Abstract
Small GTPases of the Rho family play key roles in the formation of neuronal axons and dendrites by transducing signals from guidance cues, such as neurotrophins, to the actin cytoskeleton. However, there is little insight into the mechanism by which neurotrophins regulate Rho GTPases. Here, we show the crucial role of the ubiquitous Rac1-specific guanine nucleotide exchange factor, Tiam1 (T lymphoma invasion and metastasis 1), in transducing a neurotrophin-mediated change in cell shape. We demonstrate that BDNF, acting through TrkB, directly binds and specifically activates Tiam1 by phosphorylating Tyr-829, leading to Rac1 activation and lamellipodia formation in Cos-7 cells and increased neurite outgrowth from cortical neurons. A point mutation in Tiam1, Tyr-829 to Phe-829, blocked these BDNF-induced changes in cellular morphology. The findings are evidence of a previously uncharacterized mechanism for the activation of Tiam1 and of a role for this effector in neurotrophin-mediated signal transduction leading to changes in cellular morphology.
Keywords: actin, cortical neuron, neurotrophin, Rho guanine nucleotide exchange factors, Rho GTPases
Mammalian neurotrophins, consisting of nerve growth factor, BDNF, neurotrophin 3, and neurotrophin-4/5, are known to support critical aspects of neuronal development, including differentiation, process growth and guidance, synaptogenesis, and survival (1, 2). The secreted neurotrophins bind to two different classes of plasma membrane receptors, the tyrosine kinase receptors (Trks) (TrkA, TrkB, and TrkC) and the p75 neurotrophin receptor (NTR). Trk receptors show selectivity for neurotrophin binding, whereas p75NTR binds all neurotrophins with similar affinity.
The growth of axons and dendrites is a highly regulated process that is fundamental to the development of the nervous system. It is mediated through changes in the actin cytoskeleton. Pointing to the conservation of mechanisms across cell types, neuronal growth cones, which mediate the growth of neuronal processes, feature filopodia and lamellipodia that are structurally analogous to those seen in fibroblasts (3). Neurotrophins have significant effects on the growth cone and axonal morphology (3). For example, BDNF causes the formation of filopodia and lamellipodia in Xenopus spinal cords (4), and neurotrophin 3 binding to p75NTR increases filopodial formation and neurite length in cortical subplate neurons (5). Despite the demonstrated relevance of neurotrophin signaling in process growth, the molecular basis by which neurotrophins modify the actin cytoskeleton and cellular morphology is poorly defined.
Several lines of evidence indicate that small GTPases of the Rho family are critical regulators of the organization of the actin cytoskeleton (6, 7). The best-characterized Rho GTPases, RhoA, Rac1, and Cdc42, are implicated in the formation of actin stress fibers, lamellipodia, and filopodia of fibroblasts, respectively. Rho GTPases also play pivotal roles in regulating the structure of actin in neuronal growth cones (3). Pointing to a role in neurotrophin-mediated actions, Rac and Rho regulate neurotrophin-induced axon growth in trigeminal brainstem whole-mount cultures (8). Also, BDNF activates Cdc42 to stimulate neurite extension and growth cone filopodial activity in cultured Xenopus spinal neurons (9). Given the probability that Rho GTPases mediate the growth and guidance signals induced by neurotrophins, it is important to decipher the pathways that link neurotrophin signal transduction to the Rho GTPases and their downstream actions on the actin cytoskeleton.
Like Ras GTPases, Rho GTPases cycle between active GTP-bound and inactive GDP-bound states. Rho GTPases are regulated positively by guanine nucleotide exchange factors (GEFs), which catalyze the exchange of GDP for GTP, and negatively by GTPase-activating proteins, which enhance intrinsic GTPase activity. To date, ≈70 GEFs have been identified in human genomes (10). Certain GEFs are known to activate several Rho GTPases, whereas several GEFs are specific for each Rho GTPase. For example, whereas Vav acts as the GEF for RhoA, Rac1, and Cdc42, Tiam1 (T lymphoma invasion and metastasis 1) (11) and FRG (FGD-1-related Cdc42-GEF) (12) function as specific GEFs for Rac1 and Cdc42, respectively. Tiam1, which is widely expressed, is involved in the promotion of invasion in lymphocytes and fibroblasts and adhesion in epithelial cells (13). We noted that Tiam1 is located within growth cones and promotes the formation of axons (14, 15). The NMDA receptor controls the growth and morphological changes of dendritic arbors and spines through Tiam1-dependent Rac1 activation (16). These reports create interest in a role for this GEF in mediating neurotrophin actions on the cytoskeleton.
This study examines a role for BDNF and its receptor, TrkB, in regulating the actin cytoskeleton. We show that BDNF, acting through TrkB and Tiam1, activates Rac1 and enhances the formation of lamellipodia. Surprisingly, we found that TrkB directly binds to and phosphorylates Tiam1. Tiam1 is phosphorylated at Tyr-829 to enhance the Rac1-GEF activity in vitro. The results are consistent with a model in which BDNF binding and activation of TrkB leads directly to activation of Tiam1/Rac1 signaling, leading to changes in the actin cytoskeleton. The findings provide evidence of a previously uncharacterized mechanism by which a receptor tyrosine kinase acts to induce signaling of Rac1, a regulator of the actin cytoskeleton.
