Abstract
Microarray based gene expression studies allow simultaneous analysis of relative amounts of messenger RNA (mRNA) for thousands of genes using fluorescently labeled nucleic acid targets. Most common methods use enzymatic techniques, such as oligo-dT primed reverse transcription to produce labeled cDNA. These labeling methods have a number of shortcomings, including enzyme- introduced labeling and sequence bias, laborious protocols, high experiment-to-experiment variability and an inability to detect small changes in expression levels. Here, we describe a novel labeling methodology that uses platinum-linked cyanine dyes to directly chemically label mRNA from as little as 2 µg of total RNA. We show that the gene expression data produced using the labeled mRNA method has very high precision, low error, no labeling bias and a dynamic range over several orders of magnitude. This allows a greater accuracy in the identification of differentially expressed genes and cuts down on the need for running too many replicate assays. Small changes in gene expression can now be detected in large-scale gene expression profiling assays using this simple, easy and quick procedure.
INTRODUCTION
A key component of microarray assays is the step used for fluorescent labeling of target nucleic acids, which is currently performed using multi-step enzymatic processes (1). Fluorescent label is introduced using either dye-labeled or amine-labeled nucleotides during the cDNA synthesis step (2,3). The labeled material is then purified and hybridized onto a microarray for detection. But, the copying process introduces errors due to enzymatic labeling bias (4). Additionally, different mRNA sequences in a starting mRNA mixture may not be represented at the same level in the final labeled cDNA mixture due to selective and non-linear target amplification (5). Another consequence of oligo-dT primed reverse transcription is that many of the mRNA species are represented with their 3′ ends only in the labeled cDNA, due to limited processivity of the reverse transcriptase (6). Incomplete denaturation of RNA secondary structure during cDNA synthesis step can also halt the polymerase, resulting in shorter cDNA copies of target mRNA. These limitations affect the precision and quality of the resulting data. Multiple replicates can be used to gain confidence in the results for such experiments, but that is not possible when the samples are limiting or rare.
Although use of random primers or indirect cDNA labeling may circumvent some of the issues with enzymatic labeling methods, labeling mRNA itself may be more advantageous. Here we present a novel labeling technology that uses a chemical reagent to directly label mRNA with fluorescent dyes. The mRNA is labeled in total RNA mixture in a one- step non-enzymatic reaction reducing errors in prior methodologies due to multiple steps. The resulting labeled mRNA has no labeling bias, provides reproducible gene expression profile and allows a greater amount of precision in the identification of differentially expressed genes.
MATERIALS AND METHODS
Microarrays
MICROMAX™ Human cDNA Microarray I (PerkinElmer Life Sciences, Boston, MA), pre-spotted with 2398 known human genes (spotted in duplicate, total 4800 spots), were used in all of these experiments. The microarrays are spotted with genes obtained from diverse tissue sources that cover a broad spectrum of functionality.
RNA extraction
Total RNA from human Jurkat and HL-60 cells were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions.
Synthesis, labeling and purification of cDNA
Synthesis and labeling of cDNA was carried out using the MICROMAX™ Direct Labeling Kit (PerkinElmer Life Sciences) according to the manufacturer’s instructions. Typically, 50 µg total RNA was used in each of the labeling reactions. Combined Cyanine 3 and Cyanine 5 reactions were purified using Microcon YM-100 columns (Millipore, Bedford, MA). Purified target was dried in a speed-vac and stored at –70°C.
Labeling and purification of mRNA
Labeling of mRNA was carried out using the MICROMAX™ ASAP RNA Labeling Kit (PerkinElmer Life Sciences) according to the manufacturer’s instructions. Typically, 10 µg total RNA was used in each of the labeling reactions. Total RNA was diluted in the ASAP labeling buffer, on ice, to a final volume of 18 µl. Two microlitres of either Cyanine 3 or Cyanine 5 chemical-labeling reagent was added and the mixture was incubated at 85°C for 15 min in a thermal cycler (MJ Research). The reaction was then cooled to 4°C for 5 min and added 5 µl of stop solution. Combined Cyanine 3 and Cyanine 5 reactions were purified using Oligotex mRNA purification columns (Qiagen, Valencia, CA). Purified target was dried in a speed-vac and stored at –70°C.
