Abstract
The ASPM (abnormal spindle-like microcephaly-associated) protein has previously been implicated in the determination of human cerebral cortical size, but the cell biological basis of this regulation has not been studied. Here we investigate the role of Aspm in mouse embryonic neuroepithelial (NE) cells, the primary stem and progenitor cells of the mammalian brain. Aspm was found to be concentrated at mitotic spindle poles of NE cells and to be down-regulated with their switch from proliferative to neurogenic divisions. Upon RNA interference in telencephalic NE cells, Aspm mRNA is reduced, mitotic spindle poles lack Aspm protein, and the cleavage plane of NE cells is less frequently oriented perpendicular to the ventricular surface of the neuroepithelium. The alteration in the cleavage plane orientation of NE cells increases the probability that these highly polarized cells undergo asymmetric division, i.e., that apical plasma membrane is inherited by only one of the daughter cells. Concomitant with the resulting increase in abventricular cells in the ventricular zone, a larger proportion of NE cell progeny is found in the neuronal layer, implying a reduction in the number of NE progenitor cells upon Aspm knock-down relative to control. Our results demonstrate that Aspm is crucial for maintaining a cleavage plane orientation that allows symmetric, proliferative divisions of NE cells during brain development. These data provide a cell biological explanation of the primary microcephaly observed in humans with mutations in ASPM, which also has implications for the evolution of mammalian brains.
Keywords: asymmetric division, brain size evolution, cleavage plane, neurogenesis
The size of the mammalian neocortex is thought to be principally determined by the number of radial units generated during development (1). This lateral expansion largely reflects the number of proliferative divisions of progenitor cells (one progenitor→two progenitors). The primary progenitor cells of the central nervous system are the neuroepithelial (NE) cells, which characteristically exhibit apical–basal polarity (2). A key feature of proliferative divisions of NE cells and of the radial glial cells they transform into (3) is that cleavage occurs along their apical–basal axis, i.e., perpendicular to the ventricular surface of the neuroepithelium, which ensures the symmetric distribution of polarized cell fate determinants to the daughter cells (2, 4, 5). The switch of NE and radial glial cells from symmetric, proliferative divisions to asymmetric, neurogenic divisions (one progenitor→one progenitor plus one neuron/neuronal precursor), which limits the lateral expansion of the neocortex, is accompanied by a deviation of the cleavage plane from the apical–basal orientation (4). This deviation often is only relatively small but nonetheless results in the apical plasma membrane of NE cells being bypassed (rather than bisected) by the cleavage furrow and, hence, being inherited by only one of the daughter cells (5, 6).
NE and radial glial cells are characterized by a highly elongated shape. Consequently, given that junctional complexes at the apical-most end of the lateral plasma membrane separate the apical and basolateral surface domains of these polarized cells, the apical domain constitutes only a minute (1–2%) fraction of their entire cell surface (5, 7), which, in turn, implies that when these cells undergo symmetric, proliferative divisions, the mitotic spindle must be oriented exactly perpendicular to their apical–basal axis to ensure that the cleavage furrow will bisect (rather than bypass) the apical domain. It follows that NE and radial glial cells, in particular, should express proteins that maintain this spindle orientation and do so especially during symmetric, proliferative divisions and, given that the cleavage furrow ingresses in the basal-to-apical direction (5), until the very end of M phase.
The Aspm (abnormal spindle-like microcephaly-associated) protein (8) is an interesting candidate for such a role for several reasons. First, the Drosophila homologue of Aspm, Asp, exerts a critical role at spindle poles during mitosis (9). Specifically, Asp is thought to focus microtubules, including those of the central spindle (10), a key structure for the positioning of the cleavage furrow (11). Similar to Asp, ASPM in nonneural human cells has recently been shown to localize to spindle poles (12–14), suggesting that it may be involved in some aspect of mitotic spindle function in mammalian cells.
Second, mutations in ASPM are the most common cause of primary microcephaly in humans (8, 15), indicating a direct role for this protein in regulating cerebral cortical size. Interestingly, the macroscopic structure of the cerebral cortex in these humans (8) suggests a reduction in the generation of radial units during cortical development, which raises the possibility that ASPM is required for the maintenance of symmetric, proliferative divisions of NE cells. Consistent with a role of Aspm in the lateral expansion of the neocortex, the primate and human lineages show strong positive selection for evolutionary change in the Aspm protein (16, 17).
Together, these data suggest that Aspm may regulate cerebral cortical size by controlling an aspect of mitotic spindle function that is crucial for maintaining symmetric, proliferative divisions of the highly elongated, polarized NE cells (2), thereby allowing the lateral expansion of the neocortex. In the present study, we have investigated a possible role of Aspm in regulating cleavage plane orientation and symmetric, proliferative divisions versus asymmetric, neurogenic divisions of NE cells in the mouse embryonic telencephalon.
Results and Discussion
Down-Regulation of Spindle Pole-Associated Aspm in NE Cells Undergoing Neurogenic Divisions.
