Abstract
Rab3A small G protein is a member of the Rab family and is most abundant in the brain, where it is localized on synaptic vesicles. Evidence is accumulating that Rab3A plays a key role in neurotransmitter release and synaptic plasticity. Rab3A cycles between the GDP-bound inactive and GTP-bound active forms, and this change in activity is associated with the trafficking cycle of synaptic vesicles at nerve terminals. Rab3 GTPase-activating protein (GAP) stimulates the GTPase activity of Rab3A and is expected to determine the timing of the dissociation of Rab3A from synaptic vesicles, which may be coupled with synaptic vesicle exocytosis. Rab3 GAP consists of two subunits: the catalytic subunit p130 and the noncatalytic subunit p150. Recently, mutations in p130 were found to cause Warburg Micro syndrome with severe mental retardation. Here, we generated p130-deficient mice and found that the GTP-bound form of Rab3A accumulated in the brain. Loss of p130 in mice resulted in inhibition of Ca2+-dependent glutamate release from cerebrocortical synaptosomes and altered short-term plasticity in the hippocampal CA1 region. Thus, Rab3 GAP regulates synaptic transmission and plasticity by limiting the amount of the GTP-bound form of Rab3A.
Keywords: Rab3A, Rab3 GAP p130, neurotransmitter release, synaptic plasticity, Warburg Micro syndrome
The Rab family small G proteins appear to be key regulators of intracelluar vesicle transport (1, 2). The Rab3 subfamily, which consists of Rab3A, -3B, -3C, and -3D, is involved in regulated exocytosis (1, 3, 4). Rab3A is the most abundant isoform in the brain, where it is localized on synaptic vesicles, and is involved in Ca2+-dependent neurotransmitter release (1, 3–5). Electrophysiological studies of Rab3A-deficient mice revealed that Rab3A is not essential for synaptic transmission but performs a modulatory function that acts at the Ca2+-triggered fusion step (6). Rab3A is also required for hippocampal CA3 mossy fiber long-term potentiation (LTP) (7).
Like other Rab family members, Rab3A cycles between the GDP-bound inactive and GTP-bound active forms (1, 3, 4). This cycling is regulated by three types of regulators: Rab GDP dissociation inhibitor (GDI), Rab3 GDP/GTP exchange protein (GEP), and Rab3 GAP (1, 3, 4). Because the cyclical activation is coupled with membrane association and allows both spatial and temporal control of Rab3A activity, these regulators are thought to be important for the proper functioning of Rab3A in synaptic vesicle transport. The significance of Rab GDIα, a neuron-specific isoform, and Rab3 GEP in synaptic transmission and plasticity has been shown by our knockout studies in mice (8–10) and by the identification of mutations in the Rab GDIα gene that cause human X-linked nonspecific mental retardation (11).
Then, what is the significance of Rab3 GAP? Because GTP hydrolysis catalyzed by GAP converts Rab proteins to their inactive states, GAP is expected to serve as a timer to determine the timing of the termination of Rab function. Genetic studies in yeast, however, demonstrated that GAP and GTP hydrolysis are not necessary for the function of Rab proteins in budding yeast, although these Rab proteins are essential for secretion (12). In contrast, Rab GAP-5, a GAP for Rab5, was recently shown to regulate endocytic transport through the inactivation of Rab5 (13). Thus, the significance of Rab GAP has not yet been established.
Rab3 GAP, which is specific for the Rab3 subfamily, consists of two subunits: the catalytic subunit p130 and the noncatalytic subunit p150 (14, 15). Rab3 GAP is ubiquitously expressed and enriched in the synaptic soluble fraction of brain (16). This observation suggests that Rab3 GAP inactivates Rab3A in nerve terminals and thereby regulates neurotransmitter release and synaptic plasticity. Consistent with this prediction, mutations in p130 have recently been reported to cause Warburg Micro syndrome with severe mental retardation (17).
To clarify the physiological role of Rab3 GAP, we generated Rab3 GAP p130-deficient mice and found that Rab3 GAP regulates synaptic transmission and plasticity by limiting the amount of the GTP-bound form of Rab3A.
Results
Generation of Rab3 GAP p130-Deficient Mice.
