Abstract
Adaptor protein 3 (AP-3) is a vesicle-coat protein that forms a heterotetrameric complex. Two types of AP-3 subunits are found in mammalian cells. Ubiquitous AP-3 subunits are expressed in all tissues of the body, including the brain. In addition, there are neuronal AP-3 subunits that are thought to serve neuron-specific functions such as neurotransmitter release. In this study, we show that overexpression of neuronal AP-3 in mouse chromaffin cells results in a striking decrease in the neurotransmitter content of individual vesicles (quantal size), whereas deletion of all AP-3 produces a dramatic increase in quantal size; these changes were correlated with alterations in dense-core vesicle size. AP-3 appears to localize in the trans-Golgi network and possibly immature secretory vesicles, where it may be involved in the formation of neurosecretory vesicles.
Keywords: amperometry, electron microscopy, quantal content, vesicle size, chromaffin cells
Adaptor protein (AP) complexes play an important role in protein trafficking. They mediate the sorting of membrane proteins among numerous intracellular compartments (1–4). Importantly, they aid in the vesiculation of donor membranes through direct association with membrane cargo and other vesicle-coat proteins (5). Four different types of adaptor protein have been identified (AP-1–AP-4) (1, 2, 4). These adaptor proteins are heterotetrameric complexes composed of a large variable subunit (uniquely named for each AP complex), a second large subunit (β subunit), a medium-sized subunit (referred to as μ), and a small subunit (σ subunit) (2, 5, 6). Although a few APs sort the same or similar cargo, some of the specificity of different AP functions may arise from their selective recruitment to the correct membrane compartment (3, 5).
AP-2 has been studied extensively in synaptic-vesicle formation and receptor trafficking from the plasma membrane (endocytosis), but the role of AP-3 is less well known and is proposed to serve in vesicle formation from various organelles. AP-3 is known to be involved in lysosomal cargo sorting (7, 8), and it may be involved in the normal function of some synaptic vesicles (9). Ubiquitous AP-3 is composed of δ, β3A, μ3A, and σ3 subunits expressed throughout the body (10, 11). Neuronal AP-3 contains β3B and μ3B subunits that are expressed only in neurons (12, 13). Recent studies indicate that ubiquitous and neuronal AP-3 have distinct functions. In mocha mice, the complete loss of all forms of AP-3 leads to defects in neurological and hematopoietic cell function (14); these animals exhibit balance problems and deafness, and they are hyperactive and undergo seizures (14, 15). Selective ablation of ubiquitous AP-3 observed in pearl mice results in animals with defects in hematopoietic cells but no neurological symptoms (2) [with the possible exception of night blindness (16)]. Animals lacking either neuronal AP-3 subunit, β3B (17) or μ3B (5, 9), exhibit neurological deficits that are less severe than those in mocha animals. Taken together, these data suggest that neurological deficits result primarily from alterations in neuronal AP-3 subunits.
This study addresses the role of AP-3 in neurotransmitter release from neurosecretory vesicles to determine whether alterations in secretion could be involved in the neurological problems observed in mutant mice. Adrenal chromaffin cells secrete catecholamine neurotransmitters and neuropeptides from dense-core vesicles (DCVs) in response to elevations in intracellular Ca2+. Immature, large DCVs are thought to originate directly from the trans-Golgi network (TGN). Maturation of these vesicles, which takes ≈30 min, is thought to be complex and to involve changes in vesicle diameter (18). Maturation may impart unique morphological and physiological features to the vesicles (18–20).
In the past few years, considerable evidence has accumulated for kiss-and-run release in chromaffin cells (21–23). In this type of release, the fusion pore opens transiently, releasing neurotransmitter. Under certain conditions, vesicles appear to be able to release a fraction of their neurotransmitter content (21, 24). Although changes in quantal content have previously been described, our study shows that alterations in vesicle size can lead to changes in quantal content. In chromaffin cells, overexpression of AP-3 produced a large number of small-volume vesicles that released relatively small amounts of neurotransmitter, whereas deleting AP-3 produced large-diameter vesicles that released large amounts of neurotransmitter. Our results suggest that regulation by AP-3 takes place at the vesicle budding or in a subsequent maturation step. The interaction of AP-3 with vesicles may represent an important regulatory step for large DCV neurotransmitter content.
Results
Chromaffin Cells Express Neuronal AP-3 Subunits.
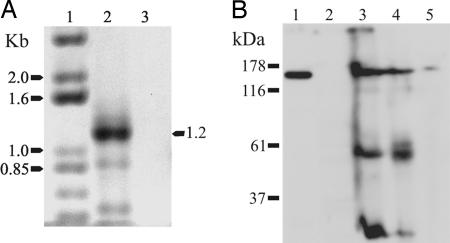
Previous studies have demonstrated the expression of neuronal AP-3 subunits in the CNS (13) without exploring expression in chromaffin cells. Fig. 1 shows that the neuronal β3B subunit of AP-3 is expressed in chromaffin cells at both the levels of mRNA and protein. Bovine chromaffin cells and mouse adrenal glands were used for these experiments because it was not feasible to prepare sufficient numbers of cultured mouse chromaffin cells. RT-PCR with RNA isolated from bovine chromaffin cells yielded a product of the expected size (1.2 kb). This product was absent in the control reaction, which was performed without the addition of reverse transcriptase (Fig. 1A). The 1.2-kb PCR product was cloned and sequenced. The sequence aligned well with that of human β3B (GenBank entry NM_004644; bovine β3B is not available in GenBank), and this region is not present in the ubiquitous β3A subunit. To determine whether β3B protein is expressed in chromaffin cells, immunoblots were carried out with a monoclonal antibody against β3B and protein samples from mouse and bovine brain, mouse adrenal glands, and bovine chromaffin cells. A protein of the appropriate size for β3B (≈160 kDa) was observed in samples from bovine brain (Fig. 1B, lane 1), mouse brain (lane 3), whole mouse adrenal gland (lane 4), and bovine chromaffin cells (lane 5), but not in HEK293 cells (lane 2). These data suggest that neuronal AP-3 subunits are found in chromaffin cells in addition to the CNS.