In situ hybridization (Fig. 5, which is published as supporting information on the PNAS web site) confirmed previous observations (8, 18) that Aspm mRNA is expressed in the ventricular zone (VZ) of the murine embryonic forebrain. Interestingly, Aspm expression in NE cells was highest when these cells underwent proliferative (rather than neurogenic) divisions and exhibited a highly (rather than only moderately) elongated shape. Specifically, Aspm expression (i) was still comparatively rare at early developmental stages [embryonic day (E)8.5], when NE cells are not yet highly elongated (7); (ii) was detected in most, if not all, VZ cells around the onset of neurogenesis (E9.5–E11.5), when symmetric, proliferative divisions of NE cells prevail (19); and (iii) declined progressively at later stages of neurogenesis (E13.5–E17.5), when an increasing proportion of VZ cells have switched to neurogenic divisions (19). Given these observations, we decided to explore the possibility that Aspm may function specifically in the proliferative divisions of NE cells.
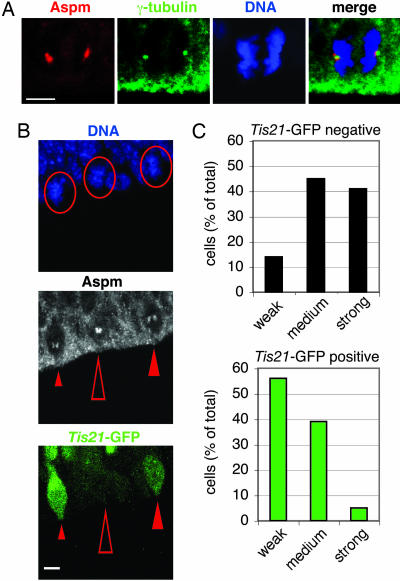
To this end, we first determined the subcellular localization of Aspm in NE cells. Immunostaining of mitotic NE cells in the E12.5 mouse telencephalon using an affinity-purified antibody raised against recombinant mouse Aspm150–263 showed that Aspm was concentrated at the poles of the mitotic spindle (Fig. 1A; see also Fig. 6, which is published as supporting information on the PNAS web site). Specifically, Aspm immunoreactivity was clustered in the immediate vicinity of, but did not overlap with, the γ-tubulin staining of centrosomes. Association of Aspm with the spindle poles was observed during all phases of mitosis, with an apparent decrease in the intensity of immunostaining in telophase (Figs. 1A and 6). However, Aspm did not appear to be associated with centrosomes during interphase (Fig. 2B, arrows). The association of Aspm with the spindle poles of NE cells is consistent with previous observations on the subcellular localization of Asp in mitotic Drosophila neuroblasts (9) and of ASPM in mitotic nonneural human cells in culture (12–14).
Fig. 1.
Aspm is concentrated at the spindle poles during mitosis and is down-regulated in NE cells undergoing neurogenic divisions. (A) The dorsal telencephalon of E12.5 Tis21–GFP knockin mice was stained with DAPI (DNA, blue) to reveal NE cells in anaphase and immunostained for Aspm (red) and γ-tubulin (green). All cells analyzed were Tis21–GFP-negative (data not shown). Note the concentration of Aspm in the immediate vicinity of the γ-tubulin-stained centrosomes. (Scale bar, 5 μm.) (B) Comparison of Aspm immunoreactivity at spindle poles in metaphase Tis21–GFP-negative NE cells (proliferative divisions, open arrowheads) versus Tis21–GFP-positive NE cells (neurogenic divisions, filled arrowheads) in the dorsal telencephalon of an E14.5 Tis21–GFP knockin mouse. (Top) DNA staining using DAPI (blue); circles indicate the three metaphase cells analyzed. (Middle) Aspm immunoreactivity (white); the small, medium, and large arrowheads indicate weak, medium, and strong Aspm immunoreactivity at spindle poles, respectively. (Bottom) Tis21–GFP fluorescence (green). (Scale bar, 5 μm.) (A and B) The apical surface of the VZ is down. (C) Quantification in the dorsal telencephalon of E14.5 Tis21–GFP knockin mice of prophase or metaphase Tis21–GFP-negative (black bars, 22 cells) and Tis21–GFP-positive (green bars, 18 cells) NE cells showing weak, medium, or strong Aspm immunoreactivity at spindle poles, expressed as a percentage of total (weak plus medium plus strong). Data are from 19 cryosections that originated from at least four brains.
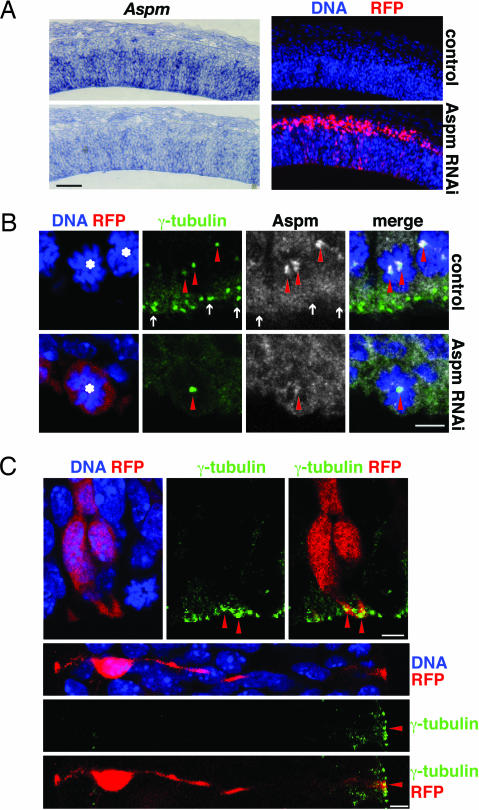
Fig. 2.