Hybridization and wash conditions
Hybridization reactions were performed using the manufacturer recommended buffers and conditions. Twenty microlitres of MICROMAX™ Q hybridization buffer was added to dried labeled-cDNA and the mixture was denatured at 90°C for 2 min. Twenty microlitres of hot (70°C) MICROMAX™ ASAP hybridization buffer was added to dried labeled-mRNA and the mixture was denatured at 70°C for 3 min. After quickly spinning-down the reaction vials in a centrifuge, the mixtures were separately pippetted onto microarrays. Coverslips were applied (22 × 22 mm from VWR Scientific) and the slides were placed in a hybridization chamber (Corning Inc., Corning, NY). Arrays were incubated at 65°C for 14–16 h, and subsequently washed with 0.5× sodium citrate sodium chloride buffer (SSC) (150 mM sodium chloride, 15 mM sodium citrate, pH 7.0) 0.01% SDS, followed by 0.06× SSC, 0.01% SDS, followed by 0.06× SSC at room temperature with shaking for 15 min each. Slides were next placed in 50 ml conical tubes (Corning) and spun dried for 3 min at 200 g at room temperature.
Microarray scanning
Arrays were scanned at 10 µm resolution and variable photo multiplier tube (PMT) settings to obtain optimum signal intensities. The resulting images were used to generate gpr data files.
Data analysis
Data analysis was done using GeneSpring 4.2 (Silicon Genetics, CA) and Microsoft Excel (Microsoft Corp., WA). Typically, data was normalized to the median. Only those genes with a minimum signal intensity of 100 or 150 fluorescence units in both channels were allowed in the analyses. Furthermore, for combined multiple experiments, only those signals with t-test P-values <0.05 were considered.
Note about oligonucleotide arrays
Most of the commercially available oligonucleotide arrays may not be suitable for detecting directly chemically labeled mRNA targets (sense strands). This is because the immobilized oligonucleotide probes have incorrect orientation for binding to mRNA as these probes are designed against anti-sense cDNA or aRNA targets.
RESULTS AND DISCUSSION
We have developed a novel non-enzymatic system for fluorescently labeling nucleic acids. The technology uses a platinum reagent to directly chemically label mRNA with cyanine dyes (7). The mRNA is labeled in the total RNA mixture and there is no enzymatic step involved. The mono-valent platinum reagent, which has cyanine fluorophores synthetically attached via a linker arm, reacts with the N7 of guanine residues in the RNA chain to form a stable coordinate bond. Remarkably, the guanine modification does not alter the stability of nucleic acid chains, unlike its modification with alkylating reagents that result in depurination and subsequent strand-scission (8,9). It also does not affect the ability of modified single-strands to hybridize, as the modification is placed in the major grove of the resulting double-stranded complex.
The reaction is initiated by combining total RNA with the platinum labeling reagent and takes only 15 min to go to completion. Unincorporated dye and other non-mRNA species are simultaneously removed in quick single-step mRNA purification using oligo-dT columns, without any loss in the yield of purified mRNA. We find that only a fraction of the amount of total RNA that is required to prepare enzymatically labeled cDNA is sufficient to obtain comparable data with chemically labeled mRNA. We performed two-color ratiometric microarray experiments using chemically labeled mRNA and the conventional enzymatically dye-labeled cDNA (Fig. 1A). The results show that the average signal-to-noise ratios and expression profiles were similar between the two methods despite the 5-fold less total input RNA used with the new method. The signal intensities were found to be comparable between the two methods when the experiments are performed starting with the same mass of total RNA target (data not shown). The new method is also linear over a dynamic range of >104 (see Supplementary Material for more details). In fact, we find that with the new method as little as 2 µg of total RNA generates comparable expression profiles with similar signal-to-noise ratios (Fig. 1B). That is 25-fold less than the 50 µg typically used in a cDNA labeling assay. Analysis of the gene expression profiles shows that only about 55 genes (7 differential) out of a total of 2398 genes were not detected when 2 µg total RNA was used in the new labeling method compared with the optimum 10 µg of total RNA target. These 55 genes are likely to be low abundance genes and had weak signal, no higher than 200 fluorescence units, when using 10 µg total RNA. Additionally, we analyzed outliers in the two conditions. Upon comparing the Cy3/Cy5 ratios of filtered data (signal intensity >100) between the two data sets, we found that only 7.4% genes disagreed (using a typical variance allowance of 30%). A careful inspection of these genes showed that only 1.8% have divergent expression ratios (i.e.; the up-regulated genes under one condition were neutral or down-regulated in the other condition) and the rest generated similar expression profile.