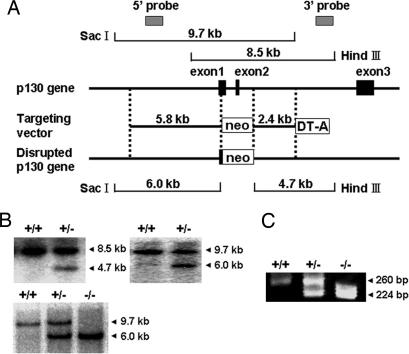
To disrupt Rab3 GAP p130 in ES cells, gene targeting was used to replace the 3′ half of the exon 1, the exon 2, and the 5′ portion of the downstream intron with an MC1-neomycin-resistance cassette (Fig. 1A). The targeting vector was electroporated into RW4 ES cells, which were then selected with G418 (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). The genotypes of the G418-resistance colonies were confirmed by Southern blot hybridization (Fig. 1B Upper). Cells from two independent ES clones were used to generate chimeric mice and successfully contributed to germ-line transmission. Rab3 GAP p130 heterozygous mice were intercrossed to produce homozygous mutant offspring. The genotypes of mice homozygous for the disrupted allele were confirmed by Southern blot hybridization (Fig. 1B Lower) and PCR (Fig. 1C). Mice homozygous for the disrupted allele expressed no intact p130 (Fig. 2A). The p130-deficient mice were viable and fertile.
Fig. 1.
Targeted disruption of the Rab3 GAP p130 gene. (A) The structure of the mouse Rab3 GAP p130 gene with the first, second, and third coding exons is shown. A targeting vector was designed to remove the 3′ half of exon 1, exon 2, and the 5′ portion of the downstream intron. The construct contained 5.8 kb of the 5′ flanking sequence and 2.4 kb of the 3′ flanking sequence. The diphtheria toxin DT-A cassette was inserted at the 3′ end of the construct. In the targeted allele, the MC1-neo cassette replaces 1.2 kb of the genomic DNA. Homologous recombination in ES cells was verified by using informative restriction fragments and diagnostic probes as indicated. (B) Southern blot hybridization using the digested DNA extracted from ES cells (Upper) or mouse tails (Lower) and the 3′ external or the 5′ internal probe shown in A. The 3′ probe hybridized to the 8.5-kb wild-type or the 4.7-kb mutant fragments digested with HindIII, and the 5′ probe hybridized to the 9.7-kb wild-type or the 6.0-kb mutant fragments digested with SacI, respectively. (C) Genotyping 21-day-old mice by PCR analysis. PCR primers were selected to generate a 260-bp product indicative of the wild-type allele or a 224-bp product resulting from the disrupted p130 allele.
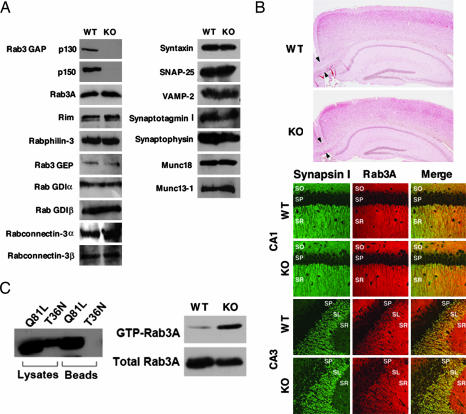
Fig. 2.
Biochemical and morphological analyses of the p130-deficient mice. (A) Expression of synaptic proteins in the wild-type (WT) and p130-deficient (KO) mice was analyzed by Western blotting. (B) Normal synaptic architecture in the p130-deficient brains. (Upper) Sagittal sections of hippocampi and cortices stained with hematoxylin and eosin. Arrowheads indicate the corpus callosum. (Lower) Confocal immunofluorescence analysis of frozen sections of the hippocampal CA1 and CA3 regions. Each section was double-labeled with a polyclonal antibody against synapsin I (green) and a monoclonal antibody against Rab3A (red). SO, stratum oriens; SP, stratum pyramidale; SR, stratum radiatum; SL, stratum lucidum. (C) Pull-down assays. (Left) BHK cells were transfected with Myc-tagged Rab3AQ81L or Rab3AT36N. After 48 h, the mutants were isolated by using a GST-Rim1αN-affinity column and visualized by Western blotting using an anti-Myc antibody. (Right) Wild-type or p130-deficient mouse brains were homogenized. GTP-Rab3A was then purified from the homogenates with a GST-Rim1αN-affinity column and visualized by Western blotting using an anti-Rab3A antibody.
Among the synaptic proteins examined, the expression of p150 was severely attenuated in the p130-deficient mice, presumably because p150, forming a stable complex of heterodimer with p130, becomes destabilized in the absence of p130 (Fig. 2A and Supporting Materials and Methods). No significant changes were observed in the expression levels of Rab3A or other synaptic proteins (Fig. 2A). Although Micro syndrome results in ocular and neurodevelopmental defects, including hypoplasia of the corpus callosum (17), the mice did not show such morphological abnormalities (Fig. 2B and data not shown). Moreover, the layered structures of the cerebral cortex and the hippocampus were not different between the wild-type and mutant brains (Fig. 2B). In the hippocampal CA1 and CA3 regions, Rab3A colocalized with synapsin I at presynaptic terminals in the p130-deficient mice as well as in the wild-type mice.