Knockdown of Aspm results in its loss from centrosomes in mitosis but does not affect the apical localization of centrosomes of interphase NE cells. (A) RNAi of Aspm mRNA. Mouse E10.5 dorsal telencephalon was coelectroporated with Aspm esiRNAs and mRFP plasmid followed by 24 h of whole-embryo culture, and consecutive cryosections were analyzed by in situ hybridization for Aspm mRNA (Left) and RFP fluorescence (Right). Nontargeted (control; Upper) and targeted (Aspm RNAi; Lower) hemispheres are from the same cryosections. The apical surface of the VZ is down. (Scale bar, 100 μm.) (B) Knockdown of Aspm. Mouse E12.5 dorsal telencephalon was coelectroporated with Aspm esiRNAs and mRFP plasmid followed by 24 h of in utero development, and cryosections were analyzed for mitotic NE cells (asterisks) by DNA staining using DAPI (blue), RFP fluorescence (red), and γ-tubulin (green) and Aspm (white) immunofluorescence. Single optical sections are shown. (Upper) Nontargeted hemisphere serving as control. (Lower) Targeted hemisphere subjected to Aspm RNAi. Note the loss of Aspm immunoreactivity from the spindle poles (arrowheads) upon Aspm knockdown. The cell shown is representative of all targeted cells of the electroporated hemisphere. Arrows indicate centrosomes of adjacent NE cells in interphase, which lack Aspm. The apical surface of the VZ is down. (Scale bar, 5 μm.) (C) Interphase centrosomes. Mouse E10.5 dorsal telencephalon was coelectroporated with Aspm esiRNAs and mRFP plasmid followed by 24 h of whole-embryo culture, and cryosections were analyzed for DNA staining with DAPI (blue), RFP fluorescence (red), and γ-tubulin immunofluorescence (green). Note the apical localization of centrosomes (arrowheads) in the targeted NE cells in interphase. (Upper) Two NE cells in G1/G2. The apical surface of the VZ is down. (Scale bar, 5 μm.) (Lower) S-phase NE cell. The apical surface of the VZ is to the right. (Scale bar, 5 μm.)
If Aspm has a critical function in maintaining spindle orientation during symmetric, proliferative NE cell divisions, one might expect that it is more highly expressed in proliferating than neuron-generating NE cells. To address this issue, we made use of Tis21–GFP knockin mouse embryos (19) in which GFP in the developing central nervous system is selectively expressed in progenitors undergoing neurogenic divisions but not in progenitors undergoing proliferative divisions. Indeed, comparative analysis of the telencephalon of E14.5 Tis21–GFP knockin mice revealed that Tis21–GFP-negative NE cells tended to exhibit more intense Aspm immunostaining at spindle poles than did Tis21–GFP-positive NE cells (Fig. 1C), as exemplified for three neighboring metaphase NE cells in Fig. 1B. In no case did we observe more intense Aspm staining in Tis21–GFP-positive than in Tis21–GFP-negative cells. These data indicate that Aspm is down-regulated in NE cells concomitant with their switch from proliferative to neurogenic divisions.
Knockdown of Aspm Perturbs Vertical Cleavage Plane Orientation.
Given these observations, we explored whether Aspm contributes to maintaining the mitotic spindle positioned perpendicular to the apical–basal axis of NE cells, thus ensuring bisection of their apical plasma membrane and, hence, their symmetric divison (5). Aspm was knocked down by RNA interference (RNAi) elicited by electroporation of endoribonuclease-prepared, short interfering RNAs (esiRNAs; a mixture of short interfering RNAs) along with a monomeric red fluorescent protein (mRFP) plasmid into one telencephalic hemisphere of E10.5 or E12.5 mice followed by development for 24 h in whole-embryo culture or in utero, respectively (20, 21). This protocol indeed resulted in a reduction in Aspm mRNA levels (Fig. 2A) and the loss of spindle pole-associated Aspm protein (Fig. 2B, arrowheads). Aspm knockdown did not perturb the localization of centrosomes in NE cells in interphase, which remained associated with the apical cell cortex (Fig. 2C, arrowheads), consistent with previous observations (22).
However, Aspm knockdown had severe effects on centrosome localization in M-phase NE cells. Centrosomes were frequently seen detached from the sister chromatids (Fig. 3A Lower, arrowheads). This phenotype was particularly evident in telophase cells and was not observed before anaphase, suggesting that centrosome detachment due to loss of Aspm may occur during sister chromatid separation.