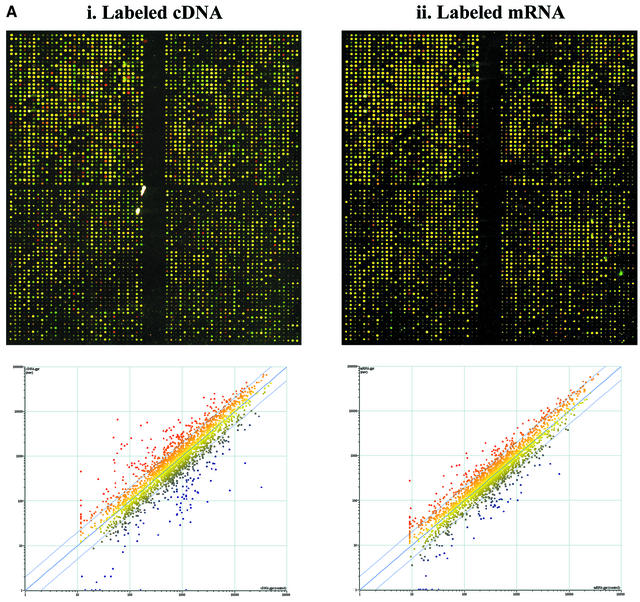
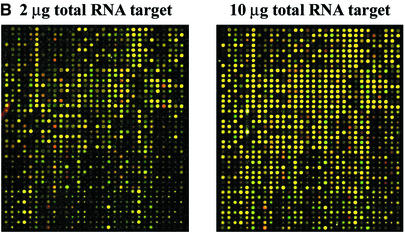
Figure 1.
(Next page) (A) Comparison of microarray results obtained with the conventional cDNA labeling and the new mRNA chemical labeling methodologies. Two-color differential expression assay overlay images of Cyanine 3 and Cyanine 5 scans are shown along with their respective scatter-plots. In both experiments, the target from HL-60 cells was labeled with Cyanine 3 and the target from Jurkat cells was labeled with Cyanine 5 and hybridized to 4800-element cDNA arrays. It shows that the two methods produce very similar data. (i) Total RNA (50 µg) was used to produce labeled cDNA in a standard cDNA labeling assay with each Cyanine dye. Median S/N was 2.6 for Cyanine 3 and 2.4 for Cyanine 5. (ii) Total RNA (10 µg) was used to produce labeled mRNA in the new labeling assay with each Cyanine dye. Median S/N was 3.1 for Cyanine 3 and 4.4 for Cyanine 5. (B) Overlay scans (at same laser power and PMT settings 780 and 710 for the Cyanine 3 and Cyanine 5 channels respectively) from one quadrant of a microarray comparing expression patterns obtained from 2 µg of total RNA versus 10 µg of total RNA in a RNA labeling assay. It shows that 2 µg of total RNA produces data (median S/N was 3.2 for Cyanine 3 and 4.0 for Cyanine 5) similar to 10 µg of total RNA.
Next we compared the precision and reproducibility of the two methodologies by performing multiple independent microarray assays. Results from each of the assay sets were autonomously combined and are presented as scatter plots with error bars in Figure 2. It shows that the data obtained with the newly developed method are considerably more precise than the conventional method. The new method has a much lower error associated with each measurement. For the combined cDNA target assays, the cumulative variation (CV) of ratios is 27.2% with a median P-value of 0.058. For the combined labeled-mRNA assays, the CV of ratios is 17.9% with a median P-value of 0.010.
