Abstract
ES cells represent a valuable model for investigating early embryo development and hold promise for future regenerative medicine strategies. The self-renewal of pluripotent mouse ES cells has been shown to require extrinsic stimulation by the bone morphogenetic protein (BMP) and leukemia inhibitory factor signaling pathways and the expression of the transcription factors Oct4 and Nanog. However, the network of interactions among extrinsic and intrinsic determinants of ES cell pluripotency is currently poorly understood. Here, we show that Nanog expression is up-regulated in mouse ES cells by the binding of T (Brachyury) and STAT3 to an enhancer element in the mouse Nanog gene. We further show that Nanog blocks BMP-induced mesoderm differentiation of ES cells by physically interacting with Smad1 and interfering with the recruitment of coactivators to the active Smad transcriptional complexes. Taken together, our findings illustrate the existence of ES cell-specific regulatory networks that underlie the maintenance of ES cell pluripotency and provide mechanistic insights into the role of Nanog in this process.
Keywords: pluripotency, T (Brachyury), self-renewal, mesoderm differentiation, leukemia inhibitory factor
Mouse ES cells are self-renewing, pluripotent cell lines derived from preimplantation embryos (1, 2). Strict culture conditions must be followed to maintain the self-renewal of pluripotent mouse ES cells. Two extrinsic culture requirements, a feeder layer of fibroblasts and the addition of FBS, have been identified as necessary to sustain proliferation of undifferentiated mouse ES cells and their activities pinpointed to specific molecules (reviewed in ref. 3). Thus, self-renewal of mouse ES cells can be sustained in feeder-free conditions by supplementing the culture media with the cytokine leukemia inhibitory factor (LIF) (4, 5). In the absence of LIF, ES cell colonies flatten and form epithelium-like sheets (4, 5). More recently, the self-renewal promoting activity of animal serum has been identified as being mediated by ligands of specific families of the TGF-β superfamily, including the bone morphogenetic protein (BMP) family members BMP2 and BMP4 and the growth and differentiation factor (GDF) family member GDF6 (6). In the absence of BMP/GDF signals, LIF is not sufficient to prevent the neural differentiation of ES cells, whereas the absence of both BMP/GDF and LIF stimulation results in a flattened cell phenotype similar to that observed during LIF withdrawal (6).
The intracellular signaling cascades initiated by both LIF and BMP/GDF that sustain self-renewal of mouse ES cells have been worked out to a significant degree of detail (reviewed in ref. 3). In summary, binding of LIF to its cognate LIF receptor results in the recruitment of gp130 and the formation of a ternary complex that catalyzes the tyrosine phosphorylation, dimerization, and nuclear translocation of the downstream signal transducer STAT3. BMP/GDF, in turn, promotes ES cell self-renewal by inducing the expression of members of the inhibitor of differentiation (Id) family of negative transcriptional modulators, most likely mediated by activation of the TGF-β downstream signal transducer Smad1 (6).
In addition to extrinsic requirements, the pluripotency of mouse ES cells has been shown to depend on intrinsic determinants, such as the expression of the POU transcription factor Oct4 (7) and the divergent homeodomain-containing factor Nanog (8, 9). Both factors are absolutely required for ES cells to maintain their pluripotent identity. Thus, the lack (7) or down-regulation (10) of Oct4 expression induces trophoectoderm differentiation, whereas ES cells lacking Nanog function differentiate to endoderm lineages (8). The relationships among extrinsic and intrinsic determinants of ES cell identity are only recently beginning to be understood. The maintenance of pluripotent ES cell self-renewal by Oct4 requires functional LIF/STAT3 and BMP/GDF/Id signaling cascades (6, 10), but the function of LIF/STAT3 does not seem to be the maintenance of Oct4 expression (10). Overexpression of Nanog, in turn, circumvents the necessity of either LIF or BMP/GDF stimulation (6, 9), although synergism between Nanog function and LIF/STAT3 signaling has been noted (9).