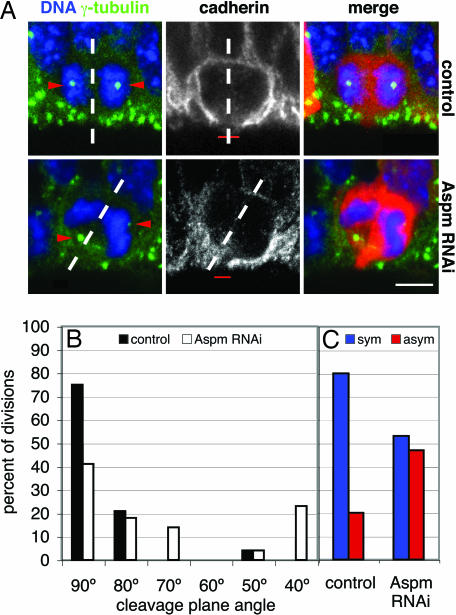
Fig. 3.
Knockdown of Aspm alters the cleavage plane of NE cells, resulting in their asymmetric division. (A) Cleavage plane and cadherin hole analysis. Mouse E12.5 dorsal telencephalon was either electroporated with mRFP plasmid only (Upper) or coelectroporated with Aspm esiRNAs and mRFP plasmid (Lower), followed by 24 h of in utero development, and cryosections were analyzed for NE cells in anaphase or telophase by DNA staining with DAPI (blue), RFP fluorescence (red), and γ-tubulin (green) and cadherin (white) immunofluorescence. The cleavage plane (dashed lines) was deduced from the orientation of the sister chromatids (5, 6). Note the aberrant position of the γ-tubulin-stained centrosomes (arrowheads, stack of optical sections) in the targeted cell and the oblique cleavage plane, which bypasses the cadherin hole (red bar, single optical section), upon Aspm knockdown. The apical surface of the VZ is down. (Scale bar, 5 μm.) (B and C) Quantification of cleavage plane orientation (B) and cadherin hole distribution (C). Dorsal telencephalon of E10.5 Tis21–GFP knockin mice was electroporated with mRFP plasmid only (control) or coelectroporated with mRFP plasmid and Aspm esiRNAs (Aspm RNAi), followed by 24 h of whole-embryo culture. Tis21–GFP-negative, mRFP-expressing NE cells in anaphase or telophase were analyzed for cleavage plane orientation and the position of the cadherin hole as in A. (B) Orientation of the cleavage plane relative to the radial, apical–basal axis of the neuroepithelium (defined as 90°), expressed as a percentage of all divisions for the control (black bars; n = 24) or Aspm RNAi (white bars; n = 22) condition. Groups of cleavage plane angle are ±5°. (C) Cleavage planes bisecting [symmetric (sym), blue bars] or bypassing [asymmetric (asym), red bars] the cadherin hole, expressed as a percentage of all divisions for the control (n = 15) or Aspm RNAi (n = 15) condition.
A related aspect of this phenotype was a significant alteration in the orientation of the cleavage plane of Tis21–GFP-negative NE cells, which normally is vertical, i.e., perpendicular or nearly perpendicular to the ventricular surface of the neuroepithelium (compare with Fig. 3A Upper, dashed lines) (5). Deduction of the cleavage plane from the orientation of the sister chromatids of NE cells in anaphase/early telophase (5) revealed that, in agreement with these previous observations, nearly all mitotic Tis21–GFP-negative NE cells in the control condition showed a vertical cleavage plane orientation (Fig. 3B, black bars). Upon Aspm knockdown, however, almost half of the cleavage planes of Tis21–GFP-negative NE cells deviated significantly from the normal, vertical orientation (Fig. 3B, white bars).
Our observation that Aspm-knocked-down NE cells progressed through telophase (Fig. 3A Lower) implies that, unlike asp mutations in Drosophila, which cause neuroblasts to arrest in metaphase (23), Aspm knockdown in mouse NE cells does not appear to block mitosis in metaphase. This conclusion is in line with the notion that, in vertebrates, mitotic spindles can exist in the absence of centrosomes (24). Accordingly, we did not detect any increase in mitotic NE cells (identified by phosphohistone H3 immunostaining) upon Aspm knockdown (see the legend to Fig. 7, which is published as supporting information on the PNAS web site).
Lack of Aspm Promotes Asymmetric Cell Division.
At the onset of neurogenesis, NE cells, which characteristically show apical–basal polarity, have a highly elongated shape such that only a subtle deviation in cleavage plane from the normal vertical orientation suffices to result in an asymmetric rather than symmetric distribution of their apical plasma membrane and adjacent adherens junctions to the daughter cells (5–7). Upon immunostaining for cadherin, a constituent of the lateral NE cell plasma membrane, the apical plasma membrane of mitotic NE cells, identified by the presence of the apical marker prominin-1 (25), appears as a small, unstained segment of the cell surface, referred to as “cadherin hole” (5). Symmetric versus asymmetric distribution of the apical plasma membrane to the daughter cells can be predicted from the orientation of the cleavage plane relative to the cadherin hole (5).