Accumulation of the GTP-Bound Form of Rab3A in the p130-Deficient Mouse Brains.
Because p130 was shown to activate Rab3A GTPase activity in vitro (14), we examined whether p130 exhibits this activity in vivo. For this purpose, we performed pull-down assays using a GST fusion protein that included the N terminus of the Rab3 effector protein rat Rim1α (amino acids 1–200; GST-Rim1αN). This fusion protein contained the Rab3A-binding domain and specifically interacted with GTP-Rab3A (18). First, we confirmed that this assay could detect GTP-Rab3A in BHK cells expressing dominant-active Rab3AQ81L or dominant-negative Rab3AT36N (Fig. 2C). We then assessed the amount of GTP-Rab3A in mouse brains. As expected, the GTP-Rab3A level was increased in the p130-deficient brains compared with the wild-type brains (Fig. 2C). This indicates that p130 functions as a GAP for Rab3A in vivo and that GTP-Rab3A accumulates in the p130-deficient mouse brains.
Inhibition of Glutamate Release from the p130-Deficient Synaptosomes.
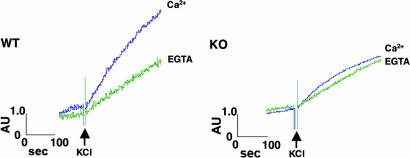
We next examined how the deletion of Rab3 GAP affects neurotransmitter release. We measured the release of endogenous glutamate from synaptosomes that were prepared from cerebral cortex by differential and Percoll-gradient centrifugations. For synaptosomes prepared from the wild-type mice, KCl-induced depolarization led to Ca2+-dependent glutamate release (Fig. 3). Under the same conditions, the p130-deficient synaptosomes showed weak Ca2+-dependent release (Fig. 3), although cytoplasmic free Ca2+ concentration was elevated to a similar extent as that seen in the wild-type synaptosomes (Fig. 7, which is published as supporting information on the PNAS web site). In contrast, α-latrotoxin, a potent excitatory neurotoxin, induced massive release of neurotransmitter from both the wild-type and p130-deficient synaptosomes to a similar level (Fig. 8, which is published as supporting information on the PNAS web site), indicating that the p130-deficient synaptosomes were viable and healthy. It was reported that α-latrotoxin induces neurotransmitter release from Munc13-1-deficient hippocampal cells, in which normal release is not observed (19). Munc13-1 is considered to be a regulator of the priming step of exocytosis, and our present results suggest that Rab3 GAP acts upstream of or in the same step as Munc13-1.
Fig. 3.
Glutamate release from the p130-deficient synaptosomes. Glutamate release from the wild-type (WT) or p130-deficient (KO) synaptosomes by KCl-induced depolarization was measured spectrofluorometrically after the conversion of glutamate by glutamate dehydrogenase. Incubations were performed in the presence of 1.3 mM CaCl2 (Ca2+) or 0.5 mM EGTA.
Altered Short-Term Plasticity at the Hippocampal CA1 Synapse of the p130-Deficient Mice.
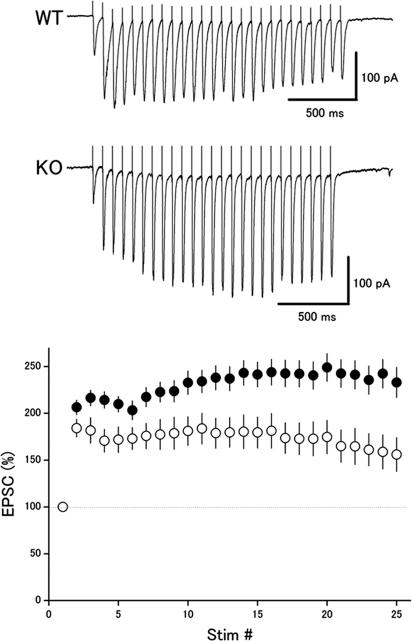
We also examined whether the deletion of Rab3 GAP affected synaptic transmission as well as several forms of activity-dependent plasticity at the hippocampal CA1 synapse. In the previous studies, the Rab3A- and Rim1α-deficient synapses displayed altered responses to repetitive stimulation at moderate frequency, such as 14 Hz (20, 21). Therefore, we first examined changes in this form of short-term plasticity. In the wild-type mice, repetitive stimuli (25 times at 14 Hz) elicited an initial facilitation, followed by a slight depression of excitatory postsynaptic current (EPSC) amplitude (Fig. 4). In contrast, the mutant synapses showed larger facilitation with almost no depression of the EPSC amplitude. The relative EPSC amplitudes at the end of the train were 156 ± 18% (n = 33) and 233 ± 16% (n = 50) of the first EPSCs in the wild-type and mutant mice, respectively. The difference between the two groups was significant (P < 0.01, t test).