During the investigation of the early events of ES cell specification toward mesoderm lineages, we have recently shown that ES cell cultures normally contain a population of cells expressing T (Brachyury) [T encodes one of the earliest markers of mesoderm differentiation (11, 12)] that we have termed early mesoderm-specified (EM) progenitors (13). Interestingly, in the presence of LIF, the mesoderm-specification of EM progenitors is reverted to generate fully pluripotent ES cells by a mechanism involving Nanog and T, prompting the possibility that T regulates Nanog expression in EM progenitors (13). Here, we show that T and STAT3 coordinately bind to a regulatory element in the mouse Nanog promoter, resulting in increased Nanog expression in EM progenitors. Furthermore, we provide evidence from gain- and loss-of-function experiments demonstrating that Nanog prevents the progression of BMP-induced mesoderm differentiation of ES cells by directly binding to Smad1 and interfering with the recruitment of coactivators, thus blocking the transcriptional activation of downstream targets, including that of T.
Results
T- and STAT-Binding Sites in the Mouse Nanog Gene.
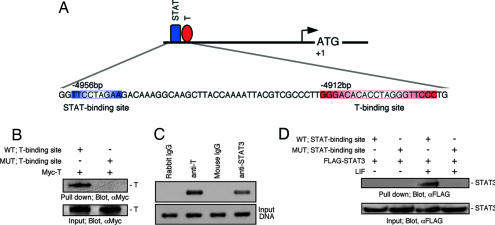
To gain insights into the regulation of Nanog expression by T, we analyzed the mouse Nanog gene in search of regulatory sequences. At 4.91 kb upstream of the translation start site of Nanog, we identified a 20-bp sequence forming an imperfect palindrome that shares homology with the proposed binding site for T (14) (Fig. 1A). We tested the ability of T to bind to oligonucleotides representing this sequence, but not to mutated versions thereof, by performing in vitro pull-down assays of biotin-labeled oligonucleotides incubated with lysates of NIH 3T3 cells expressing Myc-tagged T (Fig. 1B). We also investigated the ability of endogenous T to bind the region of interest in the Nanog promoter in vivo by chromatin immunoprecipitation (ChIP) assays of ES cells with a T-specific antibody (Fig. 1C). Our search for putative regulatory elements in the Nanog promoter also identified a predicted STAT-binding site 44 bp upstream of the T-binding site (Fig. 1A). We tested the ability of STAT3 to bind to this site in vitro (Fig. 1D) and in vivo (Fig. 1C) using experimental approaches similar to the ones used to characterize the T-binding site. These results uncover the presence of functional binding sites for T and STAT3 in the mouse Nanog promoter.
Fig. 1.
Presence of STAT- and T-binding sites in the mouse Nanog promoter. (A) Schematic representation of the 5′ upstream regulatory region of the mouse Nanog gene. Putative STAT- and T-binding sites are indicated. (B) T binds to the putative T-binding site in the Nanog regulatory region, as shown by pull-down assays. WT and mutated (MUT) versions of double-strand oligonucleotides representing the putative T-binding site were used as probes. Input lysates were also blotted with anti-Myc antibody. (C) Chromatin immunoprecipitation (ChIP) assays for the putative T- and STAT-binding sites in the Nanog regulatory region demonstrate specific binding of T and STAT3 to the regulatory region. (Lower) A PCR amplification of input DNA before immunoprecipitation. (D) LIF-dependent binding of STAT3 to the putative STAT-binding site in the Nanog regulatory region. WT and mutated versions of double-strand oligonucleotides for the putative STAT-binding site were used as probes. Input lysates were also blotted with anti-FLAG antibody.
Functional Characterization of the Nanog EM Enhancer.
We next analyzed the significance of the T- and STAT3-binding sites in the Nanog promoter for the biology of EM progenitors. We generated two constructs driving the expression of luciferase, one comprising 5.2 kb of the Nanog genomic sequence upstream of the translation start (−5203Nanog-Luc, which included both STAT3- and T-binding sites) and the other lacking the 5′-most 1 kb (and thus both STAT3- and T-binding sites, −4191Nanog-Luc). Transient transfection of ES cells with either reporter construct resulted in a similar, ≈40-fold transcriptional induction (compared with a promoterless luciferase construct) when ES cells were cultured in medium containing 1,000 units/ml LIF (Fig. 2A), a condition in which EM progenitors are generated at very low frequency (13). These results indicate that the regulatory elements responsible for the constitutive expression of Nanog in ES cells are located in the first 4.2 kb of the mouse Nanog gene upstream of the translation start, consistent with the recent identification of functional Oct4/Sox2-binding sites in the proximal mouse Nanog promoter (15, 16).
