Abstract
Ciliary neurotrophic factor (CNTF) receptor controls a pathway supporting the differentiation and survival of a wide range of neural cell types during development and in adulthood. Cardiotrophin-like cytokine (CLC)–cytokine-like factor 1 (CLF) composite cytokine is a second ligand for the CNTF α-component receptor (CNTFRα). This composite cytokine is built on the structural model of IL-12, with a complex formed by a four-helix bundle type I cytokine, CLC (also referred to as CLCF1), bound to a soluble receptor subunit, CLF (also known as CRLF1). We have reported mutations in the chaperone soluble receptor CLF, causing cold-induced sweating syndrome (CISS). In this study, we studied the CLC-mutated alleles in a patient suffering from a similar disease. This patient was compound heterozygous for two different CLC mutations. The first allele was inactivated by a stop codon at position 107 (Y107X). In the second allele, a R197L mutation in the CLC-predicted binding site to the CNTFRα was detected. Functional analysis of the mutated protein revealed an incapacity for R197L CLC to bind to CNTFRα and activate the subsequent signaling events. Structural and docking interaction studies showed that the R197L substitution destabilized the contact site between CLC and CNTFRα.
Keywords: gene inactivation
The ciliary neurotrophic factor (CNTF) receptor pathway is essential for the development and maintenance of motor neurons during embryonic life and adulthood (1). The CNTF binding chain, CNTF α-component receptor (CNTFRα), is expressed early during neuronal precursor development in both neuroepithelium and neural-crest-derived progenitors (2). In adults, CNTF has been shown to attenuate motor deficits found in different strains of mice developing neuromuscular deficiencies (3, 4). Beside its activities in the nervous system, CNTF also displays trophic effects on skeletal muscle and is a regulator of muscular strength during aging (5, 6).
To display its activities, CNTF uses a tripartite membrane receptor and interacts first with CNTFRα through a site 1 before recruiting gp130 signal-transducing and the leukemia inhibitory receptor (LIFR) through sites 2 and 3, respectively (7, 8). CNTF binding also leads to a gp130–LIFR dimerization and downstream signaling events involving an activation of the STAT3 signaling pathway (9).
Newborn mice that are deficient in CNTFRα fail to initiate feeding, die shortly after birth, and show a dramatic loss of motor neurons (10). In contrast, mice homozygous for null mutations in the CNTF gene display only a mild loss of motor neurons, leading to minor muscle weakness with age (11). Of humans, ≈2% are homozygous for a mutation inactivating the CNTF gene (12). Individuals lacking CNTF are healthy and do not develop neurological abnormalities. The striking difference between the effects of CNTF and CNTFRα gene inactivation suggested the existence of a second CNTFRα ligand that is important for neural cell survival during development. We identified neuropoietin (NP) and CLC as two additional ligands for the CNTF tripartite receptor (13, 14). NP is expressed concomitantly and colocalizes with CNTFRα during the mouse development. However, an 8-nt deletion inactivates human NP, indicating that NP has evolved into a pseudogene in our species (13).
CLC, also known as CLCF1, NNT-1, or BSF-3, is a cytokine identified based on its homology with the IL-6 family (15–17). CLC supports the survival of developing motor neurons in vitro, and CLC transcripts are present in skeletal muscle during the period of massive motor neuron cell death (14, 18). Although CLC contains a putative signal peptide, association with the soluble cytokine receptors cytokine-like factor (CLF) 1 (also known as CRLF1) is required for efficient secretion (14, 19, 20). Therefore, it belongs to a growing family of composite cytokines, which also comprises IL-12, IL-23, and IL-27 (21).
Mice lacking the CLF gene are unable to suckle and die from starvation shortly after birth (22). A detailed phenotypic analysis of CLF-deficient mice revealed a motor neuron loss (18). The phenotype was similar to that observed in CNTFRα gene-inactivated mice, indicating that CLF is important for neuronal development (10, 22). In ref. 23, mutations in the CLF gene causing cold-induced sweating syndrome (CISS) are reported in humans. CISS was described by Sohar et al. (24) in two Israeli sisters. These patients had CLF missense mutations and responded to low environmental temperatures with profuse sweating on large segments on their back and chest. They display additional abnormalities, including a high-arched palate, nasal voice, depressed nasal bridge, and inability to fully extend their elbows. However, patients with CLF stop mutations showed a more severe phenotype, including a progressive kyphoscoliosis, some defects in pain and temperature sensitivity, and a severe lack of appetite from birth (23).
In this article, we report the functional consequences of mutations in the gene coding for CLC in a patient with a CISS phenotype. Whereas a number of pathological situations have been attributed to mutations in type I cytokine receptors, no deleterious mutations have been yet identified in the corresponding cytokine genes. This absence of yet reported inactivated ligand in pathology is likely to reflect a large functional redundancy within families of cytokines. The phenotype of this CISS patient is direct evidence for a nonredundant function for a type I cytokine in human development, whose inactivation leads directly to a disease phenotype.
Results
Clinical Feature of the CISS Patient.