Fig. 4.
Altered activity-dependent plasticity during repetitive stimulation in the p130-deficient mice. (Upper) EPSCs in response to 25 stimuli at 14 Hz were recorded from CA1 neurons in hippocampal slices in the presence of 100 μM picrotoxin. (Lower) Relative amplitudes of EPSCs plotted as a function of stimulus number in the wild-type mice (open circles, n = 33) and mutant mice (filled circles, n = 50).
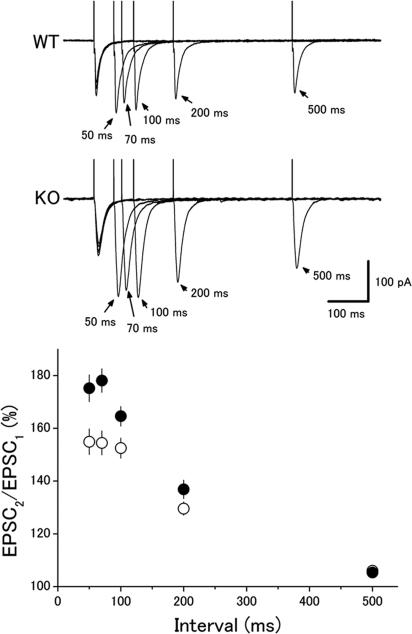
Next, we examined paired-pulse facilitation (PPF), another form of short-term plasticity, at the CA1 synapse. As was reported for the Rab3A- and Rim1α-deficient mice (6, 21), the magnitude of PPF was larger in the p130-deficient mice (Fig. 5). Ratios of the second to the first EPSC amplitudes at a 50-ms interstimulus interval were 155 ± 5% (n = 54) and 178 ± 4% (n = 56) in the wild-type and mutant mice, respectively. The difference between the two groups was significant at 50-, 70-, and 100-ms intervals (P < 0.05, t test).
Fig. 5.
Enhanced paired-pulse facilitation in the p130-deficient mice. (Upper) EPSCs recorded from CA1 neurons in response to paired stimuli with different interstimulus intervals (50–500 ms). (Lower) Ratios of the second to the first EPSC amplitude in the wild-type mice (open circles, n = 54) and mutant mice (filled circles, n = 56) are plotted as a function of the interstimulus interval.
Because the magnitude of facilitation depends on the initial quantal contents, the results suggest that release probability (Pr) is suppressed in the mutant synapse. To directly test this possibility, we compared time courses of use-dependent block of NMDA receptor-mediated EPSC (EPSCNMDA) in the presence of MK-801 between the wild-type and mutant mice. MK-801 irreversibly blocks NMDA receptors only when they are activated by synaptically released glutamate, and, therefore, difference in Pr would result in different time courses of use-dependent block of EPSCNMDA (8, 22). In fact, it was demonstrated that knockout mice of Rim1α, but not of Rab3A, displayed a slower decay time course of blocking by MK-801 (6, 21), suggesting that Pr was decreased in the Rim1α-deficient synapses. Contrary to our expectation, consecutive stimulation in the presence of MK-801 led to a decrease in amplitudes of EPSCNMDA, but the time course of blocking was indistinguishable between the wild-type and mutant mice (see Fig. 9, which is published as supporting information on the PNAS web site). The decay time constants of the peak amplitudes of EPSCNMDA were 205 ± 32 sec (n = 5) and 201 ± 16 sec (n = 9) in the wild-type and mutant mice, respectively (P = 0.89, t test). This result suggests that basal Pr is not affected significantly by deletion of p130, although this experiment may not be sensitive enough to detect a small reduction in Pr under our conditions.
Normal LTP at the Mossy Fiber Synapse in the p130-Deficient Mice.