Fig. 2.
Regulation of Nanog expression by LIF/STAT3 signaling and T. (A and B) Analysis of transcriptional activities of the Nanog regulatory region by luciferase reporter assay in mouse ES cells (1,000 or 400 units/ml LIF). (A) Both −5203Nanog-Luc and −4191Nanog-Luc showed a similar activation with 1,000 units/ml LIF, whereas −5203Nanog-Luc activity was further increased in cultures maintained with 400 units/ml LIF. (B) Both the T- and STAT3-binding sites were required for activation of Nanog EM enhancer activity in ES cells cultured with 400 units/ml LIF. WT indicates base pairs −5203 to −4192. MUT/T, MUT/S, and MUT/TS indicate the mutation in T-, STAT3-, or both T- and STAT3-binding sites, respectively, in the Nanog EM enhancer. Bars show mean ± SD (n = 4). (C) T and STAT3 physically interact inside cells. T and STAT3 were coimmunoprecipitated when STAT3 was activated by LIF. IP, immunoprecipitation.
Importantly, the transcriptional activity of the −5203Nanog-Luc was increased by ≈4-fold with respect to that of the −4191Nanog-Luc in ES cells adapted to grow in medium supplemented with 400 units/ml LIF (Fig. 2A), in which the EM progenitor population represents ≈20% of the culture (13) (see also Fig. 3). These findings suggest that the up-regulation of Nanog expression in EM progenitors is controlled by regulatory sequences located between base pairs −5,203 and −4,192 upstream of the translation start of the mouse Nanog gene, a region containing the functional STAT3- and T-binding sites. To address whether this region could function as a transcriptional enhancer, we cloned it into a luciferase reporter construct driven by a minimal promoter. This construct increased transcription levels by ≈4.5-fold when transiently transfected into ES cells cultured with 400 units/ml LIF (Fig. 2B). Moreover, the enhancer activity of this region was lost when either or both the STAT3- and the T-binding sites were mutated (Fig. 2B). These results demonstrate the existence of an enhancer of Nanog expression located between base pairs −5,203 and −4,192 in the mouse Nanog gene that is active in conditions that promote the appearance of EM progenitors and to which we refer as the Nanog EM enhancer.
Fig. 3.
The Nanog EM enhancer is active in EM progenitors. Fluorescent images of T-EGFP and Nanog EM enhancer DsRed2 expression in ES cell colonies formed in culture with 400 units/ml LIF. The coexpression of EGFP with DsRed2 in WT (A), but not when T- and/or STAT3-binding sites are mutated (B–D), indicates that the activity of the Nanog EM enhancer in EM progenitor cells is regulated by T and STAT3. (Scale bar: 10μm.)
Because the binding sites for STAT3 and T are located in close proximity to one another in the Nanog EM enhancer and because both T box transcription factors (17–19) and STAT3 (20, 21) have been described as physically interacting with other transcription factors for the regulation of specific promoters, we decided to analyze whether T and STAT3 could interact inside the cell. We tested this possibility in NIH 3T3 cells by cotransfecting expression vectors encoding tagged versions of STAT3 and T (FLAG-STAT3 and Myc-T) and carrying out immunoprecipitation assays. Interestingly, we found an association of T with STAT3 only when nuclear translocation of STAT3 was activated by stimulation with LIF (Fig. 2C).
The Nanog EM Enhancer Is Active in EM Progenitors.
We next investigated whether the activity of the Nanog EM enhancer was restricted to EM progenitors. For this purpose, we used transgenic reporter ES cells that express EGFP under the regulatory sequences of the mouse T gene (T-EGFP ES cells). The maintenance of these cells in culture medium supplemented with 400 units/ml LIF increased the size of the EM progenitor population, which reached a plateau of ≈20% of the cells after 15 passages and was readily detected by the activity of the T-EGFP reporter (13) (see also Fig. 3). To visualize the activity of the Nanog EM enhancer in specific cells, we used it to drive the expression of a red fluorescent protein (DsRed2). T-EGFP ES cells stably expressing this second reporter showed activity of the Nanog EM enhancer only in EM progenitors, as evaluated by the colocalization of EGFP and DsRed2 signals in these cells (Fig. 3A). Consistent with the results of the luciferase reporter assays, mutation of either or both STAT3- and T-binding sites in the Nanog EM enhancer abrogated the activity of this reporter in EM progenitors (Fig. 3 B–D). Our results thus far demonstrate that the up-regulation of Nanog expression in EM progenitors depends on the binding of STAT3 and T to specific sites in the EM enhancer in the mouse Nanog gene.