Such analysis revealed that the alteration in cleavage plane orientation upon loss of Aspm (Fig. 3B) had marked consequences for the distribution of the apical membrane upon division of Tis21–GFP-negative NE cells. Whereas, in the control condition, consistent with previous observations (5), 80% of the cleavage planes were predicted to bisect the apical membrane, resulting in a symmetric distribution to the daughter cells (Fig. 3C, left blue bar; see also Fig. 3A Upper Center, dashed line), almost half of the cleavage plane orientations observed upon Aspm knockdown were predicted to bypass the apical membrane, resulting in an asymmetric distribution (Fig. 3C, right red bar; see also Fig. 3A Lower Center, dashed line). Such effects on cleavage plane orientation have not been noticed upon RNAi using esiRNAs for various other proteins expressed in NE cells (F. Calegari, L. Farkas, A. Grzyb, A.-M. Marzesco, and W.B.H., unpublished data), indicating that the present phenotype was specifically due to the loss of Aspm.
Increased Non-NE Fate of NE Cell Progeny After Aspm Knockdown.
In normal mouse brain development, an asymmetric distribution of the apical plasma membrane upon division of NE cells is highly correlated with these divisions switching from being proliferative to becoming neurogenic (5), as reflected by Tis21 (26) or Tis21–GFP (19) expression. A corollary of this switch is that NE cell expansion ceases. Given the increase in cleavage plane orientations leading to an asymmetric apical membrane distribution in dividing NE cells upon Aspm knockdown (Fig. 3), we explored whether this effect would indeed reduce proliferative divisions of NE cells, as reflected by an increase in non-NE progeny.
In contrast to NE cells (and the related radial glial cells), whose centrosomes [as in other epithelial cells (27)] are located at the ventricular (apical) surface (22, 27), one of the characteristics of their non-NE progeny, which lack apical plasma membrane, is the abventricular localization of their centrosome. For example, in neurons, centrosomes are located in the vicinity of the nucleus when they are migrating through the VZ (28) (A. Attardo, W. Haubensak, F. Calegari, and W.B.H., unpublished data) and after they have reached the neuronal layers (Fig. 4A, control). Consistent with these observations, cells in the VZ showing abventricular centrosomes in the vicinity of the nucleus often lacked BrdU incorporation (Fig. 8, which is published as supporting information on the PNAS web site). We therefore used abventricular centrosomes in the VZ as a marker of the generation of non-NE progeny.
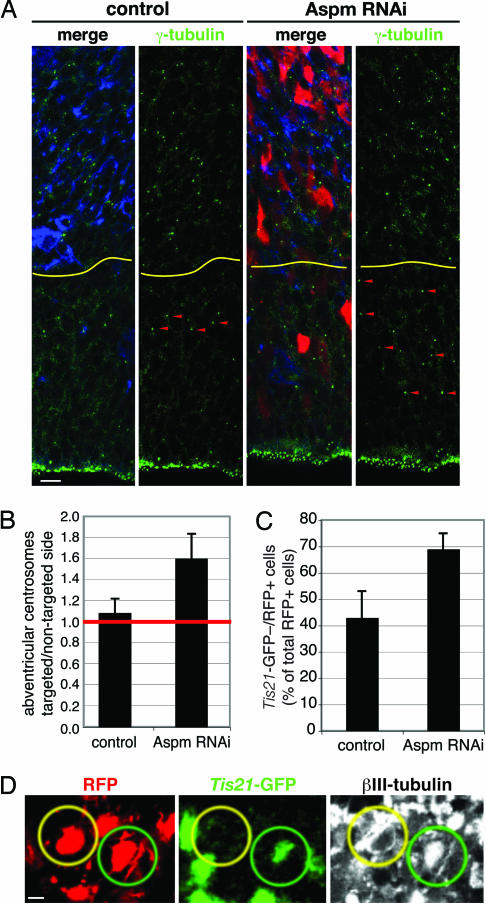
Fig. 4.