Fig. 1.
The neuronal β3B subunit of AP-3 is expressed in chromaffin cells. (A) RT-PCR was used to demonstrate the expression of mRNA for the neuronal β3B subunit. Total RNA isolated from bovine chromaffin cells was reverse-transcribed (lane 2), and the negative control reaction was performed without reverse transcription (lane 3). The resulting cDNA was then used for PCR with degenerate primers designed specifically to amplify a region of β3B as described in Materials and Methods. Lane 1 contains DNA size markers. The PCR product obtained with chromaffin-cell cDNA was of the expected size of 1.2 kb, and partial sequencing of the PCR product established that it was 91% identical to human β3B (nucleotides 2734–3126 of GenBank AF022152). (B) Immunoblot of β3B protein. Protein samples from brain and chromaffin cells were separated by SDS/PAGE and immunoblotted with a monoclonal antibody to β3B (610892; BD Biosciences). Lane 1, crude membranes from bovine brain; lane 2, HEK293 cells (negative control); lane 3, whole mouse brain; lane 4, whole mouse adrenal gland; and lane 5, bovine chromaffin cells. All samples except the negative control HEK293 cells displayed an immunoreactive band of β3B of ≈160 kDa. The additional bands in the mouse samples (lanes 3 and 4) are mouse immunoglobulins recognized by the anti-mouse IgG secondary antibody.
AP-3 and DCV Markers Overlap in TGN and Post-Golgi Regions.
Both small-molecule neurotransmitter transporters (25) and peptide cargo precursors are acquired (26, 27) during the budding of large DCVs from the TGN. In mammalian cells, AP-3 is reported to localize in the TGN (11) and immature granules (28). There, AP-3 may sort out cargo by pinching off small transport vesicles from the larger immature secretory vesicles (ISVs) (18, 29), thereby regulating the final DCV size. Using confocal microscopy, we found that AP-3 and the vesicular monoamine transporter 1 (VMAT-1), a marker of DCVs in chromaffin cells (30), had overlapping staining in TGN and post-TGN regions of mouse chromaffin cells as well as around the perimeter of the cell (see Fig. 6, which is published as supporting information on the PNAS web site). Syntaxin 6 (Syn-6) is a target-membrane SNARE reported to reside on membranes of immature granules (31), and it is often used to label post-TGN regions. AP-3 colocalized with Syn-6 and VMAT-1 in what appears to be Golgi–TGN regions, which showed the strongest Syn-6 staining (see Fig. 7, which is published as supporting information on the PNAS web site). A similar staining pattern has been reported for Syn-6 (32). Our data are consistent with the hypothesis that AP-3 may interact with granule markers around the TGN. If our hypothesis is correct, then the maximum overlap of AP-3 with these markers should occur during budding, and it may be transient.
Quantal Secretion Is Reduced with the Overexpression of AP-3.
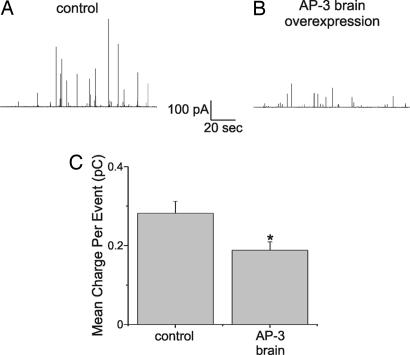
To determine whether AP-3 influences secretion from DCVs, stimulated catecholamine release was assayed with single-cell amperometry 48–72 h after transfection of chromaffin cells derived from C57BL/6J mice. The neuronal-specific AP-3 subunits (referred to as AP-3B: δ, β3Β, μ3Β, and σ3) and green fluorescent protein (GFP) were overexpressed in chromaffin cells. Secretion was initiated by exposing digitonin-permeabilized cells to a fixed, high concentration of Ca2+ (100 μM) (33). The amperometric traces show that a typical control cell (Fig. 2A) exhibited many more large amperometric events than did an AP-3B-overexpressing cell (Fig. 2B), but the total number of events was unaltered. The amount of transmitter detected per individual release event was measured as charge (Q, referred to as quantal size), and it was averaged per cell (34). Fig. 2C shows that the average charge for control cells (0.282 ± 0.03 pC, 21 cells, 1,719 events) was significantly larger (P = 0.017) than that in neuronal AP-3-overexpressing cells (0.188 ± 0.02 pC, 35 cells, 1,516 events). Wild-type cells overexpressing GFP alone had 28 ± 3 amperometric events per 2-min stimulation epoch, and cells overexpressing GFP and AP-3 had an identical 28 ± 3 amperometric events per stimulation epoch.
Fig. 2.