Nanog Blocks Mesoderm Induction by BMPs.
Because BMPs are potent inducers of mesoderm differentiation in the context of embryo development (22–24), as well as in mouse ES cells (25–28), we next tested whether the generation of EM progenitors was modified by increasing or decreasing BMP signaling in cultures of ES cells. After three passages in medium containing 400 units/ml LIF, the size of the EM progenitor population reached ≈6% in cultures of T-EGFP ES cells (Fig. 4A). This percentage almost doubled when cells were incubated in the presence of recombinant BMP2, BMP4, or BMP7 and was reduced by half upon incubation with noggin (Fig. 4A), a secreted factor that blocks BMP signaling (29, 30). We then tested whether BMP signaling also regulated the maintenance of EM progenitors. When pure populations of EM progenitors were plated in culture medium containing 400 units/ml LIF, ≈75% of the resulting cells underwent a transition to ES cells, whereas the remaining ≈25% maintained EM progenitor identity (Fig. 4B). When the cultures were supplemented with BMPs, the maintenance of EM progenitors increased by ≈2-fold, whereas it was decreased by half upon incubation with noggin (Fig. 4B). Interestingly, overexpression of Nanog in EM progenitors resulted in a decrease in their maintenance similar to that induced by noggin (Fig. 4B). These results indicate that the generation and maintenance of EM progenitors depends, at least in part, on the differentiation-promoting activity of BMPs and suggest that Nanog’s ability to reduce the numbers of EM progenitors may depend on the blockade of BMP signaling.
Fig. 4.
BMP signaling promotes the mesoderm specification of ES cells. (A) Flow-cytometric analysis of T-positive cells in T-EGFP ES cells cultured for three passages with 400 units/ml LIF under conditions of inhibition (noggin) or activation (BMP2, BMP4, and BMP7) of BMP signaling. Bar shows mean ± SD (n = 4). (B) Flow-cytometric analysis of T-positive cells produced from purified T-positive cells cultured with 400 units/ml LIF under conditions of inhibition or activation of BMP signaling. Inhibition of endogenous BMP signaling by noggin decreased the percentage of T-positive cells at a similar level of Nanog overexpression, whereas BMP activation increased the percentage of T-positive cells. Bar shows mean ± SD (n = 4). (C) RT-PCR analyses in ES cells cultured with 400 units/ml LIF with or without addition of BMP7 show that BMP-dependent Id1 expression is negatively regulated by overexpressing Nanog and enhanced when Nanog function is down-regulated. (D) A reporter construct of −1147Id1-Luc containing the Smad-binding sites, but not −927Id1-Luc, was activated in a BMP-dependent manner in ES cells cultured with 400 units/ml LIF. Nanog and inhibitory Smads (Smad6 and Smad7) down-regulated −1147Id1-Luc activity in a similar manner. Bars show mean ± SD (n = 4).
Signaling by BMPs is intracellularly transduced by receptor-regulated Smads (Smad1, Smad5, and Smad8) and the comediator Smad4 and is antagonized by inhibitory Smads [Smad6 and Smad7 (reviewed in ref. 31)]. To characterize the mechanism by which Nanog blocks BMP signaling, we first analyzed the effects of Nanog overexpression and down-regulation in the BMP-induced expression of Id1, a well characterized transcriptional target of BMP signaling (32). Gain of Nanog function in ES cells induced decreased basal levels of Id1 expression and greatly impaired the ability of BMP signaling to up-regulate Id1 expression (Fig. 4C). Conversely, down-regulating Nanog function with Nanog-specific short hairpin RNAs (shRNAs) resulted in increased levels of Id1 expression (Fig. 4C). Next, we used Id1-luciferase reporter constructs containing (−1147Id1-Luc) or lacking (−927Id1-Luc) the Smad-binding sites (33) and analyzed their activity in ES cells. Transient transfection of these reporters in ES cells resulted in a ≈4.5-fold activation of the −1147Id1-Luc reporter when compared with the −927Id1-Luc reporter (Fig. 4D), indicating the existence of a significant level of endogenous BMP signaling associated with our culture conditions (see Discussion). Addition of BMP to the culture medium resulted in a strong up-regulation of the −1147Id1-Luc reporter as compared with the −927Id1-Luc reporter (Fig. 4D). That the activation of the −1147Id1-Luc reporter was due to BMP signaling was further confirmed by the fact that cotransfection of ES cells with cDNAs encoding inhibitory Smads drastically reduced the transcriptional activity of the reporter induced by endogenous or exogenous BMPs (Fig. 4D). Interestingly, Nanog overexpression in ES cells closely mimicked the effect of inhibitory Smads (Fig. 4D), suggesting that Nanog may block BMP signaling by interfering with the formation of activated Smad complexes.