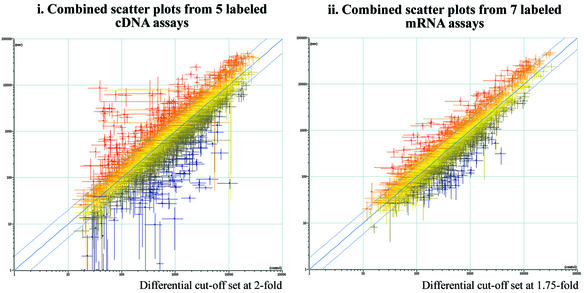
Figure 2.
Precision of the data obtained using novel RNA labeling technology. We performed five assays with Cyanine labeled cDNA target and seven assays with Cyanine labeled mRNA targets. The combined scatter plots from each set are shown. Since the probes were spotted in duplicate on these microarrays, each data point in the scatter plot has the benefit of 10 separate measurements for the cDNA target assays and 14 separate measurements for the mRNA target assays. Error bars representing the standard deviations for each gene have been added to the scatter plots for the respective experiments to demonstrate the amount of variability in each of the labeling assays. (i) Scatter plot for microarray experiments using standard labeled cDNA target (five experiments). (ii) Scatter plot for microarray experiments using labeled mRNA target (seven experiments). Note that the differential expression axes are set at 1.75× in the labeled mRNA assays as compared with the traditional 2× in labeled cDNA assays. An implication from these experiments is that fewer microarray experiments are necessary to determine with a high degree of statistical rigor that a given gene is truly up, down or non-differentially expressed.
Although the overall expression profiles obtained with chemically labeled mRNA and enzymatically labeled cDNA were very similar, they were not identical. One difference between the resulting data from the two methodologies is that the scatter plots from labeled mRNA assays appear ‘tighter’ and closer to the median. Since the error associated with mRNA assays was so low, we wondered if this would allow us to identify genes that differ <2-fold in expression between two conditions. Currently, it is very difficult to detect 2-fold different gene expression and one can only reliably describe gene expression differences >3-fold (10). But the expression of some of the most interesting genes may only change a little, not many-fold, between different conditions. To determine the minimum fold-change that the new labeling methodology can detect, we performed multiple same versus same expression assays using total RNA from Human HL-60 cells. Figure 3 clearly shows that chemically labeled mRNA can reliably detect >1.6-fold gene expression differences, compared with at least >2-fold changes that the current best conditions can provide. The new mRNA labeling methodology showed a 0.7% labeling bias as compared with a 2.7% bias with conventional enzymatic cDNA labeling methodology at a differential cut-off of 2.0. When we compared replica mRNA labeling experiments, we found 0.1–0.5% genes as outliers. Although these outliers differed from experiment to experiment, we feel that this percentage is quite small and within experimental error. We now routinely use a differential cut-off value of 1.75 with the new labeling methodology. These experiments also show that while direct enzymatic cDNA labeling has some dye bias, the new methodology has almost no labeling bias.
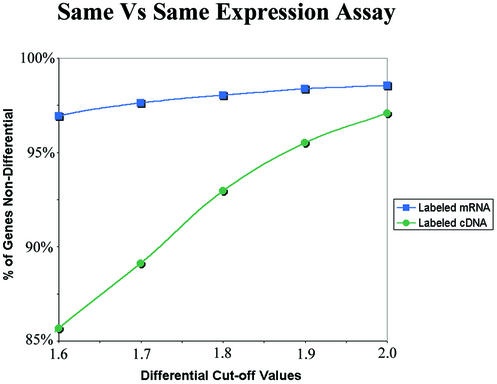
Figure 3.
Comparison of the labeling bias between conventional cDNA labeling and the new mRNA labeling technologies. RNA from HL-60 cells was labeled both with Cyanine 3 and Cyanine 5 (same versus same assay). Microarray data scatter plots of the intensity of the Cyanine 3 signals versus those observed for Cyanine 5 should ideally lie in a perfectly straight 45° line. Deviations from this line indicate both the inherent noise in the assay as well as any preference to incorporate one Cyanine dye over the other. The amount of labeling bias can be quantified by determining the percentage of the genes spots on the scatter plot that lie between the differential cut-off values. This figure shows that the percentage of non-differential genes drops off significantly for targets that were labeled enzymatically with the labeled cDNA assay as the differential cut-off intervals were narrowed. This indicates that there is significantly less (∼80% less) labeling bias observed with the labeled mRNA assay. Note that the differential expression axes are set at 1.75× in the labeled mRNA assays as compared with the traditional 2× in labeled cDNA assays. It shows that 97.3% of all genes are within the 1.75 differential axes (99.3% fall within the 2× differential axes). In comparison, only 95.9% genes fall within the 2× differential axes in a labeled cDNA assay.