Results and Discussion
Neurotrophins are diffusible trophic factors that are best known for their ability in developing neurons to regulate survival and differentiation, including the growth of axons and dendrites (1, 2). Given the evidence suggesting that Rho GTPases mediate the process of growth in developing neurons (3), we asked whether neurotrophins act by influencing the activities of endogenous Rho GTPases. Using a transient transfection system in Cos-7 cells, the active GTP-bound form of Rac1 was detected by using an affinity-precipitation assay with the GST-tagged Cdc42·GTP/Rac1·GTP interactive binding domain (CRIB) of αPak (17). As shown in Fig. 1A and B, Rac1 was activated in a time-dependent manner after stimulation with BDNF in Cos-7 cells transfected with a plasmid encoding TrkB. At 45 min, the activity of Rac1 reached a maximum of 2.0–3.0 times the pretreatment levels; by 90–120 min, it returned to the pretreatment baseline. The effect was blocked by pretreatment with K252a, an inhibitor of the Trks (Fig. 6A, which is published as supporting information on the PNAS web site). Similarly, BDNF increased GTP-bound Cdc42, but the time course was different from that of Rac1 (Fig. 7, which is published as supporting information on the PNAS web site). In contrast, BDNF had no effect on RhoA (data not shown). We conclude that through the activation of TrkB, BDNF increases the levels of the GTP-bound forms of Rac1 and Cdc42.
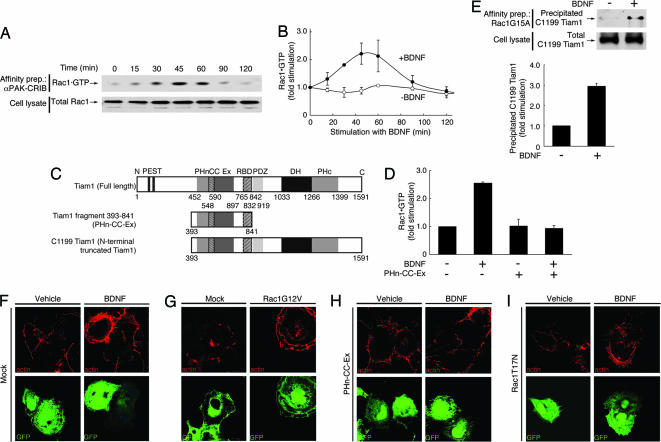
Fig. 1.
Tiam1 is essential for Rac1 activation and the formation of lamellipodia induced by BDNF. (A and B) Rho GTPase Rac1 is activated after stimulation with BDNF. (A) The activity of endogenous Rac1 was measured after the addition of BDNF (10 ng/ml for 0–120 min) with an affinity-precipitation assay by using recombinant αPak-CRIB. The total Rac1 in the cell lysates was immunoblotted with an anti-Rac1 antibody. (B) The levels of Rac1 activation with (filled circles) or without (open circles) the addition of BDNF were quantified and normalized against the total cell lysates. (C) Schematic structures of Tiam1. (D) Cos-7 cells were transfected with FLAG-PHn-CC-Ex and TrkB. The amount of GTP-bound Rac1 was assessed after the addition of BDNF (10 ng/ml for 45 min). (E) After stimulation with BDNF, increased Tiam1 was detected by an affinity-precipitation assay using GST-Rac1G15A from the lysates of Cos-7 cells after incubation with BDNF. Cells were transfected with FLAG-C1199 Tiam1 and TrkB. Active FLAG-C1199 Tiam1 was precipitated with GST-Rac1G15A. The total FLAG-C1199 Tiam1 in the cell lysates is also shown. (F–I) Tiam1 and Rac1 are required for the formation of BDNF-induced lamellipodia. Cos-7 cells were transfected with TrkB and EGFP (F), Rac1G12V and EGFP (G), FLAG-PHn-CC-Ex, TrkB, and EGFP (H), or Rac1T17N, TrkB, and EGFP (I), incubated with BDNF (10 ng/ml for 30 min) (F, H, and I), and stained with rhodamine phalloidin for F-actin.
Because activation of Rac1 and Cdc42 followed different time courses (Figs. 1 and 7), we reasoned that different GEFs might regulate the activities of Rac1 and Cdc42. Although it has been reported that TrkC activates the Rac1-GEF Tiam1 (18) and the Cdc42-GEF Dbs (19), less is known about the relationships between TrkB and Rho family GEFs. At first, we focused on Rac1 in this study. To identify a Rac1-specific GEF that may be responsible for BDNF-induced Rac1 activation, a search of public databases for known mammalian Rac1-specific GEFs led us to consider the Rac1-GEF Tiam1. Tiam1 is expressed at high levels in the developing brain (20). Overexpression of Tiam1 promotes the formation of lamellipodial structures in fibroblasts and neurites in neuroblastoma cells (14). STEF/Tiam2, another Rac1-GEF, is highly homologous with Tiam1 and also increases neurite outgrowth (21). Because endogenous Tiam1 was detected at high levels in Cos-7 cells, we set out to study whether Tiam1 mediated BDNF-induced Rac1 activation in these cells.