Neuronal AP-3 overexpression results in amperometric events with smaller quantal size. Catecholamine release was elicited by permeabilizing cells for 10 s with 20 μM digitonin followed by a 2-min exposure to 100 μM Ca2+. Single-cell amperometry was used to assay release. (A) Amperometric recording from control (C57BL/6J-derived) chromaffin cells transfected with GFP and empty vector. (B) Amperometric recording under conditions similar to those in A except that cells were transfected with neuronal AP-3 (δ, β3B, μ3B, and σ3 subunits) and GFP. (C) Average charge per release event recorded from control C57BL/6J chromaffin cells (n = 21) and neuronal AP-3-overexpressing cells (n = 35). Error bars represent SEM. The average charge was calculated for each individual cell and then averaged. All events were recorded 48–72 h after transfection. The quantal size recorded in response to digitonin was similar to that obtained in cells stimulated with nicotine (33). Asterisk indicates that the groups are significantly different.
Selective Deletion of the Ubiquitous Form of AP-3 Does Not Alter Secretion Properties.
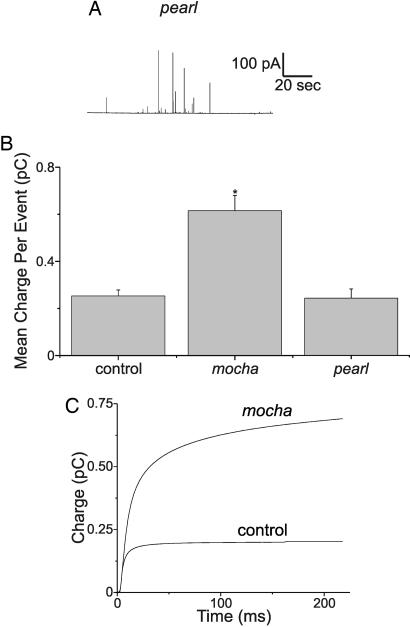
A different approach was taken to probe the possibility that ubiquitous AP-3 participates in neurosecretory vesicle formation. The pearl mouse (Ap3b1pe) carries a spontaneous mutation in the Ap3b1 gene that codes for β3A, which leads to degradation of the ubiquitous AP-3 tetramer (35). Quantal secretion measurements (Fig. 3A) from pearl-derived chromaffin cells showed no difference in mean quantal size compared with wild-type animals on the same genetic background (C57BL/6J) (Fig. 3B, C57BL/6J mice: 14 cells, 1,272 events, mean quantal size 0.275 ± 0.039 pC; Fig. 3C, pearl mice: 20 cells, 1,842 events, mean quantal size 0.242 ± 0.038 pC; P = 0.396). Therefore, the pathway for making neurosecretory vesicles does not appear to be sensitive to perturbations in ubiquitous AP-3. In these experiments, pearl mice had 35 ± 4 amperometric events per stimulation epoch, whereas control cells had 37 ± 6 amperometric events per stimulation epoch.
Fig. 3.
Deletion of neuronal AP-3 results in amperometric events with larger quantal events. (A) amperometric recording from a pearl mouse chromaffin cell. pearl mice lack the β3A subunit of AP-3, and therefore they make no ubiquitous AP-3. (B) Average charge observed for amperometric events from control (grizzled/untyped-derived, see below) chromaffin cells (n = 26), mocha-derived cells (n = 31), and pearl-derived cells (n = 20 cells). mocha mice lack the δ subunit of AP-3, and therefore they make no AP-3 of any kind. Asterisk indicates that the groups are significantly different. (C) Events from individual mocha and control cells were time-aligned and averaged, and then the current integrals were plotted. mocha heterozygote animals produce mocha homozygote, grizzled, and untyped mice when mated. grizzled and untyped chromaffin cells were used as controls for mocha.
Because overexpressing AP-3B decreased quantal size, and manipulation of ubiquitous AP-3 produced no difference, it follows that the absence of AP-3 might give rise to larger quantal events. The mocha mouse (Ap3dmh) has a mutation in the Ap3d gene that codes for the nonredundant δ subunit. The truncated δ subunit produced by the Ap3d mutation is degraded, which results in the loss of all forms of AP-3 (15). Secretion from mocha and appropriate control animals was compared (mocha mice were derived on a grizzled background; grizzled and untyped are appropriate control animals for mocha). The average quantal size for mocha cells (0.60 ± 0.11 pC, 31 cells, 2,691 events) was dramatically larger than that measured for control cells (0.25 ± 0.02 pC, 26 cells, 1,729 events). This dramatic 2.4-fold difference in quantal size was significant (P < 0.029). In these experiments, mocha mice had 36 ± 4 amperometric events per stimulation epoch, whereas grizzled cells had 33 ± 7 amperometric events per stimulation epoch.
Another way to express the difference between mocha and control amperometric events is shown in Fig. 3D, which plots the amperometric current integral as a function of time. For this plot, all of the amperometric events from individual mocha and control cells were aligned in time, averaged, and then plotted as a current integral. The amount of neurotransmitter released from an average secretory event from mocha cells is much larger than that of control cells. Fig. 3D also indicates that mocha cells exhibited a slow component in the release process that was not observed in wild-type cells. A detailed kinetic analysis is not presented because the large amperometric events observed in mocha had significant variability in spike kinetics.