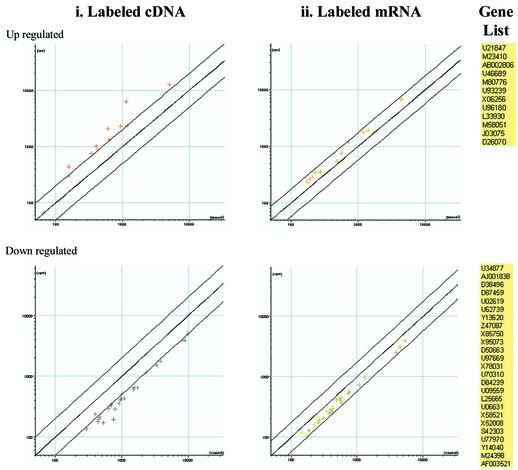
The second difference is that the relative gene expression pattern of a few genes was dissimilar between labeled cDNA assays and the labeled mRNA assays, though there were no divergent expression ratios (i.e. none of the genes were up-regulated in one protocol and down-regulated in the other). For example, out of the 187 up-regulated genes in the labeled cDNA assay, 27 were found to be neutral in the labeled mRNA assay. However, 15 of these 27 genes had overlapping error bars, meaning that their expression ratios were within the experimental error. Similarly, out of the 164 down-regulated genes in the labeled cDNA assays, 38 were found to be neutral in the labeled mRNA assays. 13 of these 38 genes had overlapping error bars. Thus, overall, the differences were not significant—6.4% (up-regulated) and 15.2% (down-regulated) of all differential genes had different expression pattern in the labeled mRNA assays as compared with the conventional labeled cDNA assays. Figure 4 shows the scatter plots of dissimilar genes along with their GenBank accession numbers. Results from labeled cDNA assays were used as the standard in these comparisons. Only those genes with good reproducibility (P-value of the t-test <0.05) and signal intensity of >100 in all of the 10 data points and above the differential cut-off value of 2 were considered differential. A sub-set of these genes that appeared non-differential in labeled mRNA assays went through a second data filtration to ensure reproducibility (P-value <0.05, signal intensities of >100 in both channels) in all of the 14 data points.
Figure 4.
Gene expression profile results from labeled cDNA assays as compared with the results from the newly developed labeled mRNA assays. Scatter plots show dissimilar genes along with their GenBank accession numbers.
It is not uncommon to find differences in expression profiles when divergent labeling methods and target molecules are used (11–13). Some of the responsible factors include: (i) different hybridization kinetics between RNA and DNA molecules; (ii) enzyme introduced labeling and copying biases; (iii) differences in the length of labeled RNA and DNA targets; and (iv) other inherent differences in the two methods including their labeling efficiencies.
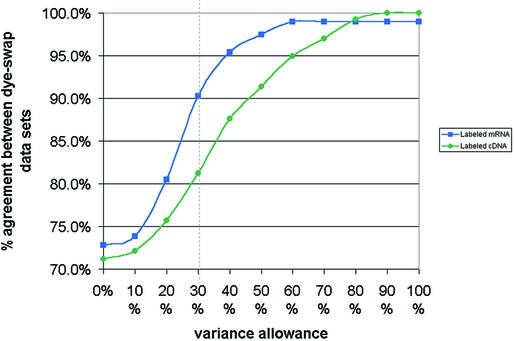
To rule out that the new labeling method was causing any assay bias, we performed a dye-swap assay using total RNA from HL-60 and Jurkat cells. Relative signal intensities from the results were then used in calculating the percentage of all signals that matched between a set of dye-swapped assays. This calculation was performed with an increasing variance allowance, to account for assay-to-assay and slide-to-slide variations. Data from labeled cDNA assays were compared with the data from labeled mRNA assays. Results are presented as a graph in Figure 5. It clearly shows that labeled mRNA targets have a much better inversion correlation than the labeled cDNA targets. Using a typical variance allowance of 30% overall agreement between the dye-swapped assay sets for labeled mRNA generated data was 9% better than for labeled cDNA generated data. We believe that this is due to both the higher dye bias and variability inherent in the enzymatic cDNA labeling system. We also verified expression of a small set of genes by northern analyses and found it to be in 100% agreement (see Supplementary Material for more details).