The Tiam1 protein is 1,591 aa long and contains several distinct domains (Fig. 1C). Tiam1 contains two N-terminal PEST sequences [rich in proline (P), glutamate (E), serine (S), and threonine (T)], an N-terminal Pleckstrin homology domain (PHn), a coiled-coil region with an adjacent sequence (named CC-Ex), a Ras-binding domain (RBD), a PDZ (PSD-95/DigA/ZO-1) domain, a Dbl homology domain, and its adjacent C-terminal PH domain (22). The PEST sequences are known to target proteins for degradation; deletion of these PEST sequences strikingly enhances the stability of the Tiam1 protein (22). The isolated PHn-CC-Ex region of Tiam1 functions as a dominant negative, because its expression inhibits Tiam1-mediated membrane ruffling in Cos-7 cells (23). To better define the role of these domains in BDNF-induced Rac1 activation, we made the following constructs: (i) C1199 Tiam1 (amino acids 393-1591), in which the N-terminal amino acids 1–392 containing two consensus PEST domains were deleted (Fig. 1C), and (ii) PHn-CC-Ex (amino acids 393–841), which contains the PHn-CC-Ex region and the RBD of Tiam1 (amino acids 393–841) (Fig. 1C).
To show whether BDNF activates Rac1 through Tiam1, we carried out affinity-precipitation assays. Transfection of the dominant-negative PHn-CC-Ex into Cos-7 cells blocked BDNF-induced Rac1 activation (Fig. 1D), suggesting that Tiam1 is involved in this signaling pathway. Next, to examine whether Tiam1 is activated after stimulation with BDNF, we carried out an affinity-precipitation assay to detect active Tiam1 by using GST-Rac1G15A, a guanine nucleotide-free form of Rac1 (24). Active GEFs preferentially interact with guanine nucleotide-free forms of the small GTPases. A point mutation of glycine to alanine of residue 15 on Rac1 decreases its nucleotide binding (24). After stimulation of Cos-7 cells cotransfected with TrkB and C1199 Tiam1 with BDNF, the level of Tiam1 that was precipitated with Rac1G15A showed a marked increase (Fig. 1E). This effect was blocked with K252a pretreatment (Fig. 6B). To determine whether BDNF induces activation of Tiam1 in neurons, affinity precipitation was used to detect the active form of endogenous Tiam1 in primary cultures of cortical neurons. There was an increase in endogenous Tiam1 precipitated with Rac1G15A upon BDNF stimulation of TrkB (Fig. 8A, which is published as supporting information on the PNAS web site). Pretreatment with K252a blocked the increase in Tiam1 precipitated with Rac1G15A (data not shown). Taken together, these results indicate that BDNF activates Rac1 through the Rac1-specific GEF Tiam1.
RhoA regulates the assembly of contractile actin-myosin filaments. In contrast, Rac1 and Cdc42 regulate the polymerization of actin to form peripheral lamellipodial and filopodial protrusions, respectively. To investigate the effect of BDNF on the actin structure in Cos-7 cells, we cotransfected constructs together with pEGFP-C1 to visualize the transfected cells by EGFP expression. The cells were also stained with rhodamine phalloidin for F-actin. BDNF induced a marked change in the distribution of actin, as demonstrated by a large increase in staining near the cell surface, a change that marks lamellipodia (Fig. 1F Right). The formation of lamellipodia was seen in ≈90% of Cos-7 cells cotransfected with TrkB and pEGFP-C1, a result that was replicated in cells transfected with a constitutively active form of Rac1 (Fig. 1G). In contrast, this pattern of staining was seen in <1% of vehicle-treated cells (Fig. 1F Left).
To investigate the involvement of Tiam1 in BDNF-induced lamellipodia, we transfected PHn-CC-Ex into Cos-7 cells. Cotransfection of PHn-CC-Ex inhibited the BDNF-induced lamellipodia of Cos-7 cells (Fig. 1H). In contrast, these structures were still observed in untransfected cells (Fig. 1H). To test whether the involvement of Tiam1 in the BDNF-induced lamellipodia was through Rac1, we next transfected a dominant-negative form of Rac1 into Cos-7 cells. Cotransfection of Rac1T17N suppressed the BDNF-induced lamellipodia, and transfected cells showed prominent filopodia, perhaps because of concurrent activation of Cdc42 (Fig. 1I). We conclude that the induction of lamellipodia by BDNF is through Tiam1 and Rac1.