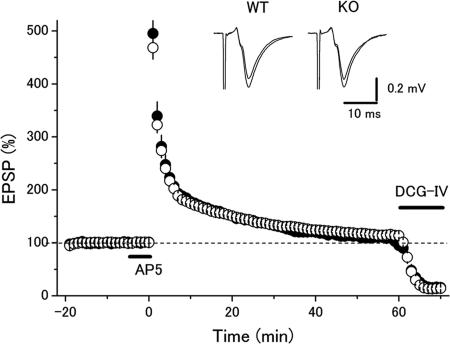
Finally, we addressed whether presynaptic LTP at the mossy fiber–CA3 synapse (MF–LTP) was affected by deletion of p130, because LTP at this synapse is absent in the Rab3A- and Rim1α-deficient mice (7, 23). Posttetanic potentiation (PTP) or LTP was not different between the two groups (Fig. 6). PTP (measured soon after the tetanus) was 468 ± 21% (n = 9) and 495 ± 45% (n = 8) in the wild-type and mutant mice, respectively (P = 0.58, t test). LTP (measured at 60 min after the tetanus) was 114 ± 3% and 109 ± 3% in the wild-type and mutant mice, respectively (P = 0.27). Thus, NMDA receptor-independent long-term synaptic plasticity at the mossy fiber–CA3 synapse was unchanged in the p130-deficient mice, suggesting that the inactivation of Rab3A is not necessary for MF–LTP, whereas Rab3A is required for MF–LTP.
Fig. 6.
Unchanged MF–LTP in the p130-deficient mice. LTP was induced by 100-Hz stimuli for 1 s, and 25 μM d-AP5 was applied 5 min before the tetanic stimulation. There was no significant difference between the wild-type mice (open circles, n = 9) and mutant mice (filled circles, n = 8) in posttetanic potentiation or LTP.
Discussion
In this study, we have generated Rab3 GAP p130-deficient mice and found that the GTP-bound form of Rab3A accumulates in the brain. We have shown that Ca2+-dependent glutamate release from cerebrocortical synaptosomes is inhibited in the mutant mice. These findings are consistent with those of the previous studies using the GTPase-deficient mutant of Rab3A in PC12, chromaffin, and insulin-secreting cells (24–26) and demonstrate that the GTP hydrolysis of Rab3A is linked to the exocytotic process and is important for the function of Rab3A. Previous studies showed that Rab3A dissociates from synaptic vesicles during Ca2+-dependent exocytosis, probably through the conversion of GTP-Rab3A to GDP-Rab3A by Rab3 GAP (27). These results suggest that the deletion of p130 may suppress this dissociation, thereby inhibiting exocytosis.
In contrast to the inhibitory effect in the synaptosome experiments, we have found that loss of p130 in mice increases two forms of short-term plasticity, paired-pulse and train facilitation, in the hippocampal CA1 region. The molecular mechanisms to explain the electrophysiological data in agreement with those of the synaptosome experiments are still obscure, but a likely interpretation is that basal synaptic transmission is suppressed in the mutant hippocampal synapses, although the MK-801 experiments are negative. Even if this interpretation would be accepted, there still seems to be a discrepancy between the mild phenotype observed in the electrophysiological experiments and the dramatic difference observed in the synaptosome experiments. However, it is very difficult to assess the extent of inhibition from the data obtained from the separate assay systems, because the experimental conditions are quite different [materials (synaptosomes or hippocampal slices), kinds of stimulation (KCl or electrical stimulation), etc.]. Then, another interpretation would not necessarily be required to explain this issue. In any case, our present results imply that Rab3 GAP contributes to maintaining a limiting amount of GTP-Rab3A by stimulating its GTPase activity during the modulation of synaptic transmission and plasticity.
The increased amount of GTP-Rab3A in the p130-deficient mice could affect the synaptic vesicle transport as follows: GTP-Rab3A localized on synaptic vesicles binds to its effector protein, Rim1, at the active zone of the presynaptic membrane, which contributes to the tethering of synaptic vesicles to the active zone. Therefore, GTP-Rab3A should be maintained during tethering process. In the p130-deficient mice, however, the increased level of GTP-Rab3A might hold the Rab3A–Rim1 complex for the longer periods of time, so that the reduced amount of free Rim1 would cause a defect in additional rounds of vesicle tethering. It is also likely that disassembly of the Rab3A–Rim1 complex is required for the process from the tethering step to the priming and fusion steps. Recent studies, based primarily on experiments with knockout mice, have shown that Munc13-1 regulates the priming step, whereas Munc18 and the SNARE proteins are involved in the priming/fusion step (5). Defective GTP hydrolysis might prevent the disassembly of the Rab3A–Rim1 complex, leading to the inhibition of the function of these proteins in these downstream steps.