Nanog Binds to Smad1.
Inhibitory Smads negatively regulate BMP signaling by binding to activated receptor-regulated Smads, hence limiting their availability to form transcriptionally active complexes with Smad4 and/or other nuclear cofactors (reviewed in ref. 31). To address whether Nanog blocked BMP signaling by a similar mechanism, we first analyzed its ability to interact with the receptor-regulated Smad1 inside the cell. Coimmunoprecipitation assays in NIH 3T3 cells revealed that Nanog was indeed able to bind Smad1 only when the latter was activated by cotransfection of a constitutively active ALK3. Similarly, the interaction of endogenous Nanog and Smad1 could be detected in ES cells and was enhanced by BMP stimulation (Fig. 6 A and B, which is published as supporting information on the PNAS web site).
Next, we mapped the interaction domain of Smad1 with Nanog. The different Smads contain two conserved domains, the N-terminal Mad homology (MH) 1 and the C-terminal MH2 domain, separated by a poorly conserved linker. The interaction of receptor-regulated Smads with Smad4 and other transcription factors and cofactors, as well as with inhibitory Smads, occurs through the MH2 domain (reviewed in ref. 31). In cells cotransfected with Nanog and expression constructs encoding the individual MH1, MH1 plus linker, or MH2 domains of Smad1, interaction with Nanog was found exclusively with the MH2 domain (Fig. 6C). These results are consistent with a negative role of Nanog in BMP signaling by interfering with the interaction of receptor-activated Smads with Smad4 and/or additional nuclear factors.
The paralogous transcriptional coactivators cAMP responsive element-binding protein (CREB) binding protein (CBP) and p300 are nuclear cofactors important for TGF-β signaling, including that of BMPs, which interact with the MH2 domain of receptor-regulated Smads and Smad4 (reviewed in ref. 31). To gain further insights into the mechanism of Nanog-mediated down-regulation of BMP signaling, we tested whether Nanog interfered with the recruitment of p300 to the complexes of activated Smads. For this purpose, Myc-tagged Smad1, hemagglutinin (HA)-tagged p300, and constitutively active ALK3 were expressed in NIH 3T3 cells with or without HA-tagged Nanog. Immunoprecipitations of cell lysates were performed with anti-Myc antibodies followed by Western blotting using anti-HA antibodies. In the absence of Nanog, Smad1 efficiently coimmunoprecipitated p300. In the presence of coexpressed Nanog, the amount of p300 bound to Smad1 decreased in a Nanog dose-dependent manner (Fig. 6D). The functional significance of these findings was further verified by the fact that overexpression of p300 completely rescued the down-regulation in the transcriptional activity of the Id promoter induced by Nanog (Fig. 5A). These results indicate that Nanog negatively regulates BMP signaling by interfering with the recruitment of the coactivator p300 to the Smad transcriptional complex.
Fig. 5.
