig. 8b). The hydrophobic core of eTAFH is populated by seven residues that are perfectly conserved across the entire TAFH family (Fig. 8 a and b). Additional conservative similarities in the hydrophobic core and the α3/α4 loop imply that all members of the TAFH family adopt a PAH-like fold resembling that described here for eTAFH. Given the extensive use of AD1-like binding motifs in transcriptional regulation and the contribution of conserved residues in AD1 binding (compare Fig. 2B), it is also credible that all TAFH domains interact with similar AD1-like target sequences at a common interface. By extension, such a capability for the TAFH domain in TAF4 would raise the interesting possibility of direct recruitment of TFIID by AD1-like peptide motifs in DNA-bound transcription factors.
The role of ETO as a corepressor in both WT and AML-fused forms is well established (1, 3). The interaction between E proteins and eTAFH represents an alternative mechanism for transcriptional repression by AML1-ETO compared with the classical model involving the disruption of AML1/CBF function. Specifically, eTAFH facilitates the repression of E-box-regulated genes in an AML1-independent fashion by directly recruiting the ETO corepressor to a promoter-bound transcription factor. The role of eTAFH in this process displays an appreciable correlation to that played by PAH domains in Sin3 corepressors. These functional similarities, combined with the structure of eTAFH presented here, the fold similarity with PAH domains, and the common amphiphatic α-helical nature of their target sequences, suggest that eTAFH, like PAH domains, may also possess the capacity to interact with multiple target proteins. Such functionality suggests that the mechanism described for E protein silencing (8) may be representative of a general mode of repression by ETO and potentially AML1-ETO. The involvement of MTG16 and E proteins in TAL-1/SCL regulation (31) is likely to occur by such a mechanism. Coupled with the recent observation that the eTAFH domain is retained in a naturally truncated form of AML1-ETO that promotes proliferation (32), the results detailed here present a strong argument for exploring the interaction profile of eTAFH in search of other aberrantly regulated transcriptional pathways.
Materials and Methods
Preparation of Protein Samples for NMR.
Recombinant eTAFH (residues A90-S192 of human ETO, GenBank accession no. AAP88873.1) was expressed in Escherichia coli BL21 Rosetta (Novagen) from an isopropyl β-d-thiogalactoside-induced pGEX-4T-1 expression vector (Amersham Pharmacia Biosciences). Unlabeled or isotope-enriched (e.g., 2H, 13C, 15N) protein was purified from crude lysate by using a Glutathione Sepharose resin (Amersham Pharmacia Biosciences). The desired protein was cleaved from a GST-fusion tag on the resin by using recombinant thrombin (Calbiochem). Fractions containing eTAFH were identified by using SDS/PAGE. Undesired thrombin was removed by using benzamidine Sepharose resin (Amersham Pharmacia Biotech). eTAFH samples were subjected to further purification by size exclusion chromatography, concentrated, and dialyzed into NMR buffer [20 mM sodium phosphate, pH 6.0/50 mM NaCl/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride/1 mM PMSF/0.25 mM NaN3/10% D2O]. Samples of AD112–28 (residues G12-S28 of human HEB, GenBank accession no. AAH50556.1) were either prepared or commercially synthesized (Dalton, Toronto). Isotope-labeled AD1 samples were purified as described above for eTAFH, except that after separation from GST, AD112–28 samples were dialyzed against water and lyophilized. Freeze-dried AD112–28 residue was resuspended in NMR buffer or samples of eTAFH. Protein samples were adjusted to 0.5–1 mM for the isotope-labeled component with complexes containing a 1.5- to 2-fold molar excess of unlabeled binding partner.
Acquisition and Analysis of NMR Data.
Except where mentioned, NMR data were recorded at 25°C on Varian Unity-Plus 500- or 600-MHz spectrometers equipped with 5-mm triple-resonance cold probes and z-axis pulse-field gradients. Backbone resonance assignments of 2H, 13C, 15N-labeled apo-eTAFH and AD112–28-bound eTAFH were obtained from HNCA, HN(CO)CA, HN(CA)CB, and HN(COCA)CB experiments (33). Side-chain aliphatic 13C and 1H resonance assignments of 13C, 15N-labeled eTAFH complexed with AD112–28 were obtained from CCC-total correlation spectroscopy (TOCSY)-NNH and HCCH-TOCSY-NNH experiments (33). Aromatic side-chain 1H resonances were obtained from HBCBCGCDHD and HBCBCGCEHE experiments (34). All side-chain assignments were confirmed by using a simultaneously acquired 13C/15N-NOESY-heteronuclear sequential quantum correlation (HSQC) experiment (35) (acquired at 800 MHz). Resonance assignments of [13C,15N]-labeled AD112–28 were obtained from HNCACB and [15N]-NOESY-HSQC spectra. Compound chemical-shift values were calculated as described (36). All data were processed by using nmrpipe/nmrdraw (37) and analyzed with azara (www.bio.cam.ac.uk/azara) and ansig (38) software packages. Data format conversion was performed with the ccpnmr package (39).