It was reported that Munc13-1 interacts with Rim1α and its isoform Rim2α (28, 29). Disrupting the Rim1/2α–Munc13-1 interaction reduces the size of the readily releasable pool of vesicles (28, 29). Similar changes are observed in Rim1α- and Munc13-1-deficient mice but not in Rab3A-deficient mice (6, 19, 30). These results suggest that GTP-Rab3A inhibits the formation of Rim1/2α–Munc13-1 complex or the function of the complex in the priming step. Recent NMR work by Dulubova et al. (29) revealed that Munc13-1, Rim2α, and GTP-Rab3A can form a tripartite complex, favoring the latter possibility. In this case, it is likely that the Rab3 GAP-induced dissociation of Rab3A from the tripartite complex is important for the function of the Rim1/2α–Munc13-1 complex in the priming step. Further studies are necessary to clarify the relationship among Rab3A, Rim1/2α, and Munc13-1 and the precise point of action of Rab3 GAP in synaptic vesicle transport.
From the point of view of the pathogenesis of Warburg Micro syndrome, it is important to know why the p130-deficient mice do not appear to show developmental abnormalities of the eye and central nervous system, which are usually observed in patients with this disease (17). One explanation is that this is because of the difference of species. The mutations of the gene that cause the disease in human do not always cause the similar symptoms in mice. In Micro syndrome, most mutations of the p130 gene were identified at the C-terminal region that has the catalytic activity (17). Therefore, it is likely that the mutated protein of p130 lacking Rab3 GAP activity is expressed in the patients, whereas p130 is not expressed in our mice. This mutated protein could interact with various proteins, such as the noncatalytic subunit p150 (15), rabconnectin-3 (31, 32), and the unidentified proteins as well as the intact protein. Another possibility is that this mutated p130 may suppress (or stimulate) the functions of these interacting proteins, resulting in the generation of abnormalities. Further insight into the pathophysiological responsibility of Rab3 GAP in Warburg Micro syndrome would be greatly aided by analyses of these interacting proteins.
Materials and Methods
Pull-Down Assays for the Measurement of the GTP-Bound Pool of Rab3A.
BHK cells (6 × 105 cells per 60-mm dish) were transiently transfected by FuGENE 6 with 5 μg of plasmids (pEFBOS-Myc-Rab3AQ81L or -Rab3AT36N). After a 48-h incubation at 37°C, the cells were washed once and scraped from the dishes in PBS. The cells were lysed in 300 μl of buffer A [10 mM Tris·HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% (wt/vol) Nonidet P-40, 10 μM p-APMSF, and 10 μM leupeptin]. The cell lysates were centrifuged at 4°C for 5 min at 16,100 × g. An aliquot of the supernatant was saved and used to verify the total amount of mutant Rab3A expressed in the cells. The rest of the supernatant was mixed with 4 μg of GST-Rim1αN attached to glutathione-Sepharose beads and incubated for 1 h at 4°C. The beads were then washed with buffer A four times and resuspended in SDS sample buffer. Comparable amounts of the proteins that remained associated with the beads were separated by SDS/PAGE. The fraction of mutated Rab3A bound to the affinity column was determined by Western blotting using a monoclonal antibody against the myc epitope tag (9E10; American Type Culture Collection).
Wild-type or p130-deficient mouse brains were homogenized in buffer A, followed by centrifugation at 4°C for 7 min at 500 × g. The supernatant was mixed with 4 μg of GST-Rim1αN attached to glutathione-Sepharose beads and incubated for 1 h at 4°C. After the incubation, the beads were washed four times in buffer A and resuspended in SDS sample buffer. Comparable amounts of the proteins that remained associated with the beads were separated by SDS/PAGE. The fraction of Rab3A bound to the affinity column was determined by Western blotting using a monoclonal antibody against Rab3A.
Synaptosomal Secretion Assay.
Synaptosomes were prepared as described in ref. 33. Glutamate release was assayed by on-line fluorometry as described (34). Briefly, a solution containing 1.0 mg of synaptosomes in 1.5 ml of buffer B (140 mM NaCl, 5 mM KCl, 10 mM Hepes/NaOH at pH 7.4, 5 mM NaHCO3, 1.2 mM NaH2PO4, 1 mM MgCl2, and 10 mM glucose) was prepared and prewarmed for 15 min at 37°C. A 1.5-ml aliquot was transferred to a stirred cuvette in a FP-6300 Spectrofluorometer (Jasco, Tokyo). Either 1.3 mM CaCl2 or 0.5 mM EGTA was added together with 1 mM NADP+ (Wako Pure Chemical, Osaka) to the cuvette and incubated under stirring for 2 min at 37°C, followed by addition of 50 units of l-glutamate dehydrogenase (Wako Pure Chemical). After further incubation for 3 min at 37°C, 30 mM KCl was added and the incubation was extended for an additional 5 min. Generation of NADPH was measured at excitation and emission wavelengths of 340 and 460 nm, respectively.