Knockdown of Aspm promotes VZ cells to adopt a non-NE fate and increases the appearance of neuron-like NE cell progeny in the neuronal layer. (A and B) Mouse E12.5 dorsal telencephalon was either coelectroporated with Aspm esiRNAs and mRFP plasmid (A and B) or electroporated with mRFP plasmid only (B), followed by 48 h in utero development. (A) Cryosections of the nontargeted (control) and targeted (Aspm RNAi) hemispheres were analyzed for RFP fluorescence (red) and γ-tubulin (green) and βIII-tubulin (blue) immunofluorescence. Note the increase in abventricular centrosomes (arrowheads) upon Aspm knockdown. The yellow lines indicate the boundary between the VZ/sub-VZ and the neuronal layers. The apical surface of the VZ is down. (Scale bar, 10 μm.) (B) Numbers of centrosomes per 10,000 μm2 counted in γ-tubulin-immunostained cryosections are expressed as a ratio of targeted/nontargeted hemispheres for electroporation with mRFP plasmid only (control) and mRFP plasmid plus Aspm esiRNAs (Aspm RNAi). Data are the mean of five cryosections from three different embryos each; error bars indicate SD. (C and D) Dorsal telencephalon of E10.5 Tis21–GFP knockin mice was coelectroporated with Aspm esiRNAs and mRFP plasmid (Aspm RNAi) or electroporated with mRFP plasmid only (control), followed by 24 h of whole-embryo culture. (C) Cryosections were analyzed for the proportion of RFP-positive cells in the neuronal layer that lacked Tis21–GFP fluorescence. Data are the mean of three cryosections from two to three different embryos each (control, 70 cells; Aspm RNAi, 74 cells); error bars indicate SD. (D) Representative example of two neighboring RFP-positive cell bodies (red) in the neuronal layer, with one being positive (green circles) and one being negative (yellow circles) for Tis21–GFP fluorescence (green) but both exhibiting βIII-tubulin immunofluorescence (white). (Scale bar, 5 μm.)
We observed a significant increase in abventricular centrosomes in the VZ of the targeted hemisphere (Fig. 4A, Aspm RNAi) compared with the nontargeted hemisphere (Fig. 4A, control) 48 h after initiation of Aspm knockdown in E12.5 mice. This increase was 1.6-fold over the abventricular centrosomes observed in the VZ under control conditions (Fig. 4B), which is what would be expected, given (i) the efficiency of the targeting of NE cells upon in utero electroporation (<50%), (ii) the normal abundance of abventricular centrosomes (compare with Fig. 4A, control), (iii) the proportion of NE cells undergoing proliferative divisions at this developmental stage (19), and (iv) the increase in asymmetric NE cell divisions upon Aspm knockdown (Fig. 3C), among other parameters. The increase in abventricular centrosomes was due to the presence of Aspm esiRNAs, because no such increase was observed upon electroporation of the mRFP reporter plasmid alone (Fig. 4B, control). Because Aspm knockdown did not perturb the apical localization of centrosomes in interphase NE cells (Fig. 2C), we conclude that knockdown of Aspm causes NE cells to increasingly generate progeny with abventricular centrosomes, i.e., non-NE progeny.
Increased Neuron-Like Fate of NE Cell Progeny After Aspm Knockdown.
Does the non-NE progeny generated by Aspm knockdown exhibit characteristics of neurons? In contrast to the increase in abventricular centrosomes (Fig. 4 A and B), we did not observe an obvious increase, after Aspm knockdown, in VZ cells expressing βIII-tubulin (data not shown), a marker of young neurons. This finding, however, does not necessarily mean that there is no increase in neuronal progeny upon Aspm knockdown. Live imaging results for telencephalic neuroepithelium of transgenic mouse embryos expressing GFP under the control of the βIII-tubulin promoter imply that βIII-tubulin expression in the majority of cells born at the ventricular surface occurs relatively late (>5 h after birth), that is, when these cells have left the VZ (A. Attardo, W. Haubensak, F. Calegari, and W.B.H., unpublished data). We therefore analyzed the neuronal layer of the targeted hemisphere of Tis21–GFP knockin mouse embryos subjected to Aspm knockdown at E10.5 followed by 24 h of whole-embryo culture.
Consistent with previous observations (19), in the control condition (electroporation of RFP only), less than half of the young neurons (identified by βIII-tubulin immunofluorescence) in the neuronal layer adjacent to the VZ that derived from electroporated NE cells, as indicated by RFP fluorescence, lacked GFP fluorescence (Fig. 4C, control). Aspm knockdown resulted in a significant increase in the proportion of GFP-negative/RFP-positive cells in the neuronal layer (Fig. 4C, Aspm RNAi), suggesting an increasing contribution of progeny derived from NE cells lacking Tis21–GFP expression. Remarkably, almost all of the GFP-negative/RFP-positive cells in the neuronal layer observed upon Aspm knockdown showed βIII-tubulin expression, as exemplified in Fig. 4D. We conclude that at least some of the non-NE progeny generated by Aspm knockdown migrate to the neuronal layer and express neuronal markers.
In the telencephalon, neurons arise from NE and radial glial cells dividing at the ventricular surface and from progenitors dividing basally in the VZ and sub-VZ (19, 29, 30). Given that the centrosomes of the basal progenitors are abventricular, we examined whether the increase in abventricular centrosomes in the VZ upon Aspm knockdown reflected an increase in basal progenitors. However, no obvious increase in abventricular mitotic cells was observed upon Aspm RNAi (Fig. 7). This lack of increase in basal progenitors in turn suggests that the neuron-like cells observed in the neuronal layer after Aspm knockdown (Fig. 4 C and D) were generated directly by NE cells.