Structure Calculation.
The 3D structure of eTAFH (in complex to AD112–28) was calculated by cyana (40) using standard protocols. NOE-based restraints were obtained from a combination of manual {from (15N]-NOESY-HSQC data sets) and cyana-based automatic NOE assignment (from [13C/15N]-NOESY-HSQC data sets) procedures (41). Estimation for φ/ψ torsion angle restraints were derived from Cα, Cβ, N, and C(O) chemical shifts using talos (13). Hydrogen bond constraints were generated for protected NH groups in H2O/D2O solvent exchange experiments. The associated carbonyl groups were estimated from the predicted locations of secondary structure.
Docking of TAFH and AD1.
The lowest-energy NMR structure for eTAFH was used as a template for haddock (25) docking experiments. eTAFH-bound AD112–28 was predicted to be predominantly α-helical based on CD results (data not shown), secondary structure predictions (42), chemical shift perturbations on eTAFH binding, and previous structural analysis of complexes containing LxxLL motifs in the literature (23). A model of AD112–28 was generated by using swissmodel in which residues 16–26 adopted standard α-helical backbone geometry. “Active” and “passive” residues and active segments for both molecules were selected from mutagenesis results [for eTAFH from this study and for AD1 from those reported by Zhang et al. (8)] and chemical-shift changes (for eTAFH only) following published guidelines (25). For eTAFH active residues were defined as 93, 95, 99, 101, 102, 110, 113, 143, 148, 150, and 154, and passive residues were defined as 89–91, 94, 98, 105, 106, 108, 109, 111–113, 146, 147, 149, 153, and 157. For AD112–28 residues 17, 19, 20–23, 26, and 27 were defined as active, with all other residues defined as passive. Docking calculations were performed by using standard methods (25). Side-chain reorientations were permitted for active segments of eTAFH and all of AD112–28 during the semirigid docking step.
Site-Directed Mutagenesis.
Mammalian expression vectors for HEB, ETO, and eTAFH have been described (8). Site-directed mutagenesis was performed with a QuikChange Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's instructions. All mutations were verified by DNA sequencing.
Cell Culture and Luciferase Assay.
293T cells were maintained in DMEM with 10% FBS. Reporter assays were carried out as described (8). Equal total amounts of plasmids were transfected for each well by adjusting empty vectors. Luciferase reporters contained four copies of an E box (CAGATG) site. Luciferase units were normalized to β-gal activity, which served as an internal control for transfection efficiency. Fold activation values were relative to empty vector controls. Figures show the mean and standard error of duplicate samples in representative experiments unless otherwise indicated.
GST Pull-Down Assay.
GST pull-down assays were carried out as described (8). In vitro-translated 35S-labeled proteins were incubated with bacterially expressed GST fusions immobilized on Glutathione-Sepharose beads (Amersham Pharmacia). After extensive washing with buffer BC300/0.1% Nonidet P-40, bound polypeptides were resolved on SDS/PAGE and visualized by autoradiography. Input lanes show a fraction of total input.
Supplementary Material
Acknowledgments
This work was supported by a Canadian Institutes of Health Research operating grant (to M.I.), a National Institutes of Health grant (to R.G.R.), and University of Cincinnati College of Medicine start-up funding (to J.Z.). M.J.P. is a recipient of a Canadian Institutes of Health Research postdoctoral fellowship, J.Z. is a Leukemia and Lymphoma Society Fellow, and M.I. holds a Canada Research Chair in Cancer Structural Biology.
Abbreviations
- AD
activation domain
- AML
acute myeloid leukemia
- CBF
core binding factor
- ETO
eight-twenty-one
- TAFH
TATA box-binding protein-associated factor homology
- eTAFH
ETO TAFH
- HEB
HeLa E-box-binding protein
- mSin3
mammalian Sin3
- NOE
nuclear Overhauser effect
- PAH
paired amphipathic helix
- SID
Sin3 interaction domain.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: Resonance assignments for eTAFH have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 7147). Structural coordinates for eTAFH have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2H7B).