Nanog down-regulates the expression of BMP targets. (A) Overexpression of p300 rescues the down-regulation of −1147Id1-Luc activity induced by Nanog. Bars show mean ± SD (n = 4). (B) The −396T-Luc, but not the −204T-Luc, reporter construct is activated in a BMP-dependent manner in ES cells cultured with 400 units/ml LIF. Nanog and inhibitory Smads (Smad6 and Smad7) down-regulate −396T-Luc activity in a similar manner. The down-regulation of −396T-Luc activity induced by Nanog is rescued by overexpression of p300. Bars show mean ± SD (n = 4). (C) Sequence of the 5′ upstream regulatory region of the mouse T gene. Three putative BMP-responsive Smad-binding sites are indicated by boxes. (D) Schematic representation of the negative feedback mechanism by which Nanog blocks BMP-induced T expression in the presence of LIF/STAT3 signaling. Black arrows depict positive direct transcriptional regulation, gray arrows depict positive posttranslational regulation, and red lines represent inhibitory regulation. See Results for details.
Nanog Blocks the Induction of T Expression by BMPs.
Finally, the finding that the expression of Xbra, the homologue of T in Xenopus, is regulated by TGF-β signals (34) prompted us to investigate whether T could be a transcriptional target of BMP signaling in ES cells, and, if so, whether Nanog could directly block the induction of T by BMPs. In a preliminary analysis, we identified a BMP-responsive element in the ≈1.2-kb region upstream of the translation initiation site of the mouse T promoter (data not shown). We then generated a series of luciferase reporter constructs covering this region. We transfected these constructs into ES cells cultured in medium containing 400 units/ml LIF and supplemented with BMP7 and further mapped the BMP-responsive element to a region located between base pairs −396 and −204 of the mouse T gene (Fig. 5C). Under these conditions, the activity of the −396T-Luc reporter was ≈5-fold that of the −204T-Luc reporter and decreased by half upon coexpression of inhibitory Smads or Nanog (Fig. 5B). Interestingly, the down-regulation of the −396T-Luc activity induced by Nanog could be completely rescued by the coexpression of p300 (Fig. 5B). The analysis of this region in the mouse T promoter detected three motifs with homology to the reported consensus of BMP-responsive Smad-binding sites (35). These results indicate that T is a direct transcriptional target of BMP signaling and that Nanog down-regulates T expression by inhibiting BMP signaling at the level of the formation of active Smad/p300 complexes.
Discussion
Differentiation-Promoting Activity of BMPs.
The results from our analyses indicate that the generation of EM progenitors from ES cells depends on the direct mesoderm-inducing ability of BMP stimulation (Figs. 4 and 5). This finding is consistent with reported roles of BMP signaling during embryo development (22–24) and with previous studies of ES cell differentiation in vitro (25–28). However, the mesoderm-differentiating activity of BMPs seems to be at odds with their role in maintaining the self-renewal of pluripotent ES cells (6). Indeed, BMP signaling seems to have contrasting effects on the maintenance of ES cell pluripotency. On the one hand, BMPs are necessary to prevent ES cell differentiation toward neural fates (6, 26, 36). On the other hand, signaling by BMPs induces a loss of ES cell pluripotency by promoting their differentiation toward nonneural fates such as mesoderm-derived lineages (refs. 25 and 27 and this work). These opposing effects of BMPs can be partially explained by differences in the experimental conditions used in those studies. Thus, in the absence of LIF, low concentrations of BMPs (≈0.25–10 ng/ml) promote mesoderm differentiation (25–27) at the expense of neural fates (26). In the presence of LIF, however, similarly low concentrations of BMPs prevent neural differentiation of ES cells (6, 36) and maintain their pluripotency with no signs of mesoderm differentiation (6). Consistent with this notion, we did not detect increased generation of EM progenitors with BMP concentrations <100 ng/ml in the presence of LIF (data not shown). Thus, LIF seems to render ES cells refractory to the mesoderm-inducing activity of BMPs. Our studies demonstrate that this resistance depends, at least in part, on a negative feedback mechanism mediated by Nanog and T.
A Negative Feedback That Blocks Mesoderm Specification in ES Cells.
Our results also provide mechanistic insights into the relationships among ES cell pluripotency determinants. Thus, we characterize a negative feedback mechanism that prevents mesoderm specification of ES cells in the presence of LIF (Fig. 5D). In this mechanism, mesoderm differentiation of ES cells is initiated by BMP signaling. Possible sources of BMP activity in our culture conditions include FCS (25), the fibroblast feeder layer, and/or ES cells themselves (6). Consistent with this, we detect a significant activation of the −1147Id1-Luc reporter even in the absence of exogenous BMP supplements (Fig. 4C). As a direct consequence of BMP signaling, ES cells undergoing mesoderm specification activate the expression of T (Fig. 5B). In the presence of LIF, activated STAT3 interacts with T and binds to the Nanog EM enhancer, thus resulting in the up-regulation of Nanog expression in these cells. Increased levels of Nanog, in turn, directly block the mesoderm-differentiation activity of BMPs, thereby limiting the progression of mesoderm specification and down-regulating T expression, ultimately regenerating pluripotent ES cells from EM progenitors.