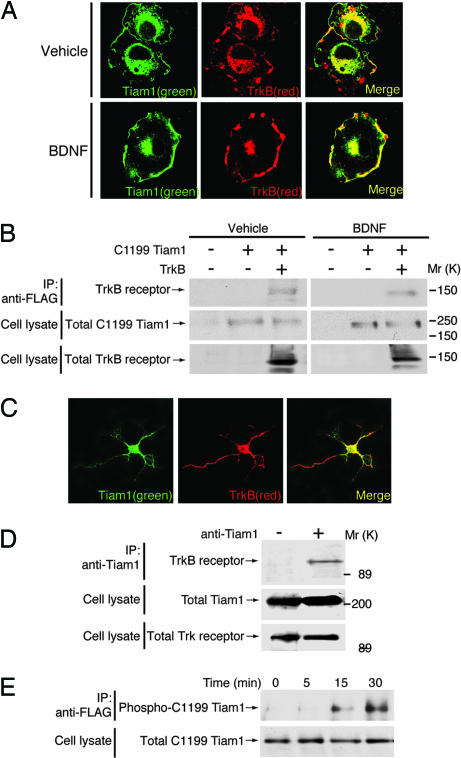
To establish the subcellular localization of TrkB and C1199 Tiam1 before and after BDNF treatment, we performed immunostaining experiments. TrkB and C1199 Tiam1 were widely distributed in vehicle-treated cells, with staining at or near the cell surface as well as in the perinuclear regions; there was partial colocalization in both sites, with more evident in the perinuclear region (Fig. 2A). After treatment with BDNF, there was a change in the distributions of both molecules. Both were increased at or near the cell surface, where they displayed significant colocalization (Fig. 2A). To further investigate the localization of endogenous TrkB and Tiam1, we used primary cortical neurons from embryonic day-16 rat embryos. TrkB was present in the cell bodies and in neurites; Tiam1 was also present in cell bodies and in some neurites (Fig. 2C). TrkB and Tiam1 were colocalized principally in cell bodies and to a lesser extent in neurites (Fig. 2C). Stimulation with BDNF had no detectable effect on the distribution of TrkB and Tiam1 (data not shown). Next, to examine whether TrkB formed a complex with Tiam1, we carried out coimmunoprecipitation studies. TrkB was coimmunoprecipitated with C1199 Tiam1 in both vehicle- and BDNF-treated cells (Fig. 2B). This result was confirmed in experiments using brain lysates (Fig. 2D). The TrkB receptor was coimmunoprecipitated with Tiam1 by its specific antibody but not by a preimmune IgG. In contrast, BDNF stimulation caused the increase of the association between Tiam1 and TrkB in cortical neurons, and the association was reduced with K252a pretreatment (Fig. 8B). These results suggest that TrkB has the ability to form a complex with Tiam1 in coexpressed Cos-7 cells but that, at the endogenous levels, their association can be observed after activation of TrkB.
Fig. 2.
BDNF increases the colocalization of Tiam1 and TrkB in the membrane structures. (A) Cos-7 cells were transfected with FLAG-C1199 Tiam1 and TrkB, incubated with BDNF (10 ng/ml for 30 min), and stained with antibodies against TrkB (red) and FLAG peptide (green). Stimulation with BDNF increased the colocalization in the membrane structures. (B) After stimulation with BDNF (10 ng/ml for 30 min), the immunoprecipitates of FLAG-C1199 Tiam1 were immunoblotted with an antibody against TrkB. The total C1199 Tiam1 and TrkB are also shown. (C) Cortical neurons were stained with antibodies against TrkB (red) and Tiam1 (green). (D) The immunoprecipitates of endogenous Tiam1 were immunoblotted with an antibody against TrkB. The total Tiam1 and TrkB are also shown. (E) After stimulation with BDNF (10 ng/ml for 0–30 min), tyrosine-phosphorylated FLAG-C1199 Tiam1 was increased in a time-dependent manner. The total FLAG-C1199 Tiam1 is also shown.
Recently, it has become apparent that some GEFs can be phosphorylated by tyrosine kinases (12, 19). To clarify whether Tiam1 was tyrosine-phosphorylated after the activation of TrkB, we examined the phosphorylation of Tiam1 by stimulating Cos-7 cells transfected with C1199 Tiam1 and TrkB with BDNF. Our results confirmed that within 15 min of BDNF treatment, C1199 Tiam1 was tyrosine-phosphorylated; there was a further increase at 30 min (Fig. 2E). Consistent with a role for TrkB in this activity, pretreatment with K252a inhibited tyrosine phosphorylation of C1199 Tiam1 (data not shown). In cortical neurons, endogenous Tiam1 was also tyrosine-phosphorylated in a manner that depended on BDNF stimulation of TrkB (Fig. 8C).