Electrophysiology.
Transverse hippocampal slices (400 μm thick) were prepared from 4- to 8-week-old mice. All experiments were performed in accordance with the animal welfare guidelines of the Physiological Society of Japan. Standard artificial cerebrospinal fluid (ACSF; 127 mM NaCl, 1.5 mM KCl, 1.2 mM KH2PO4, 1.3 mM MgSO4, 2.4 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose) was equilibrated with 95% O2 and 5% CO2. Electrical stimulation (test pulses at 0.1 Hz, 0.1 ms duration, ≈200 μA intensity) was delivered through a concentric bipolar electrode inserted into the stratum radiatum of the CA1 region. All recordings were made at room temperature. For whole-cell experiments, picrotoxin (100 μM) was added to block GABAergic inhibition. An incision was made between the CA1 and CA2 regions to block the propagation of burst activities from CA2/3 areas. In the MK-801 experiments, EPSCNMDA was monitored in 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) and 100 μM picrotoxin at a holding potential of −40 mV. Patch pipettes were filled with an internal solution (pH 7.2) containing 150 mM Cs gluconate, 0.2 mM EGTA, 8 mM NaCl, 10 mM Hepes, and 2 mM Mg2+ATP. Recordings were made with an Axopatch 1D or Multiclamp 700B amplifier (Molecular Devices). Signals were filtered at 1 kHz, digitized at 10 kHz, and analyzed with pclamp 9 software (Molecular Devices). For LTP experiments with MF–CA3 synapses, MFs were stimulated at the stratum granulosum in the dentate gyrus, and the field EPSPs were recorded from the stratum lucidum of the CA3 region. High-frequency stimulation (100 Hz for 1 s) was given to induce LTP after 25 μM d-AP5 was applied for 5 min to prevent contamination of NMDA receptor-dependent potentiation at the CA3 recurrent collaterals. The amplitudes of the EPSCs or the field EPSPs were measured, and the data are expressed as means ± SEM as a percentage of the baseline. Statistical analysis was performed by using Student’s t tests, and P < 0.05 was accepted as statistical significance.
Supplementary Material
Acknowledgments
We thank Dr. Toshiya Manabe (University of Tokyo, Tokyo) for helpful discussions and critical reading of our manuscript. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (2003-2005) and the Takeda Science Foundation (2002).
Abbreviations
- EPSC
excitatory postsynaptic current
- EPSCNMDA
NMDA receptor-mediated EPSC
- GAP
GTPase-activating protein
- LTP
long-term potentiation
- MF–LTP
LTP at the mossy fiber–CA3 synapse
- Pr
release probability.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Takai Y., Sasaki T., Matozaki T. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Zerial M., McBride H. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 3.Takai Y., Sasaki T., Shirataki H., Nakanishi H. Genes Cells. 1996;1:615–632. doi: 10.1046/j.1365-2443.1996.00257.x. [DOI] [PubMed] [Google Scholar]
- 4.Geppert M., Südhof T. C. Annu. Rev. Neurosci. 1998;21:75–95. doi: 10.1146/annurev.neuro.21.1.75. [DOI] [PubMed] [Google Scholar]
- 5.Südhof T. C. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 6.Geppert M., Goda Y., Stevens C. F., Südhof T. C. Nature. 1997;387:810–814. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- 7.Castillo P. E., Janz R., Südhof T. C., Tzounopoulos T., Malenka R. C., Nicoll R. A. Nature. 1997;388:590–593. doi: 10.1038/41574. [DOI] [PubMed] [Google Scholar]