However, it is clear that the functions of LIF and Nanog in the maintenance of ES cell pluripotency are not restricted to participating in the negative feedback mechanism characterized here. Thus, the complete lack of Nanog function promotes differentiation of ES cells to endoderm lineages (8), indicating the existence of roles of Nanog other than that of preventing mesoderm differentiation. Indeed, the up-regulation of Nanog expression by T and STAT3 takes place only in EM progenitors (Fig. 3A), whereas the constitutive expression of Nanog in ES cells is regulated by more proximal regions of the Nanog promoter (Fig. 2A).
Mouse ES cells, consistent with their developmental origin in the embryo epiblast, have the ability to give rise to derivatives of all three primary germ layers. However, unlike cells in the epiblast, in which pluripotency is very transient, mouse ES cells can be maintained in culture indefinitely in a pluripotent state. The mechanism(s) whereby the adaptation to culture conditions releases epiblast cells from the loss of pluripotency remain an outstanding question in the biology of ES cells. Our results provide insights into the mechanisms by which the determinants of ES pluripotency interact to actively prevent lineage specification of ES cells.
Materials and Methods
ES Cell Culture.
Mouse ES cells (J1 line, ref. 37) were maintained on mouse embryonic fibroblasts in standard media containing 15% FBS (HyClone) and LIF (1,000 or 400 units/ml, ESGRO; Chemicon). In some cases, BMP2, BMP4, BMP7 (all 200 ng/ml), or noggin (100 ng/ml) was supplemented in the cultures [BMP7 and noggin were kind gifts from S. Choe (The Salk Institute for Biological Studies); BMP2 and BMP4 were from R & D Systems]. In each passage, 3 ×105 cells were plated in a 6-cm dish. For clonal analyses, T-positive and -negative cells were separately isolated by using FACS (MoFlo; Cytomation, Fort Collins, CO) and plated at a density of 1 × 104 cells per 6-cm dish. Before analyses, trypsinized cells were allowed to attach to gelatin-coated plates for 45 min, thus removing >95% of feeder cells and recovering >95% of ES cells.
Fluorescent Reporter Constructs.
The first 24 bp of the T coding sequence in a mouse genomic bacterial artificial chromosome (BAC) (RP23-456E5, BACPAC Resources, Oakland, CA) were replaced with an EGFP-Neo cassette by homologous recombination in bacteria (38). The bacterial strain DY380 was kindly provided by N. G. Copeland (National Cancer Institute, Frederick, MD). Transgenic ES cell lines expressing this construct (T-EGFP) were obtained by electroporation and G418 selection. DNA fragments of the mouse Nanog gene were obtained from a mouse genomic BAC clone (RP23-406B15). The Nanog EM enhancer region (base pairs −5203 to −4192) and its mutated versions, in which the T- and/or STAT-binding sites were mutated (T, GGGACGCGCCTGAATCCTAT; STAT, CCAATAGAA; italics indicate mutated nucleotides), were inserted into a c-fos minimal promoter–DsRed2 vector. Mouse ES cells were transfected with these reporter constructs by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Supporting Information.
The immunoprecipitation, Western blotting, RT-PCR, and chromatin immunoprecipitation procedures and the expression constructs used are in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Robert Benezra, Senyon Choe, Neal G. Copeland, Richard Eckner, Kohei Miyazono, and Shinya Yamanaka for sharing reagents; Dirk Buscher, Chris Kintner, Isao Oishi, Junichiro Sonoda, and Ayumu Tashiro for helpful suggestions; Harley Pineda, Timothy Chapman, and Henry Juguilon for excellent technical assistance; and May-Fun Schwarz for help in the preparation of the manuscript. A.S. was partially supported by Japan Society for the Promotion of Science (JSPS) Research Fellowships for Young Scientists; A.S., T.M., and K.N. are partially supported by JSPS Postdoctoral Fellowships for Research Abroad; and A.R. and C.R.-E. were partially supported by Fundación Inbiomed. Funding for mouse ES cell work in J.C.I.B.’s laboratory was from the G. Harold and Leila Y. Mathers Charitable Foundation and the National Institutes of Health.