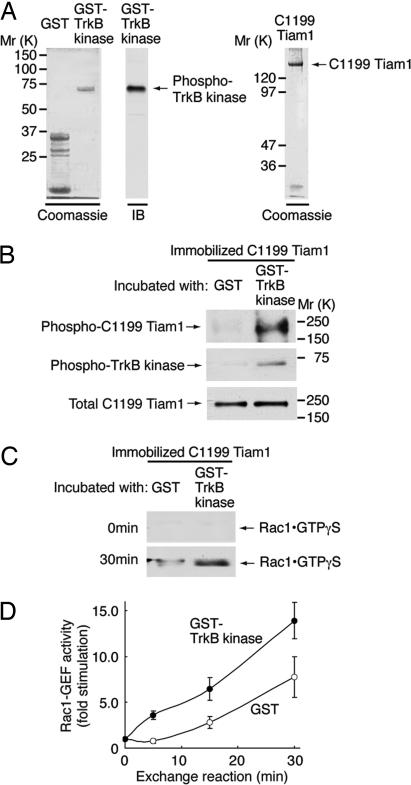
Our studies defined lamellipodia formation downstream from a signaling pathway whose elements were BDNF/TrkB/Tiam1/Rac1. Although a direct interaction exists between BDNF and TrkB, and between Tiam1 and Rac1, it was unclear how TrkB signaled to Tiam1. Because it has been shown that TrkC directly phosphorylates and activates the Cdc42-GEF Dbs (19), one possibility was that TrkB interacted directly with Tiam1 and phosphorylated Tiam1 at a tyrosine residue(s). To test this possibility, we performed in vitro kinase assays using recombinant proteins corresponding to the TrkB kinase domain and Tiam1, as described in Materials and Methods. The GST-tagged TrkB kinase domain was overexpressed in Escherichia coli and purified. The fusion protein appeared to retain its intact kinase, as indicated by immunoblotting with an anti-phosphorylated Trk antibody (Fig. 3A). FLAG-tagged C1199 Tiam1 was transiently transfected into 293T cells. The FLAG-C1199 Tiam1 protein was purified from the cell lysate by using a resin that was preabsorbed with an anti-FLAG antibody (Fig. 3A). When immobilized C1199 Tiam1 was incubated with the GST-TrkB kinase domain, the latter was found to bind (Fig. 3B Middle). Additionally, with the GST-TrkB kinase domain, but not with GST alone, there was tyrosine phosphorylation of C1199 Tiam1 (Fig. 3B Top); the input level of C1199 Tiam1 in lysates was constant (Fig. 3B Bottom). Thus, TrkB kinase was capable of directly tyrosine-phosphorylating Tiam1 in vitro.
Fig. 3.
TrkB binds, phosphorylates, and activates Tiam1 in vitro. (A) The purified GST-TrkB kinase domain (10 ng) was applied during SDS/PAGE, stained with Coomassie brilliant blue (Left), and immunoblotted (IB) with an anti-phosphorylated Trk antibody (Center). Purified FLAG-C1199 Tiam1 (100 ng) was separated by SDS/PAGE and stained with Coomassie blue (Right). (B) Immobilized FLAG-C1199 Tiam1 was incubated with 20 μM ATP and 10 ng/μl GST or a GST-TrkB kinase domain in 30 μl of reaction buffer, washed, and immunoblotted with either anti-phospho-Tyr or anti-phosphorylated Trk antibodies. (C and D) Immobilized FLAG-C1199 Tiam1 was incubated with GST or a GST-TrkB kinase domain. The supernatants were removed and mixed with 3 μM GTP-γ-S and glutathione resin preabsorbed with GST-αPak-CRIB. The GTP-γ-S-bound form of Rac1 was detected with an anti-Rac1 antibody.
To investigate whether TrkB directly stimulated tyrosine phosphorylation that led to enhanced Rac1-GEF activity of C1199 Tiam1, immobilized C1199 Tiam1 was first phosphorylated by the GST-TrkB kinase domain as described above. In a parallel experiment, GST, instead of the GST-TrkB kinase domain, was used as a control. His-Rac1·GDP was added to the reaction and incubated in a buffer containing an excess amount of GTP-γ-S. The supernatants were collected and then mixed with GST-αPak-CRIB that was preabsorbed to glutathione resin. The amount of Rac1·GTP-γ-S was assayed as before. As shown in Fig. 3 C and D, while under the condition of this experiment, some activation of Rac1 was evident in the absence of TrkB kinase domain, and C1199 Tiam1 exposed to the TrkB kinase domain markedly stimulated the exchange of Rac1·GDP to Rac1·GTP-γ-S and did so in a time-dependent manner. Taken together, the results suggest that TrkB-induced tyrosine phosphorylation of Tiam1 leads to enhanced GEF activity for Rac1.
To define the Tiam1 residues that are tyrosine-phosphorylated by TrkB, and to show whether this modification is important for the activation of Rac1, we constructed expression vectors that harbored a series of deletion constructs of Tiam1 (Fig. 9A, which is published as supporting information on the PNAS web site). We cotransfected various constructs of Tiam1 with the GST-TrkB kinase domain into Cos-7 cells. As shown in Fig. 9B, the active GST-TrkB kinase domain was able to induce the tyrosine phosphorylation only of fragment 2. As controls, these constructs were cotransfected with the GST vector; no tyrosine phosphorylation was detected for any fragment (Fig. 9B). We next investigated whether fragment 2 was directly phosphorylated by the GST-TrkB kinase domain in vitro. The immobilized fragments of Tiam1 were incubated with the GST-TrkB kinase domain as described above. In agreement with the results in the cells, fragment 2 was tyrosine-phosphorylated by the GST-TrkB kinase domain in vitro (Fig. 9C). Furthermore, this reaction required ATP; omission of ATP from the reaction mixture prevented tyrosine phosphorylation of fragment 2 (Fig. 9C). Finally, we showed that the activated GST-TrkB kinase domain was coimmunoprecipitated with fragment 2, as assessed by using an anti-phosphorylated Trk antibody (Fig. 9D). In addition to fragment 2, fragment 3 weakly associated with the TrkB kinase domain (Fig. 9D). Thus, fragment 3 might assist in binding C1199 Tiam1 to TrkB kinase. Together, these results are evidence that fragment 2 binds TrkB and provides a site(s) that is tyrosine-phosphorylated by TrkB kinase.