Figure 5.
Dye-swap microarray assay comparisons between labeled cDNA and labeled mRNA two-color ratiometric assays. Relative signal intensities were used in calculating the percentage of all signals that matched between a set of dye-swapped assays. The results were plotted as a graph against an increasing variance allowance. It clearly shows that labeled mRNA overcomes a lot of bias observed with the labeled cDNA targets.
One key advantage of this methodology is that it is simple, quick and easily automatable. This methodology has much less dye bias and experiment-to-experiment variability, especially as compared with direct enzymatic cDNA labeling methods. Another advantage is that target nucleic acids can be labeled in the presence of enzyme inhibitors, such as EDTA. Direct mRNA labeling preserves the target length and sequence, and exhibits far less bias between the 3′ and 5′ regions of the genes than the corresponding cDNA (our unpublished results). This provides researchers with the ability to study such phenomenon as alternative splicing and mRNA isoforms, where full-length gene representation is a necessity using oligonucleotide arrays. Additionally, the ease of direct RNA labeling would greatly benefit research in prokaryotic systems, where labeling and mRNA enrichment methods are very cumbersome, and RNAi research.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge H. Grevelis for preparing the RNA, K. Appasani for the northern blot data and J. Killian and L. Lacy for insightful discussions.
REFERENCES
- 1.Duggan D.J., Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Expression profiling using cDNA microarrays. Nature Genet., 21 (Suppl.), 10–14. [DOI] [PubMed] [Google Scholar]
- 2.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 3.Nimmakayalu M., Henegariu,O., Ward,D.C. and Bray-Ward,P. (2000) Simple method for preparation of fluor/hapten-labeled dUTP. Biotechniques, 28, 518–522. [DOI] [PubMed] [Google Scholar]
- 4.Stears R.L., Getts,R.C. and Gullans,S.R. (2000) A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol. Genomics, 3, 93–99. [DOI] [PubMed] [Google Scholar]
- 5.Baugh L.R., Hill,A.A., Brown,E.L. and Hunter,C.P. (2001) Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res., 29, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeakley J.M., Fan,J.-B., Doucet,D., Luo,L., Wickham,E., Ye,Z., Chee,M.S. and Fu,X.-D. (2002) Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol., 20, 353–358. [DOI] [PubMed] [Google Scholar]
- 7.Jelsma T., Linkels,E., Quint,W., Van Belkum,A. and Van De Berg,F.M. (1994) Non-isotopic labeling of DNA by newly developed hapten-containing platinum compounds. Biotechniques, 16, 148–153. [PubMed] [Google Scholar]
- 8.Baik M.-H., Friesner,R.A. and Lippard,S.J. (2002) Theoretical study on the stability of N-glycosyl bonds: why does N7-platination not promote depurination. J. Am. Chem. Soc., 124, 4495–4503. [DOI] [PubMed] [Google Scholar]
- 9.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 10.Gullans S.R. (2000) Of microarrays and meandering data points. Nature Genet., 26, 4–5. [DOI] [PubMed] [Google Scholar]
- 11.Rosenow C., Saxena,R.M., Durst,M. and Gingeras,T.R. (2001) Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res., 29, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang C.C., Kozhich,O.A., Chen,M., Inman,J.M., Phan,Q.P., Chen,Y. and Brownstein,M.J. (2002) Amine-modified random primers to label probes for DNA microarrays. Nat. Biotechnol., 20, 738–742. [DOI] [PubMed] [Google Scholar]
- 13.Mahadevappa M. and Warrington,J.A. (1999) A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat. Biotechnol., 17, 1134–1136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.