- 8.Ishizaki H., Miyoshi J., Kamiya H., Togawa A., Tanaka M., Sasaki T., Endo K., Mizoguchi A., Ozawa S., Takai Y. Proc. Natl. Acad. Sci. USA. 2000;97:11587–11592. doi: 10.1073/pnas.97.21.11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka M., Miyoshi J., Ishizaki H., Togawa A., Ohnishi K., Endo K., Matsubara K., Mizoguchi A., Nagano T., Sato M., et al. Mol. Biol. Cell. 2001;12:1421–1430. doi: 10.1091/mbc.12.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi K., Tanaka M., Mizoguchi A., Hirata Y., Ishizaki H., Kaneko K., Miyoshi J., Takai Y. Proc. Natl. Acad. Sci. USA. 2002;99:14536–14541. doi: 10.1073/pnas.212511399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Adamo P., Menegon A., Lo Nigro C., Grasso M., Gulisano M., Tamanini F., Bienvenu T., Gedeon A. K., Oostra B., Wu S. K., et al. Nat. Genet. 1998;19:134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- 12.Bernards A. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 13.Haas A. K., Fuchs E., Kopajtich R., Barr F. A. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 14.Fukui K., Sasaki T., Imazumi K., Matsuura Y., Nakanishi H., Takai Y. J. Biol. Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- 15.Nagano F., Sasaki T., Fukui K., Asakura T., Imazumi K., Takai Y. J. Biol. Chem. 1998;273:24781–24785. doi: 10.1074/jbc.273.38.24781. [DOI] [PubMed] [Google Scholar]
- 16.Oishi H., Sasaki T., Nagano F., Ikeda W., Ohya T., Wada M., Ide N., Nakanishi H., Takai Y. J. Biol. Chem. 1998;273:34580–34585. doi: 10.1074/jbc.273.51.34580. [DOI] [PubMed] [Google Scholar]
- 17.Aligianis I. A., Johnson C. A., Gissen P., Chen D., Hampshire D., Hoffmann K., Maina E. N., Morgan N. V., Tee L., Morton J., et al. Nat. Genet. 2005;37:221–223. doi: 10.1038/ng1517. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Okamoto M., Schmitz F., Hofmann K., Südhof T. C. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- 19.Augustin I., Rosenmund C., Südhof T. C., Brose N. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 20.Geppert M., Bolshakov V. Y., Siegelbaum S. A., Takei K., De Camilli P., Hammer R. E., Südhof T. C. Nature. 1994;369:493–497. doi: 10.1038/369493a0. [DOI] [PubMed] [Google Scholar]
- 21.Schoch S., Castillo P. E., Jo T., Mukherjee K., Geppert M., Wang Y., Schmitz F., Malenka R. C., Südhof T. C. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 22.Manabe T., Nicoll R. A. Science. 1994;265:1888–1892. doi: 10.1126/science.7916483. [DOI] [PubMed] [Google Scholar]
- 23.Castillo P. E., Schoch S., Schmitz F., Südhof T. C., Malenka R. C. Nature. 2002;415:327–330. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- 24.Holz R. W., Brondyk W. H., Senter R. A., Kuizon L., Macara I. G. J. Biol. Chem. 1994;269:10229–10234. [PubMed] [Google Scholar]
- 25.Johannes L., Lledo P. M., Roa M., Vincent J. D., Henry J. P., Darchen F. EMBO J. 1994;13:2029–2037. doi: 10.1002/j.1460-2075.1994.tb06476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regazzi R., Ravazzola M., Iezzi M., Lang J., Zahraoui A., Andereggen E., Morel P., Takai Y., Wollheim C. B. J. Cell Sci. 1996;109(Pt 9):2265–2273. doi: 10.1242/jcs.109.9.2265. [DOI] [PubMed] [Google Scholar]
- 27.Fischer von Mollard G., Südhof T. C., Jahn R. Nature. 1991;349:79–81. doi: 10.1038/349079a0. [DOI] [PubMed] [Google Scholar]
- 28.Betz A., Thakur P., Junge H. J., Ashery U., Rhee J. S., Scheuss V., Rosenmund C., Rettig J., Brose N. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 29.Dulubova I., Lou X., Lu J., Huryeva I., Alam A., Schneggenburger R., Südhof T. C., Rizo J. EMBO J. 2005;24:2839–2850. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calakos N., Schoch S., Südhof T. C., Malenka R. C. Neuron. 2004;42:889–896. doi: 10.1016/j.neuron.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Nagano F., Kawabe H., Nakanishi H., Shinohara M., Deguchi-Tawarada M., Takeuchi M., Sasaki T., Takai Y. J. Biol. Chem. 2002;277:9629–9632. doi: 10.1074/jbc.C100730200. [DOI] [PubMed] [Google Scholar]
- 32.Kawabe H., Sakisaka T., Yasumi M., Shingai T., Izumi G., Nagano F., Deguchi-Tawarada M., Takeuchi M., Nakanishi H., Takai Y. Genes Cells. 2003;8:537–546. doi: 10.1046/j.1365-2443.2003.00655.x. [DOI] [PubMed] [Google Scholar]
- 33.Dunkley P. R., Jarvie P. E., Heath J. W., Kidd G. J., Rostas J. A. Brain Res. 1986;372:115–129. doi: 10.1016/0006-8993(86)91464-2. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls D. G., Sihra T. S., Sanchez-Prieto J. J. Neurochem. 1987;49:50–57. doi: 10.1111/j.1471-4159.1987.tb03393.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.