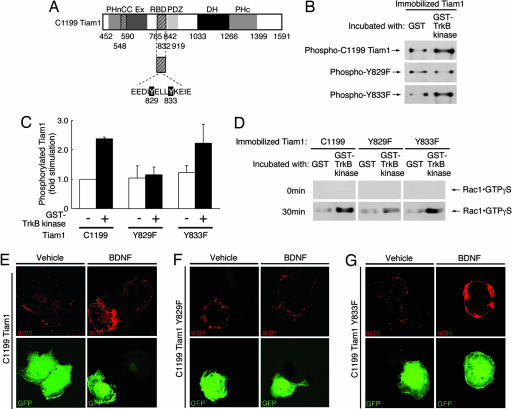
Examination of fragment 2 revealed that only two tyrosine residues, EEDIY829ELLY and YELLY833KEIE, were computationally predicted (by using the NetPhos 2.0 server) to be sites of tyrosine phosphorylation (scores were 0.986 and 0.923, respectively). To identify the TrkB tyrosine phosphorylation site(s) of Tiam1 fragment 2, we constructed point mutants of C1199 Tiam1 within fragment 2 in which either Tyr-829 or Tyr-833 was changed to phenylalanine (Y829F or Y833F, respectively) (Fig. 4A). We then conducted in vitro tyrosine phosphorylation experiments using these constructs. Immobilized Tiam1 mutants were incubated with either GST or the GST-TrkB kinase domain, as discussed above. Our results revealed that the wild-type C1199 Tiam1 was tyrosine-phosphorylated by the GST-TrkB kinase domain (Fig. 4B Top; see also Fig. 4C). A similar effect was observed for the Y833F mutant (Fig. 4B Bottom; see also Fig. 4C). However, the GST-TrkB kinase domain failed to elicit tyrosine phosphorylation in the Y829F mutant (Fig. 4B Middle; see also Fig. 4C). Importantly, the Y829F mutant lost the ability to induce Rac1-GEF activity in the guanine nucleotide-binding assay, whereas the Y833F mutant stimulated Rac1-GEF activity to the same extent as that seen in the wild-type C1199 Tiam1 (Fig. 4D). These results indicate that the phosphorylation of Tyr-829 induced by TrkB increased Rac1-GEF activity.
Fig. 4.
Tyr-829 of Tiam1 is essential for Tiam1 activation and the changes in the lamellipodia formation induced by BDNF. (A) The structures of point mutants of Tiam1. (B and C) Immobilized FLAG-C1199 Tiam1 constructs were incubated with GST or a GST-TrkB kinase domain and immunoblotted with an anti-phospho-Tyr antibody. (D) Immobilized FLAG-C1199 Tiam1 constructs were incubated with GST or a GST-TrkB kinase domain. The supernatants were incubated with GTP-γ-S and glutathione resin preabsorbed with GST-αPak-CRIB. The GTP-γ-S-bound form of Rac1 was detected with an anti-Rac1 antibody. (E–G) Tyr-829 is required for BDNF-induced lamellipodia. Cos-7 cells were transfected with FLAG-C1199 Tiam1 (E), FLAG-C1199 Tiam1 Y829F (F), or FLAG-C1199 Tiam1 Y833F (G) and also TrkB (E–G) and EGFP (E–G), incubated with BDNF (10 ng/ml for 30 min), and stained with rhodamine phalloidin. Using the calcium-phosphate method for transfection, >98% of the cells expressing Tiam1 constructs also expressed TrkB.
To determine the physiological significance of TrkB-induced tyrosine phosphorylation of Tiam1 at Tyr-829, C1199 Tiam1 or the mutants of C1199 Tiam1 were cotransfected with TrkB and pEGFP-C1 into Cos-7 cells. We monitored the effects of these constructs on the actin structure after stimulation with BDNF. As expected, stimulation with BDNF significantly increased the lamellipodia formation (≈90%) in C1199 Tiam1-expressed cells (Fig. 4E Right) as compared with that (<1%) in vehicle-treated cells (Fig. 4E Left). With respect to C1199 Tiam1, there was no decrease in BDNF-mediated induction in lamellipodia in cells expressing the Y833F mutant (Fig. 4G Right). In contrast, when the Y829F mutant was cotransfected with TrkB and pEGFP-C1, the formation of BDNF-induced lamellipodia was markedly inhibited (Fig. 4F Right), resulting in the presence of these structures in <1% of cells, a result that did not differ from that in vehicle-treated cells (Fig. 4F Left).