Taken together, our observations indicate that Aspm knockdown in NE cells increases the probability of their progeny adopting a non-NE fate, including a neuron-like fate (migration to neuronal layers and expression of neuronal markers). We do not know whether the neuron-like cells observed develop into functional neurons, and we cannot exclude that the progeny generated by the Aspm-knocked-down NE cells, including the neuron-like cells, eventually undergo apoptosis. Importantly, whichever fate the progeny ultimately adopt, it is a non-NE fate, which in any case implies a reduction in the NE progenitor pool relative to control.
Conclusion
In conclusion, our results provide a cell biological explanation for the function of Aspm in mammalian neocortical development. The need of the highly elongated, polarized NE cells to bisect their small apical membrane for symmetric, proliferative division (5, 7) implies not only that the mitotic spindle has to adopt an axis exactly perpendicular to the NE cell apical–basal axis by the end of metaphase, it also necessitates that this spindle axis is maintained during anaphase and telophase to ensure that the basal-to-apical ingression of the cleavage furrow (5) occurs precisely along the apical–basal NE cell axis. Our observations suggest that Aspm exerts a critical role at the spindle poles of NE cells in maintaining spindle position through mitosis and, consequently, in ensuring the precise cleavage plane orientation required for symmetric, proliferative divisions.
Loss of Aspm upon knockdown results in a deviation of spindle position and, hence, an alteration in cleavage plane orientation, thereby increasing the probability of asymmetric division of NE cells, with only one daughter cell inheriting apical membrane and adherens junctions and, thus, remaining epithelial. In other words, loss of Aspm reduces the expansion of the NE progenitor pool, which would explain why humans with mutations in ASPM suffer from primary microcephaly (8). [Similar considerations may hold true for mutations in other genes encoding centrosomal proteins that cause primary microcephaly (12, 31).] The observation that only the brain is affected in these patients (32), despite the expression of Aspm in other developing epithelia (13), likely reflects the highly elongated shape of NE cells and their small apical membrane, which make them more vulnerable to any perturbations in spindle position when undergoing symmetric, proliferative divisions (2, 7). A corollary of this is that with the increase in brain size during primate evolution, the further reduction in the apical membrane of NE cells, which predicts the need for even greater accuracy of cleavage plane orientation, offers a potential reason for the positive selection of ASPM observed in the primate lineage (16, 17).
Methods
Animals.
All experiments with Tis21–GFP knockin mice were performed on heterozygous embryos obtained from crossing homozygous males with C57BL/6J females (19). Wild-type embryos were obtained from Naval Medical Research Institute (NMRI) mice. The vaginal plug was defined as E0.5. BrdU labeling was performed for 30 min as described (33).
Aspm Antibody.
GST-tagged recombinant protein from mouse Aspm exon 3 (8), representing amino acids 150–263, was used as an immunogen in rabbits. The resulting antisera were affinity-purified. Antisera and affinity-purified Aspm antibodies were tested first in immunoblots of total protein of Cos-7 cells overexpressing a GFP–Aspm150–263 fusion protein. The specificity of the affinity-purified Aspm antibodies was further corroborated in immunoblots of total protein from mouse E12.5–E13.5 heads, in which a band of >300 kDa [and three to four bands with lower molecular weights, presumably representing splice variants (13)] were recognized.
Aspm Knockdown.
According to previously established methods (20, 21), Aspm esiRNAs (0.6 μg/μl) generated from double-stranded RNA complementary to nucleotides 585-1932 of exon 3 of mouse Aspm (8), together with an mRFP plasmid (0.75 μg/μl), were injected and directionally electroporated into one half of the dorsal telencephalon of E10.5 embryos ex utero or E12.5 embryos in utero, which were then allowed to develop in whole-embryo culture for 24 h or in utero for 24–48 h, respectively. The contralateral side of the dorsal telencephalon and/or dorsal telencephalon electroporated with mRFP plasmid only were used as controls. The mRFP plasmid was a pCAGGS vector expressing mRFP [a kind gift from Roger Tsien (University of California, San Diego)] under the control of the chicken β-actin promoter coupled to the CMV enhancer.
Immunofluorescence Confocal Microscopy.
Immunofluroescence confocal microscopy on cryosections of paraformaldehyde-fixed E10.5–E14.5 mouse brains from wild-type or heterozygous Tis21–GFP knockin mouse embryos (19) was performed according to standard procedures (5, 6). For details, see Supporting Methods, which is published as supporting information on the PNAS web site.
Analysis of Cleavage Plane Orientation and Apical Membrane Distribution.
Cleavage plane orientation and apical plasma membrane distribution were determined in optical sections of mitotic NE cells stained for DNA and cadherin as described in ref. 5. For details, see Supporting Methods.
Quantifications.
Details of (i) the assessment of Aspm immunofluorescence intensity in Tis21–GFP-negative versus Tis21–GFP-positive NE cells, (ii) the quantification of abventricular centrosomes, and (iii) the quantification of Tis21–GFP-negative versus Tis21–GFP-positive NE cell progeny in the neuronal layer are described in Supporting Methods.
In Situ Hybridization.
Nonradioactive in situ hybridization using digoxigenin-labeled cRNA antisense and sense probes corresponding to nucleotides 585-1932 of exon 3 of mouse Aspm (8) was carried out on 10-μm cryosections by standard methods.