We attempted to “rescue” chromaffin cells from mocha mice by transfecting them with neuronal AP-3 subunits. mocha cells transfected in this way were observed to have a number of very large events, similar to that of untransfected mocha cells. Our experiments by necessity are relatively brief because chromaffin cells remain viable for just a few days after dissociation of the adrenal gland. Thus, we recorded from cells 48–72 h after tissue harvest to ensure cell identity and viability. In this time frame, transfecting mocha cells with neuronal AP-3 subunits produced no significant change in the mean charge per amperometric event. Because the amperometric events recorded in mocha cells can be enormous, they skew the averages, even though the actual number of events is not that large. In retrospect, it is not surprising that the mean charge did not change because the mocha cells did not “turn over” many of these large vesicles in just 2–3 days. Excluding all events >1.5 pC (which represent <8% of total events) showed that the average charge in transfected mocha cells was approximately half that of untransfected cells. This difference was statistically significant (P < 10−5). Thus, the mocha cells transfected with neuronal AP-3 are promoting the formation of an excess of small vesicles. The huge vesicles that were still present 2–3 days after transfection obscured this difference and suggest that AP-3 may act at an earlier step. These data can be found in Fig. 8, which is published as supporting information on the PNAS web site.
Vesicle Size Is Altered in a Manner That Mirrors Quantal Size Changes.
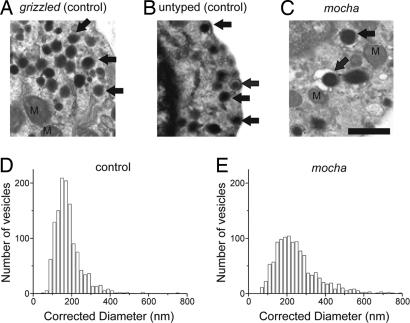
The mocha data in Fig. 3 can be explained as a change in vesicle size, a change in neurotransmitter concentration, or alternatively, more complete emptying of vesicular contents. Previously, we found a strong link between amperometric event size and vesicle size in control C57BL/6J mice under conditions similar to what we have used here (ref. 33 and unpublished observations). A morphological analysis of secretory vesicles in mocha and the appropriate control chromaffin cells was made to address the simplest scenario that vesicle size underlies the change in quantal size. Cells were prepared for electron microscopy under conditions identical to those used for amperometric recordings. Between 48 and 72 h, cells were fixed and flat embedded on glass coverslips for electron microscopy. All DCVs with circular profiles, well preserved membranes, and dense cores were included in vesicle-size analysis (36). The mocha cells (Fig. 4C) exhibited many more large secretory vesicles than did the controls (Fig. 4 A and B). Fig. 4 D and E plots corrected vesicle-size distributions (for an explanation of the correction procedure, see Materials and Methods) for control and mocha animals, respectively (data from grizzled and untyped control strains were pooled; see Fig. 4 legend). Chromaffin cells from mocha animals have many more large-diameter vesicles than control cells [in particular, look at the distribution for vesicle diameters >300 nm, which result in enormous amperometric events; 23.9% of mocha vesicles (n = 274) were >300 nm, whereas only 5.2% of grizzled and untyped vesicles (n = 67) were >300 nm].
Fig. 4.
Deletion of AP-3 results in the formation of large vesicles. (A–C) Electron-microscopic images of grizzled-, untyped-, and mocha-derived chromaffin cells, respectively. All three images are at the same magnification. Arrows indicate typical DCVs used in this analysis. (Scale bar: 500 nm.) (D and E) Vesicle-diameter histograms, after correcting for sectioning artifacts. (D) Data from control (grizzled/untyped) chromaffin cells. (E) Data from mocha cells. Because no difference in diameter was observed between grizzled (175 ± 11 nm, 13 cells) and untyped (186 ± 11 nm, 9 cells), these data were pooled. For this figure, n = 1,267 vesicles were measured in 22 grizzled and untyped chromaffin cells, and n = 1,147 vesicles were measured in 22 mocha cells. Cells from different animals appeared to contain similar numbers of vesicles.
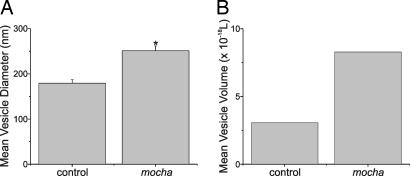
Fig. 5A shows that the average vesicle diameter was 251 ± 11 nm (22 cells, 1,147 vesicles) and 179 ± 8 nm (22 cells, 1,267 vesicles) for mocha vs. control, respectively (P < 0.00001). Expressing the vesicle-size difference as a change in volume puts the mocha average vesicle volume at ≈2.8-fold greater than control values (see Fig. 5B). The magnitude of the volume difference is strikingly similar to the difference in quantal size (2.4-fold change), which suggests that the increase in quantal size is the result of increased vesicle size.
Fig. 5.
Deletion of AP-3 results in a large increase in vesicle volume. (A) Mean vesicle diameter for control (grizzled/untyped-derived) chromaffin cells and for mocha-derived cells. Asterisk indicates that the groups are significantly different. (B) Mean vesicle volume for control (grizzled/untyped-derived) chromaffin cells and for mocha-derived cells. There was an ≈2.8-fold increase in volume.