Abbreviations
- LIF
leukemia inhibitory factor
- BMP
bone morphogenetic protein
- GDF
growth and differentiation factor
- EM
early mesoderm-specified.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Martin G. R. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans M. J., Kaufman M. H. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Chambers I., Smith A. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 4.Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 5.Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 6.Ying Q. L., Nichols J., Chambers I., Smith A. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 7.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 8.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 9.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 10.Niwa H., Miyazaki J., Smith A. G. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann B. G., Labeit S., Poustka A., King T. R., Lehrach H. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson D. G., Bhatt S., Herrmann B. G. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A., Raya A., Kawakami Y., Morita M., Matsui T., Nakashima K., Gage F. H., Rodriguez-Esteban C., Belmonte J. C. Nat. Clin. Pract. Cardiovasc. Med. 2006;3:S114–S122. doi: 10.1038/ncpcardio0442. [DOI] [PubMed] [Google Scholar]
- 14.Kispert A., Herrmann B. G. EMBO J. 1993;12:3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. Mol. Cell. Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 17.Hiroi Y., Kudoh S., Monzen K., Ikeda Y., Yazaki Y., Nagai R., Komuro I. Nat. Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 18.Stennard F. A., Costa M. W., Elliott D. A., Rankin S., Haast S. J., Lai D., McDonald L. P., Niederreither K., Dolle P., Bruneau B. G., et al. Dev. Biol. 2003;262:206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 19.Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., et al. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 20.Zhu M., John S., Berg M., Leonard W. J. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 21.Collum R. G., Brutsaert S., Lee G., Schindler C. Proc. Natl. Acad. Sci. USA. 2000;97:10120–10125. doi: 10.1073/pnas.170192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dale L., Howes G., Price B. M., Smith J. C. Development (Cambridge, U.K.) 1992;115:573–585. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]
- 23.Jones C. M., Lyons K. M., Lapan P. M., Wright C. V., Hogan B. L. Development (Cambridge, U.K.) 1992;115:639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- 24.Winnier G., Blessing M., Labosky P. A., Hogan B. L. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 25.Johansson B. M., Wiles M. V. Mol. Cell. Biol. 1995;15:141–151. doi: 10.1128/mcb.15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finley M. F., Devata S., Huettner J. E. J. Neurobiol. 1999;40:271–287. [PubMed] [Google Scholar]
- 27.Czyz J., Wobus A. Differentiation. 2001;68:167–174. doi: 10.1046/j.1432-0436.2001.680404.x. [DOI] [PubMed] [Google Scholar]
- 28.Yuasa S., Itabashi Y., Koshimizu U., Tanaka T., Sugimura K., Kinoshita M., Hattori F., Fukami S. I., Shimazaki T., Okano H., et al. Nat. Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- 29.Holley S. A., Neul J. L., Attisano L., Wrana J. L., Sasai Y., O’Connor M. B., De Robertis E. M., Ferguson E. L. Cell. 1996;86:607–617. doi: 10.1016/s0092-8674(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman L. B., De Jesus-Escobar J. M., Harland R. M. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y., Massague J. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 32.Hollnagel A., Oehlmann V., Heymer J., Ruther U., Nordheim A. J. Biol. Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima K., Takizawa T., Ochiai W., Yanagisawa M., Hisatsune T., Nakafuku M., Miyazono K., Kishimoto T., Kageyama R., Taga T. Proc. Natl. Acad. Sci. USA. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latinkic B. V., Umbhauer M., Neal K. A., Lerchner W., Smith J. C., Cunliffe V. Genes Dev. 1997;11:3265–3276. doi: 10.1101/gad.11.23.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J., Johnson K., Chen H. J., Carroll S., Laughon A. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 36.Tropepe V., Hitoshi S., Sirard C., Mak T. W., Rossant J., van der Kooy D. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 37.Li E., Bestor T. H., Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 38.Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. Proc. Natl. Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.