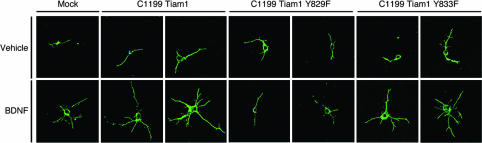
Finally, we examined the effect of C1199 Tiam1 and mutants of C1199 Tiam1 on the morphology of primary cortical neurons from embryonic day-16 rat embryos that were either stimulated with BDNF or not stimulated (Fig. 5). Cortical neurons were transfected with C1199 Tiam1, the Y829F mutant, the Y833F mutant, or FLAG alone as a control. After stimulation with BDNF, ≈45% of cortical neurons showed neurites longer than three cell bodies in length in mock-transfected cells, whereas only 20% of neurons did so without BDNF stimulation (Fig. 10A, which is published as supporting information on the PNAS web site). The number of neurites also increased in mock-transfected cells after stimulation with BDNF (Fig. 10B). Both parameters were increased with BDNF treatment of cortical neurons transfected with C1199 Tiam1 or the Y833F mutant (Fig. 10 A and B). In contrast, expression of the Y829F mutant reduced both the number of neurites (Fig. 10B) and the number of cells bearing longer neurites (Fig. 10A) in BDNF-treated cells. These findings are evidence that Tyr-829 of Tiam1 plays a critical role in BDNF-induced neurite formation. To further clarify the importance of Tiam1 in BDNF-regulated neurite extension, we used a short interfering RNA (siRNA) to knock down endogenous Tiam1 in cortical neurons. Expression of Tiam1 proteins in cortical neurons was markedly down-regulated by transfection with Tiam1 siRNA, whereas expression of β-actin was unaffected (Fig. 11C, which is published as supporting information on the PNAS web site). Knock-down of Tiam1 inhibited the BDNF-induced increase of the number of cells bearing neurites (Fig. 11A) and of neurites (Fig. 11B). Control siRNA had no effect on these parameters. In so doing, they complement and extend the results of recent studies that demonstrated an important role for STEF and Rac1 in mediating neurite outgrowth from neuroblastoma cells (21). Taken together with studies on Cos-7 cells, the observations in neuronal cells offer compelling evidence that BDNF induces changes in cell shape through a signaling pathway that depends critically on direct TrkB-mediated Tyr-829 phosphorylation of Tiam1 with resulting activation of Rac1.
Fig. 5.
Tyr-829 of Tiam1 is important for BDNF-induced neurite formation. DIV2 cortical neurons from embryonic day-16 rat embryos were transfected with FLAG-Tiam1 C1199, FLAG-Y829F mutant, FLAG-Y833F mutant, or FLAG plasmid. After transfection, cells were cultured for 24 h in medium without BDNF and then incubated with 10 ng/ml BDNF for 24 h. Cells were stained with anti-FLAG antibody. The representative confocal images of cortical neurons.
Here, we show that TrkB directly phosphorylates and activates Rac1-specific GEF Tiam1, thereby inducing changes in cellular morphology. Tyr-829 of Tiam1 is identified as the major TrkB tyrosine phosphorylation site. Importantly, concomitant with Rac1 activation, Cdc42 activation is also induced by BDNF binding to TrkB. This observation is reminiscent of coincident activation of Rac1 and Cdc42 induced by the Eph receptor tyrosine kinase. When EphB binds to the Cdc42-specific GEF intersectin 1, its activation signals the actin cytoskeleton through a neuronal Wiskott–Aldrich syndrome protein and the downstream actin-related proteins 2/3 (25). Activation of EphB also induces the translocation of Kalirin and the activation of Rac1 (26). EphB functionally interacts with different Rho GEFs, thereby activating different Rho GTPases. These integrated signals may allow EphB to induce a variety of morphological changes, although it is unclear whether EphB directly activates RhoGEFs. Identification of the Cdc42-specific GEF(s) activated by TrkB and the activation mechanism will allow us to elucidate further the mechanism by which TrkB regulates the actin cytoskeleton. Further study of the regulation of Tiam1 may help to clarify the basic molecular mechanism underlying neurotrophin-induced morphological changes in normal neurons and provide insights into neurological disorders that may be linked to disordered activation of Rho GTPases.
Materials and Methods
For details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Cell Culture of Neurons.
Cortical neurons were isolated from embryonic day-16 embryos of Sprague–Dawley rats according to the protocol of Wu et al. (27). Briefly, cerebral cortices were dissected, dissociated with 0.25% trypsin (Invitrogen), triturated, and passed through 70-μm-pore-size meshes. Cells were collected after centrifugation at 200 × g for 5 min and plated on collagen-coated dishes. Plasmid DNAs were transfected into neurons by the calcium-phosphate precipitation method. In some experiments, DIV2 cortical neurons were transfected, starved in a serum-low medium before BDNF treatment, and fixed 24 h after stimulation. For the statistical calculations (n = 5–20 in at least five microscope areas), cells bearing neurites longer than three cell bodies in length were considered as cells with neurite outgrowth. In counting, cells positive for FLAG expression were included in the analysis.
Guanine Nucleotide-Binding Assay.
The immobilized, purified fragments of Tiam1 (100 ng) were incubated in 30 μl of reaction buffer (20 mM Hepes-NaOH, pH 7.5/5 mM MgCl2/150 mM NaCl/1 mM DTT/1 mM phenylmethane sulfonylfluoride/1 μg/ml leupeptin/1 mM EDTA) containing 16 ng/μl His-Rac1, 33 ng/μl BSA, and 3 μM GTP-γ-S (Sigma) at 30°C for 0, 5, 15, and 30 min (28). The mixtures were immediately cooled on ice, and their supernatants were then mixed with glutathione resin preabsorbed with GST-αPak-CRIB in 200 μl of lysis buffer. The band intensity of Rac1·GTP in the immunoblot was semiquantified. Three to 10 separate experiments were performed, as shown in Figs. 3 and 4.