Discussion
mocha mice have a null mutation in the δ subunit of AP-3, such that they make no AP-3 at all. The mocha mice have a severe neurological phenotype (15). pearl mice have an effectively null mutation in the β3A subunit of AP-3 (2); these mice presumably make neuronal AP-3 but are deficient in ubiquitous AP-3. Although these mice are sick, they have no neurological phenotype, other than night blindness. Nakatsu and Ohno established a μ3B-deficient mouse, which shows a neurological phenotype (1) and defects in synaptic transmission (9). These mutant studies strongly suggest that only neuronal AP-3 is involved in producing the observed neurological deficits and that AP-3 may play a role in normal neurotransmission. In synaptic terminals, AP-3 may influence synaptic-vesicle formation from recycling endosomes, as has been suggested by Blumstein et al. (5). In this study, we found that overexpression of neuronal AP-3 reduced quantal size in wild-type cells. Deletion of AP-3 in mocha-derived cells produced a dramatic increase in quantal size. These results suggest that total expression levels of AP-3 influence secretion. Because selective ablation of ubiquitous AP-3 was without effect, the results suggest that the neuronal form of AP-3 is particularly important for vesicle formation. The increase in quantal size was correlated with vesicle size; the alterations in quantal and vesicle sizes were strikingly similar, 2.4- and 2.8-fold, respectively. Although it is possible that a fraction of the neurotransmitter content remained inside the vesicle, the strong correlation between vesicle size and quantal size suggests that the vesicle neurotransmitter content was completely emptied under the stimulation conditions used in this study. These findings are similar to previous studies performed in different cell types that have reported that vesicle size has a strong influence on quantal size (33, 37–39). Our studies are consistent with AP-3 participating in neurosecretory vesicle formation and secretion.
The conventional view of DCV formation assumes that as the ISV matures, small coated transport vesicles are constantly being removed, which reduces the ISV volume (40). AP-3 has been reported to coat the TGN [in these studies, the β3B subunit was probed (11, 41)] and traffic cargo from this structure (42, 43). In addition, both the neuronal and ubiquitous forms of AP-3 are recruited to ISVs in a regulated fashion (28). Hence, the results reported here may arise from the involvement of AP-3 in de novo granule maturation. The absence of AP-3, preventing removal of small cargo vesicles, would explain the larger vesicles found in mocha cells. Under these conditions, the ISVs may become arrested at a larger size and possibly in a less mature state. In addition, it is known that vesicular transporters are sorted during DCV maturation (44–46); without proper sorting, vesicle size and quantal size may be altered by neurotransmitter transporter activity, as has been suggested in other systems (47–49).
The effects seen with overexpression of neuronal AP-3 can be explained as promoting maturation of granules, possibly by accelerating cargo vesicle removal. Because vesicle size and quantal size are highly correlated, a surplus of smaller-sized vesicles may be formed in the presence of excess neuronal AP-3. Chromaffin cells, thus, may have a single population of mature secretory vesicles, possibly the small vesicles, and a separate population of ISVs, possibly the large ones similar to what Tooze (18) proposed for PC12 cells. Under the intense stimulation conditions used here, recruitment for secretion is likely to bypass subtle differences in fusogenicity that may dominate under more physiological conditions; therefore, it is possible that the release of a broad spectrum of vesicles is induced with the intense stimulation used here. An entirely different explanation is that the cells have two separate populations of vesicles, as has been proposed for chromaffin cells (19, 33) and PC12 cells (50). It is possible that AP-3 promotes the formation of smaller vesicles because overexpression of AP-3 increases the frequency of small events. The shift in the vesicle-size distribution to a higher fraction of large vesicles when all AP-3 is removed would result from fewer small vesicles being made in the absence of AP-3. A final possibility that may impact quantal size has been described by Cochilla et al. (51), who showed that pituitary lactotrophs undergo homotypic fusion, either immediately before or during exocytosis. This mechanism may alter quantal size. However, there is little evidence for homotypic fusion in chromaffin cells at this late stage of release (52). Whether chromaffin granules aggregate during recycling or during de novo vesicle synthesis was not addressed in this study, but we did not observe multiple granules in single membrane-bound vesicles in our electron-microscopic studies.
Materials and Methods
Cell Culture.
Animals were purchased from The Jackson Laboratory, and they were housed and handled as required by the Animal Resource Council (University of Chicago). Chromaffin cells were prepared similar to methods published in ref. 33. Cells were plated on glass coverslips coated with Matrigel (Discovery Labware, Bedford, MA) and maintained in a 37°C, 5% CO2 incubator. Electron microscopy and confocal and amperometry experiments were carried out 2 and 3 days after tissue harvest. Chromaffin cells were transfected with plasmids designed to express AP-3 subunits [a kind gift from Margaret Robinson (University of Cambridge, Cambridge, U.K.) using Lipofectamine 2000 (Invitrogen)]. Cells were transfected 2 h after plating, and the transfection mix was left on the cells for 16 h.
RT-PCR and Immunoblotting.
RT-PCR was used to probe for expression of β3B mRNA. Using TRIzol (Invitrogen), we isolated total bovine chromaffin cell RNA, and we performed the reverse-transcription step with total RNA, random hexamers, and the SuperScript II first-strand synthesis system (Invitrogen). Because the sequence for bovine β3B has not been reported, degenerate PCR primers were designed from amino acid sequences of β3B that are identical in human and mouse but that differed in three or four amino acids from the most closely related gene, β3A. The selected sequences are: NH2-PDWPEEA…DEYRFAG-COOH for human β3B (amino acids 642-1036 of AAB71894); NH2-PDWPEEA…DEYRFAG-COOH for mouse β3B (amino acids 35–430 of BAA92765); and NH2-SNWPEVA…NIHRFAA-COOH for human β3A (amino acids 637-1047 of NP_003655). Additional details can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
For immunoblotting, we separated proteins by SDS/PAGE and transferred them to Immobilon-P membranes (Millipore) using standard procedures (53). A monoclonal antibody against β3B (610892; BD Biosciences, Pharmingen) was used at a dilution of 1:500, followed by an anti-mouse IgG–horseradish peroxidase-coupled secondary antibody (Jackson ImmunoResearch) used at a dilution of 1:100,000. Bound antibody was detected with the ECL Advance system from Amersham Biosciences.
Amperometry.