Supplementary Material
Acknowledgments
We thank Joe Howard and Tony Hyman for discussion. W.B.H. was supported by Deutsche Forschungsgemeinschaft Grants SPP 1109 Hu275/7-3, SPP 1111 Hu275/8-3, SFB/TR13-04 B1, and SFB 655 A2 and Federal Ministry of Education and Research Nationales Genomforschungsnetz Grants SMP-RNAi 01GR0402 and PRI-S08T05.
Abbreviations
- NE
neuroepithelial
- VZ
ventricular zone
- RNAi
RNA interference
- esiRNA
endoribonuclease-prepared, short interfering RNA
- En
embryonic day n.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rakic P. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 2.Götz M., Huttner W. B. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 3.Kriegstein A. R., Götz M. Glia. 2003;43:37–43. doi: 10.1002/glia.10250. [DOI] [PubMed] [Google Scholar]
- 4.Chenn A., McConnell S. K. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 5.Kosodo Y., Röper K., Haubensak W., Marzesco A.-M., Corbeil D., Huttner W. B. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttner W. B., Kosodo Y. Curr. Opin. Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Huttner W. B., Brand M. Curr. Opin. Neurobiol. 1997;7:29–39. doi: 10.1016/s0959-4388(97)80117-1. [DOI] [PubMed] [Google Scholar]
- 8.Bond J., Roberts E., Mochida G. H., Hampshire D. J., Scott S., Askham J. M., Springell K., Mahadevan M., Crow Y. J., Markham A. F., et al. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 9.do Carmo Avides M., Glover D. M. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- 10.Wakefield J. G., Bonaccorsi S., Gatti M. J. Cell Biol. 2001;153:637–648. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bringmann H., Hyman A. A. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 12.Bond J., Woods C. G. Curr. Opin. Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Kouprina N., Pavlicek A., Collins N. K., Nakano M., Noskov V. N., Ohzeki J., Mochida G. H., Risinger J. I., Goldsmith P., Gunsior M., et al. Hum. Mol. Genet. 2005;14:2155–2165. doi: 10.1093/hmg/ddi220. [DOI] [PubMed] [Google Scholar]
- 14.Zhong X., Liu L., Zhao A., Pfeifer G. P., Xu X. Cell Cycle. 2005;4:1227–1229. doi: 10.4161/cc.4.9.2029. [DOI] [PubMed] [Google Scholar]
- 15.Bond J., Scott S., Hampshire D. J., Springell K., Corry P., Abramowicz M. J., Mochida G. H., Hennekam R. C., Maher E. R., Fryns J. P., et al. Am. J. Hum. Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J. Genetics. 2003;165:2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouprina N., Pavlicek A., Mochida G. H., Solomon G., Gersch W., Yoon Y. H., Collura R., Ruvolo M., Barrett J. C., Woods C. G., et al. PLoS Biol. 2004;2:653–663. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luers G. H., Michels M., Schwaab U., Franz T. Mech. Dev. 2002;118:229–232. doi: 10.1016/s0925-4773(02)00253-8. [DOI] [PubMed] [Google Scholar]
- 19.Haubensak W., Attardo A., Denk W., Huttner W. B. Proc. Natl. Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calegari F., Haubensak W., Yang D., Huttner W. B., Buchholz F. Proc. Natl. Acad. Sci. USA. 2002;99:14236–14240. doi: 10.1073/pnas.192559699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi M., Sato K., Nomura T., Osumi N. Differentiation. 2002;70:155–162. doi: 10.1046/j.1432-0436.2002.700405.x. [DOI] [PubMed] [Google Scholar]
- 22.Chenn A., Zhang Y. A., Chang B. T., McConnell S. K. Mol. Cell. Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- 23.Ripoll P., Pimpinelli S., Valdivia M. M., Avila J. Cell. 1985;41:907–912. doi: 10.1016/s0092-8674(85)80071-4. [DOI] [PubMed] [Google Scholar]
- 24.Khodjakov A., Cole R. W., Oakley B. R., Rieder C. L. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 25.Weigmann A., Corbeil D., Hellwig A., Huttner W. B. Proc. Natl. Acad. Sci. USA. 1997;94:12425–12430. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iacopetti P., Michelini M., Stuckmann I., Oback B., Aaku-Saraste E., Huttner W. B. Proc. Natl. Acad. Sci. USA. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinsch S., Karsenti E. J. Cell Biol. 1994;126:1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T., Serneo F. F., Higgins C., Gambello M. J., Wynshaw-Boris A., Gleeson J. G. J. Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noctor S. C., Martinez-Cerdeno V., Ivic L., Kriegstein A. R. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 30.Miyata T., Kawaguchi A., Saito K., Kawano M., Muto T., Ogawa M. Development (Cambridge, U.K.) 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 31.Feng Y., Walsh C. A. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Woods C. G., Bond J., Enard W. Am. J. Hum. Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calegari F., Haubensak W., Haffner C., Huttner W. B. J. Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.