References
- 1.Licht J. D. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- 2.Downing J. R. Leukemia. 2001;15:664–665. doi: 10.1038/sj.leu.2402035. [DOI] [PubMed] [Google Scholar]
- 3.Davis J. N., McGhee L., Meyers S. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- 4.Peterson L. F., Zhang D. E. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 5.Choi Y., Elagib K. E., Goldfarb A. N. Crit. Rev. Eukaryotic Gene Expression. 2005;15:207–216. doi: 10.1615/critreveukargeneexpr.v15.i3.30. [DOI] [PubMed] [Google Scholar]
- 6.Lutterbach B., Hiebert S. W. Gene. 2000;245:223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
- 7.Hug B. A., Lazar M. A. Oncogene. 2004;23:4270–4274. doi: 10.1038/sj.onc.1207674. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Kalkum M., Yamamura S., Chait B. T., Roeder R. G. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 9.Quong M. W., Romanow W. J., Murre C. Annu. Rev. Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 10.Murre C. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 11.Lazorchak A., Jones M. F., Zhuang Y. Trends Immunol. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Bayly R., Chuen L., Currie R. A., Hyndman B. D., Casselman R., Blobel G. A., LeBrun D. P. J. Biol. Chem. 2004;279:55362–55371. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- 13.Cornilescu G., Delaglio F., Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 14.Holm L., Sander C. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 15.Brubaker K., Cowley S. M., Huang K., Loo L., Yochum G. S., Ayer D. E., Eisenman R. N., Radhakrishnan I. Cell. 2000;103:655–665. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 16.Spronk C. A., Tessari M., Kaan A. M., Jansen J. F., Vermeulen M., Stunnenberg H. G., Vuister G. W. Nat. Struct. Biol. 2000;7:1100–1104. doi: 10.1038/81944. [DOI] [PubMed] [Google Scholar]
- 17.Nomura M., Uda-Tochio H., Murai K., Mori N., Nishimura Y. J. Mol. Biol. 2005;354:903–915. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 18.van Ingen H., Baltussen M. A., Aelen J., Vuister G. W. J. Mol. Biol. 2006;358:485–497. doi: 10.1016/j.jmb.2006.01.100. [DOI] [PubMed] [Google Scholar]
- 19.Zor T., De Guzman R. N., Dyson H. J., Wright P. E. J. Mol. Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Razeto A., Ramakrishnan V., Litterst C. M., Giller K., Griesinger C., Carlomagno T., Lakomek N., Heimburg T., Lodrini M., Pfitzner E., et al. J. Mol. Biol. 2004;336:319–329. doi: 10.1016/j.jmb.2003.12.057. [DOI] [PubMed] [Google Scholar]
- 21.Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 22.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 23.Plevin M. J., Mills M. M., Ikura M. Trends Biochem. Sci. 2005;30:66–69. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Massari M. E., Jennings P. A., Murre C. Mol. Cell. Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez C., Boelens R., Bonvin A. M. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 26.Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., Radhakrishnan I. Nat. Struct. Mol. Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- 27.De Guzman R. N., Goto N. K., Dyson H. J., Wright P. E. J. Mol. Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan I., Perez-Alvarado G. C., Parker D., Dyson H. J., Montminy M. R., Wright P. E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 29.Kokubo T., Gong D. W., Roeder R. G., Horikoshi M., Nakatani Y. Proc. Natl. Acad. Sci. USA. 1993;90:5896–5900. doi: 10.1073/pnas.90.13.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuh A. H., Tipping A. J., Clark A. J., Hamlett I., Guyot G., Iborra F. J., Rodriguez P., Strouboulis J., Enver T., Vyas P., et al. Mol. Cell. Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goardon N., Lambert J. A., Rodriguez P., Thibault P., Dumenil D., Strouboulis J., Romeo P.-H., Hoang T. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan M., Burel S. A., Peterson L. F., Kanbe E., Iwasaki H., Boyapati A., Hines R., Akashi K., Zhang D. E. Proc. Natl. Acad. Sci. USA. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sattler M., Schleucher J., Griesinger C. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 34.Yamazaki T., Forman-Kay J., Kay L. E. J. Am. Chem. Soc. 1993;115:11054–11055. [Google Scholar]
- 35.Pascal S. M., Muhandiram D. R., Yamazaki T., Forman-Kay J. D., Kay L. E. J. Magn. Reson. B. 1994;103:197–201. [Google Scholar]
- 36.Mulder F. A., Schipper D., Bott R., Boelens R. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Kraulis P. J., Domaille P. J., Campbell-Burk S. L., Van A. T., Laue E. D. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 39.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 40.Guntert P., Mumenthaler C., Wuthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann T., Guntert P., Wuthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 42.McGuffin L. J., Bryson K., Jones D. T. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.