Carbon-fiber electrodes were fabricated with 7-μm-diameter carbon fibers (Fortafil Fibers, Knoxville, TN), as described in ref. 33. The electrode was pressed gently against the cell during the recording. A newly cut surface or a new electrode was used for each cell. Recordings were made with an EPC-7 amplifier (HEKA Electronics, Lambrecht/Pfalz, Germany). The amperometric signal was low-pass filtered at 2 kHz (8-pole Bessel; Warner Instruments, Hamden, CT) and sampled into a computer at 10 kHz by using a 16-bit analog-to-digital converter (National Instruments, Austin, TX). Records with rms noise >2 pA were not analyzed. Amperometric spike features, quantal size, and kinetic parameters were analyzed with the mini analysis program (Synaptosoft, Decatur, GA). The detection threshold for an event was set at 4–5 times the baseline rms noise, and the spikes were automatically detected. Overlapping events were relatively rare, and they were rejected. The area under individual amperometric spikes is equal to the charge (pC) per release event, referred to as Q. The number of oxidized neurotransmitter molecules (N) was calculated by using the Faraday equation, N = Q/ne, with n = 2 electrons per oxidized molecule of transmitter; e is the elemental charge (1.603 × 10−19 C).
Recording Solutions and Stimulation Protocols.
Amperometric recordings were made from adherent cells that were under constant perfusion (flow rate of ≈1.0 ml/min; chamber volume, ≈150 μl). All recording solutions had the following composition: 145 mM NaCl, 2.0 mM KCl, 10 mM Hepes, and 1.0 mM MgCl2. Ca2+-free solutions contained 100 μM EGTA. All solutions used during and after cell permeabilization contained 1.0 mM Na2ATP. All experiments were performed at ambient temperature (23° ± 2°C). Cells were stimulated repeatedly by using the following protocol: (i) bathing for 2 min in a Ca2+-free solution, (ii) permeabilizing with 20 μM digitonin (Ca2+-free) for 10 s, (iii) stimulating for 2 min with a solution containing 100 μM free Ca2+, and (iv) washing for 1 min in Ca2+-free medium before repeating the cycle starting at step ii. Cells were typically stimulated for three or four cycles (at most, six cycles) or until the cell membrane changed from its initial, bright-field dark appearance to a granular texture (for details, see ref. 54). Digitonin was purchased from Calbiochem.
Electron Microscopy.
Coverslips were rinsed twice with 0.12 M cacodylate buffer (pH 7.4) to remove cell culture medium, and then they were fixed with 2% glutaraldehyde and 0.6% paraformaldehyde in 0.06 M cacodylate buffer (pH 7.4) for 1 h. The coverslips were again rinsed three times and then postfixed for 1 h in a mixture of 1% osmium tetroxide in 0.06 M cacodylate buffer (pH 7.4) containing 1.5% potassium ferricyanide that was added immediately before use. The samples were washed three times, dehydrated in a graded series of ethanol [50–100% (vol/vol)], with the 70% step containing 1% uranyl acetate (30 min), and finally they were dehydrated in propylene oxide. The samples were infiltrated with Durcupan (Fluka), and the coverslips were inverted over 15-mm round-well molds with the tissue side down, and they were polymerized for 48 h at 60°C. After polymerization, the resin plug with the coverslip was suspended in hydrofluoric acid for 10 min to dissolve the glass. Areas of interest were cut out with a jewelers’ saw, attached with cyanoacrylate to a blank BEEM capsule stub (Electron Microscopy Sciences, Fort Washington, PA), and sectioned parallel to the plane of the coverslip with a diamond knife. Sections with a silver interference color were collected on 1- × 2-mm Formvar-coated slot grids (Electron Microscopy Sciences), counterstained with uranyl acetate and lead citrate. The sections were viewed in a JEOL 1220 electron microscope at 80 kV, and images were taken on electron-micrography film (Eastman Kodak) (magnification: ×12,000).
Vesicle-Size Measurements.
Digital images of electron micrographs were made and analyzed at a final magnification of ×100,000 or ×200,000. Random, single sections from cells that possessed multiple DCVs and well fixed cellular constituents (mitochondria, nuclear material, and plasma membrane) were selected for analysis. The criteria for DCVs that were included in the size distribution are as follows: (i) the vesicle had to have an electron-dense core, (ii) an intact, continuous vesicle membrane had to surround the dense core, and (iii) the membrane had to be approximately circular in geometry. All vesicles meeting these criteria were included in the analysis. The area of each vesicle was measured with imagej software (National Institutes of Health) or sigmascan software (SPSS, Chicago) (as described in ref. 33). The two programs were compared and found to yield results that were essentially identical. The diameter was calculated from vesicle area [diameter = 2 × (area/π)0.5]. The corrected vesicle diameter was calculated by using an algorithm published in ref. 55, which we have verified with serial reconstructions. This algorithm depends on section thickness and the empirically measured vesicle diameter (referred to as apparent diameter). The section thickness was estimated to be 60 nm, based on the silver interference color of the sections (33). Apparent vesicle diameters were transformed to corrected diameters before statistical or graphical analysis of the data.
For these studies, vesicle circularity was measured, with 1 being the highest value possible and representing a perfect circle. The circularity averaged 0.94 ± 0.001 and 0.96 ± 0.001 for the gr/gr and mh/mh groups, respectively, and every vesicle included in our analysis had a circularity >0.85 (untyped was 0.96 ± 0.001). Hence, no obvious difference in vesicle shape is evident, only the overall size changes. Measurements were made with imagej, which calculates circularity as 4 × π × [area/(perimeter2)].