Supplementary Material
Acknowledgments
We thank members of W.C.M.’s laboratory for participation in helpful discussions and Ryoko Kimura for participation in helpful discussions and encouragement throughout the experiments. This work was supported by National Institutes of Health Grants AG16999, NS24054, and NS38869; the Down Syndrome Research and Treatment Foundation; The Larry L. Hillblom Foundation (W.C.M.); the Astellas Metabolic Disease Foundation (J.Y.); and the Japan Society for the Promotion of Science (Y.M.).
Abbreviations
- GEF
guanine nucleotide exchange factor
- PHn
N-terminal Pleckstrin homology domain
- CRIB
Cdc42·GTP/Rac1·GTP interactive binding domain.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Sofroniew M. V., Howe C. L., Mobley W. C. Annu. Rev. Neurosci. 2001;24:1217–1281. doi: 10.1146/annurev.neuro.24.1.1217. [DOI] [PubMed] [Google Scholar]
- 2.Huang E. J., Reichardt L. F. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 3.Luo L. Nat. Rev. Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 4.Gibney J., Zheng J. Q. J. Neurobiol. 2003;54:393–405. doi: 10.1002/neu.10149. [DOI] [PubMed] [Google Scholar]
- 5.McQuillen P. S., DeFreitas M. F., Zada G., Shatz C. J. J. Neurosci. 2002;22:3580–3593. doi: 10.1523/JNEUROSCI.22-09-03580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaibuchi K., Kuroda S., Amano M. Annu. Rev. Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville S., Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 8.Ozdinler P. H., Erzurumlu R. S. J. Comp. Neurol. 2001;438:377–387. doi: 10.1002/cne.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan X. B., Jin M., Xu X., Song Y. Q., Wu C. P., Poo M. M., Duan S. Nat. Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- 10.Rossman K. L., Der C. J., Sondek J. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 11.Habets G. G., Scholtes E. H., Zuydgeest D., van der Kammen R. A., Stam J. C., Berns A., Collard J. G. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto Y., Yamauchi J., Itoh H. J. Biol. Chem. 2003;278:29890–29900. doi: 10.1074/jbc.M301559200. [DOI] [PubMed] [Google Scholar]
- 13.Malliri A., van Es S., Huveneers S., Collard J. G. J. Biol. Chem. 2004;279:30092–30098. doi: 10.1074/jbc.M401192200. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen F. N., Kain H. E. T., van der Kammen R. A., Michiels F., Kranenburg O. W., Collard J. G. J. Cell Biol. 1997;139:797–807. doi: 10.1083/jcb.139.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunda P., Paglini G., Quiroga S., Kosik K., Caceres A. J. Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolias T. F., Bikoff J. B., Burette S., Paradis S., Harrar D., Tavazoie S., Weinberg R. J., Greenberg M. E. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Benard V., Bohl B. P., Bokoch G. M. J. Biol. Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 18.Yamauchi J., Miyamoto Y., Tanoue A., Shooter E. M., Chan J. R. Proc. Natl. Acad. Sci. USA. 2005;102:14889–14894. doi: 10.1073/pnas.0507125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamauchi J., Chan J. R., Miyamoto Y., Tsujimoto G., Shooter E. M. Proc. Natl. Acad. Sci. USA. 2005;102:5198–5203. doi: 10.1073/pnas.0501160102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habets G. G., van der Kammen R. A., Stam J. C., Michiels F., Collard J. G. Oncogene. 1995;10:1371–1376. [PubMed] [Google Scholar]
- 21.Matsuo N., Hoshino M., Yoshizawa M., Nabeshima Y. J. Biol. Chem. 2002;277:2860–2868. doi: 10.1074/jbc.M106186200. [DOI] [PubMed] [Google Scholar]
- 22.Mertens A. E., Roovers R. C., Collard J. G. FEBS Lett. 2003;546:11–16. doi: 10.1016/s0014-5793(03)00435-6. [DOI] [PubMed] [Google Scholar]
- 23.Stam J. C., Sander E. E., Michiels F., van Leeuwen F. N., Kain H. E. T., van der Kammen R. A., Collard J. G. J. Biol. Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 24.Arthur W. T., Ellerbroek M. S., Der C. J., Burridge K., Wennerberg K. J. Biol. Chem. 2002;277:42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- 25.Irie F., Yamaguchi Y. Nat. Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- 26.Penzes P., Beeser A., Chernoff J., Schiller M. R., Eipper B. A., Mains R. E., Huganir R. L. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y. J., Kruttgen A., Moller J. C., Shine D., Chan J. R., Shooter E. M., Cosgaya J. M. J. Neurosci. Res. 2004;75:825–834. doi: 10.1002/jnr.20048. [DOI] [PubMed] [Google Scholar]
- 28.Satoh T., Nakamura S., Nakafuku M., Kaziro Y. Biochim. Biophys. Acta. 1998;949:97–109. doi: 10.1016/0167-4781(88)90059-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.