Statistical analyses between experimental groups are presented as the mean ± standard error of the mean, and two-tailed P values were made by using the Mann–Whitney test for unpaired, nonparametric data (GraphPad, San Diego). All plots were performed in origin (OriginLab, Northampton, MA).
Supplementary Material
Acknowledgments
We thank Dr. Enrico Mugnaini (Northwestern University, Chicago) for recommendations on preparing electron-microscopic samples. This work was supported by a grant from the National Institute of Neurological Disorders and Stroke, National Institutes of Health (to A.P.F.), by Philip Morris USA, and by a Philip Morris International grant (to A.P.F.). C.P.G. and A.L. were also supported by the National Institutes of Health.
Abbreviations
- AP
adaptor protein
- DCV
dense-core vesicle
- ISV
immature secretory vesicle
- Syn-6
syntaxin 6
- TGN
trans-Golgi network
- VMAT-1
vesicular monoamine transporter 1.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nakatsu F., Ohno H. Cell Struct. Funct. 2003;28:419–429. doi: 10.1247/csf.28.419. [DOI] [PubMed] [Google Scholar]
- 2.Peden A. A., Rudge R. E., Lui W. W., Robinson M. S. J. Cell Biol. 2002;156:327–336. doi: 10.1083/jcb.200107140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson M. S. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Boehm M., Bonifacino J. S. Gene. 2002;286:175–186. doi: 10.1016/s0378-1119(02)00422-5. [DOI] [PubMed] [Google Scholar]
- 5.Blumstein J., Faundez V., Nakatsu F., Saito T., Ohno H., Kelly R. B. J. Neurosci. 2001;21:8034–8042. doi: 10.1523/JNEUROSCI.21-20-08034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson M. S., Bonifacino J. S. Curr. Opin. Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 7.Kyttala A., Yliannala K., Schu P., Jalanko A., Luzio J. P. J. Biol. Chem. 2005;280:10277–10283. doi: 10.1074/jbc.M411862200. [DOI] [PubMed] [Google Scholar]
- 8.Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakatsu F., Okada M., Mori F., Kumazawa N., Iwasa H., Zhu G., Kasagi Y., Kamiya H., Harada A., Nishimura K., et al. J. Cell Biol. 2004;167:293–302. doi: 10.1083/jcb.200405032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell’Angelica E. C., Ooi C. E., Bonifacino J. S. J. Biol. Chem. 1997;272:15078–15084. doi: 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- 11.Simpson F., Peden A. A., Christopoulou L., Robinson M. S. J. Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pevsner J., Volknandt W., Wong B. R., Scheller R. H. Gene. 1994;146:279–283. doi: 10.1016/0378-1119(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 13.Newman L. S., McKeever M. O., Okano H. J., Darnell R. B. Cell. 1995;82:773–783. doi: 10.1016/0092-8674(95)90474-3. [DOI] [PubMed] [Google Scholar]
- 14.Swank R. T., Reddington M., Howlett O., Novak E. K. Blood. 1991;78:2036–2044. [PubMed] [Google Scholar]
- 15.Kantheti P., Qiao X., Diaz M. E., Peden A. A., Meyer G. E., Carskadon S. L., Kapfhamer D., Sufalko D., Robinson M. S., Noebels J. L., Burmeister M. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- 16.Feng L., Seymour A. B., Jiang S., To A., Peden A. A., Novak E. K., Zhen L., Rusiniak M. E., Eicher E. M., Robinson M. S., et al. Hum. Mol. Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- 17.Seong E., Wainer B. H., Hughes E. D., Saunders T. L., Burmeister M., Faundez V. Mol. Biol. Cell. 2005;16:128–140. doi: 10.1091/mbc.E04-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tooze S. A. FEBS Lett. 1991;285:220–224. doi: 10.1016/0014-5793(91)80805-d. [DOI] [PubMed] [Google Scholar]
- 19.Duncan R. R., Greaves J., Wiegand U. K., Matskevich I., Bodammer G., Apps D. K., Shipston M. J., Chow R. H. Nature. 2003;422:176–180. doi: 10.1038/nature01389. [DOI] [PubMed] [Google Scholar]
- 20.Solimena M., Gerdes H. H. Trends Cell Biol. 2003;13:399–402. doi: 10.1016/s0962-8924(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 21.Elhamdani A., Palfrey H. C., Artalejo C. R. Neuron. 2001;31:819–830. doi: 10.1016/s0896-6273(01)00418-4. [DOI] [PubMed] [Google Scholar]
- 22.Albillos A., Dernick G., Horstmann H., Almers W., Alvarez de Toledo G., Lindau M. Nature. 1997;389:509–512. doi: 10.1038/39081. [DOI] [PubMed] [Google Scholar]
- 23.Taraska J. W., Perrais D., Ohara-Imaizumi M., Nagamatsu S., Almers W. Proc. Natl. Acad. Sci. USA. 2003;100:2070–2075. doi: 10.1073/pnas.0337526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G., Harata N. C., Tsien R. W. Proc. Natl. Acad. Sci. USA. 2004;101:1063–1068. doi: 10.1073/pnas.0307149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimer R. J., Fon E. A., Edwards R. H. Curr. Opin. Neurobiol. 1998;8:405–412. doi: 10.1016/s0959-4388(98)80068-8. [DOI] [PubMed] [Google Scholar]
- 26.Farquhar M. G., Palade G. E. J. Cell Biol. 1981;91:77S–103S. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meldolesi J., Chieregatti E., Luisa Malosio M. Trends Cell Biol. 2004;14:13–19. doi: 10.1016/j.tcb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Austin C., Boehm M., Tooze S. A. Biochemistry. 2002;41:4669–4677. doi: 10.1021/bi016064j. [DOI] [PubMed] [Google Scholar]
- 29.Tooze S. A., Martens G. J., Huttner W. B. Trends Cell Biol. 2001;11:116–122. doi: 10.1016/s0962-8924(00)01907-3. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Edwards R. H. J. Cell Biol. 1997;139:907–916. doi: 10.1083/jcb.139.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klumperman J., Kuliawat R., Griffith J. M., Geuze H. J., Arvan P. J. Cell Biol. 1998;141:359–371. doi: 10.1083/jcb.141.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer R. A., Overlease R. L., Lieber J. L., Angleson J. K. J. Cell Sci. 2004;117:2193–2202. doi: 10.1242/jcs.01093. [DOI] [PubMed] [Google Scholar]
- 33.Grabner C. P., Price S. D., Lysakowski A., Fox A. P. J. Neurophysiol. 2005;94:2093–2104. doi: 10.1152/jn.00316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colliver T. L., Hess E. J., Pothos E. N., Sulzer D., Ewing A. G. J. Neurochem. 2000;74:1086–1097. doi: 10.1046/j.1471-4159.2000.741086.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhen L., Jiang S., Feng L., Bright N. A., Peden A. A., Seymour A. B., Novak E. K., Elliott R., Gorin M. B., Robinson M. S., Swank R. T. Blood. 1999;94:146–155. [PubMed] [Google Scholar]
- 36.Thureson-Klein A., Harless S., Klein R. Cell Tissue Res. 1984;236:53–65. doi: 10.1007/BF00216513. [DOI] [PubMed] [Google Scholar]
- 37.Bruns D., Riedel D., Klingauf J., Jahn R. Neuron. 2000;28:205–220. doi: 10.1016/s0896-6273(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 38.Ales E., Tabares L., Poyato J. M., Valero V., Lindau M., Alvarez de Toledo G. Nat. Cell Biol. 1999;1:40–44. doi: 10.1038/9012. [DOI] [PubMed] [Google Scholar]
- 39.Chen G., Gavin P. F., Luo G., Ewing A. G. J. Neurosci. 1995;15:7747–7755. doi: 10.1523/JNEUROSCI.15-11-07747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez de Toledo G., Fernandez J. M. J. Cell Biol. 1990;110:1033–1039. doi: 10.1083/jcb.110.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson F., Bright N. A., West M. A., Newman L. S., Darnell R. B., Robinson M. S. J. Cell Biol. 1996;133:749–760. doi: 10.1083/jcb.133.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ihrke G., Kyttala A., Russell M. R., Rous B. A., Luzio J. P. Traffic. 2004;5:946–962. doi: 10.1111/j.1600-0854.2004.00236.x. [DOI] [PubMed] [Google Scholar]
- 43.Rehling P., Darsow T., Katzmann D. J., Emr S. D. Nat. Cell Biol. 1999;1:346–353. doi: 10.1038/14037. [DOI] [PubMed] [Google Scholar]
- 44.Krantz D. E., Peter D., Liu Y., Edwards R. H. J. Biol. Chem. 1997;272:6752–6759. doi: 10.1074/jbc.272.10.6752. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Krantz D. E., Waites C., Edwards R. H. Trends Cell Biol. 1999;9:356–363. doi: 10.1016/s0962-8924(99)01605-0. [DOI] [PubMed] [Google Scholar]
- 46.Krantz D. E., Waites C., Oorschot V., Liu Y., Wilson R. I., Tan P. K., Klumperman J., Edwards R. H. J. Cell Biol. 2000;149:379–396. doi: 10.1083/jcb.149.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pothos E. N., Larsen K. E., Krantz D. E., Liu Y., Haycock J. W., Setlik W., Gershon M. D., Edwards R. H., Sulzer D. J. Neurosci. 2000;20:7297–7306. doi: 10.1523/JNEUROSCI.20-19-07297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong L. W., Hafez I., Alvarez de Toledo G., Lindau M. J. Neurosci. 2003;23:7917–7921. doi: 10.1523/JNEUROSCI.23-21-07917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniels R. W., Collins C. A., Gelfand M. V., Dant J., Brooks E. S., Krantz D. E., DiAntonio A. J. Neurosci. 2004;24:10466–10474. doi: 10.1523/JNEUROSCI.3001-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauerfeind R., Regnier-Vigouroux A., Flatmark T., Huttner W. B. Neuron. 1993;11:105–121. doi: 10.1016/0896-6273(93)90275-v. [DOI] [PubMed] [Google Scholar]
- 51.Cochilla A. J., Angleson J. K., Betz W. J. J. Cell Biol. 2000;150:839–848. doi: 10.1083/jcb.150.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plattner H., Artalejo A. R., Neher E. J. Cell Biol. 1997;139:1709–1717. doi: 10.1083/jcb.139.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z., Currie K. P., Cahill A. L., Fox A. P. J. Neurophysiol. 2003;90:3828–3837. doi: 10.1152/jn.00617.2003. [DOI] [PubMed] [Google Scholar]
- 54.Jankowski J. A., Schroeder T. J., Ciolkowski E. L., Wightman R. M. J. Biol. Chem. 1993;268:14694–14700. [PubMed] [Google Scholar]
- 55.Parsons T. D., Coorssen J. R., Horstmann H., Almers W. Neuron. 1995;15:1085–1096. doi: 10.1016/0896-6273(95)90097-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.