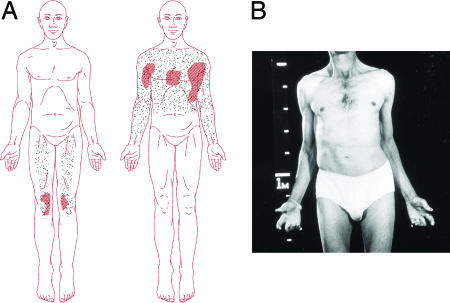
An Australian man was first examined when he was 46 years of age. He gave a history of feeding difficulties as an infant and of having suffered all his life from profuse sweating on the face, trunk, and upper limbs when cold but was unable to sweat in hot weather. Thermoregulatory sweat tests confirmed this paradoxical sweating response (Fig. 1A). Detailed autonomic studies revealed no other abnormalities. There was no family history of the condition and no parental consanguinity. His sister, four children, and grandchildren are unaffected. He displayed congenital physical abnormalities which included mild facial weakness, ears set at right angles to the skull, high arched palate, valgus deformity of the elbows and inability to extend them fully, clinodactyly of the fingers and toes, syndactyly of second and third toes of both feet, thoracolumbar scoliosis and lumbar lordosis, and degenerative disease of the cervical and lumbar spines (Fig. 1B). There was clinical and electrophysiological evidence of a mild sensorimotor peripheral neuropathy. A computed tomography scan and MRI of the brain were normal. He was reviewed neurologically at 74 years of age. He had had a surgical fusion of the lumbar spine, bilateral knee replacements, and nephrectomy for renal carcinoma. He was able to stand and walk, but he used an electric wheelchair when outdoors. There had been no progression of his congenital abnormalities. Nerve conduction studies were repeated and confirmed the presence of a mild, predominantly sensory, neuropathy.
Fig. 1.
Induced sweating test and patient description. (A) Induced sweating test. When the sublingual temperature was raised from 36.3°C to 36.9°C, mild sweating occurred only on his knees and thighs. (Left) The patient was distressed and tachypnoeic. (Right) On lowering of the sublingual temperature to 35.5°C, profuse sweating occurred on the upper trunk and arms accompanied by piloerection. (B) Patient at 46 years of age. Note the cubitus valgus and contractures of elbows, flexion contractures of fingers, and thoracolumbar scoliosis.
Identification of Mutations in Both Alleles of the CLC Gene.
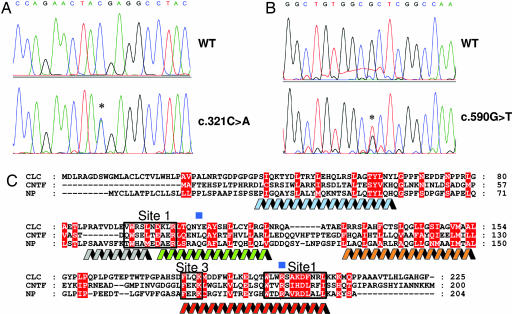
By using DNA sequencing, we analyzed the genomic DNA of the patient for the presence of CLF mutations, but no abnormalities could be detected in this gene. We next undertook a screening for mutations in the CLC gene and two different sequence variants were identified (Fig. 2 A and B). The c.590G→T mutation affects codon 197, leading to the substitution of an Arg by a Leu (R197L). The R197L mutation is located at the C-terminal part of the D-helix of CLC, a region contributing to the contact site 1 between CNTFRα with its ligands (Fig. 2C). The c.321C→A mutation predicts a stop in codon 107 (Y107X) leading to a truncated cytokine (ΔCLC) lacking its contact site 1 with CNTFRα but unaffected in the contact site 3 for CLF (25). No disease related mutations were found in additional genes involved in the CNTF receptor pathway (CNTF, LIF, CNTFRα, LIFR, and gp130). R197L, Y107X mutations were searched in the coding areas of CLC gene in 140 chromosomes from normal individuals. We failed to detect these mutations in the tested samples, indicating that the prevalence of CLC variants is low in the human population.
Fig. 2.
Identification of mutations in the CLC gene. (A) Chromatogram representing the stop mutation identified in codon 107. (B) Chromatogram representing the mutation detected in codon 197. (C) Multiple sequence alignment of CLC with CNTF and NP. Identical or similar residues are highlighted in red. Residues that are implicated in the cytokine site 1 and site 3 are boxed in black and gray, respectively. The locations of the αA-helix (blue), αA′-helix (gray), αB-helix (green), αC-helix (orange), and αD-helix (red) are indicated. Blue squares show the positions of the mutated residues in the patient.
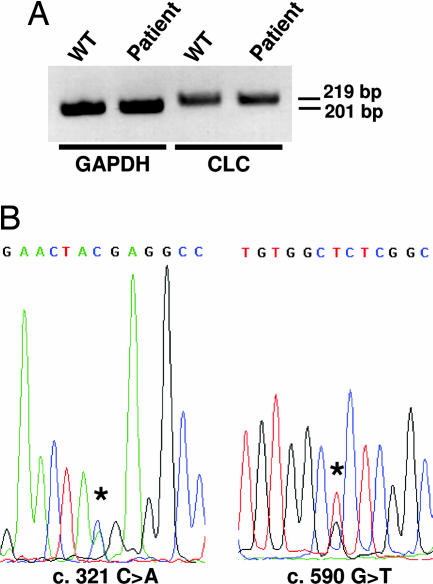
We then assessed whether the patient’s cells were able to synthesize the mutated forms of CLC. Because no Ab allowing a sensitive detection of the corresponding protein is available, the experiments were carried out at the transcript level, and peripheral blood cells were used as a source of CLC transcript (15). RT-PCR analyses show that the patient’s cells expressed a normal level of CLC transcript (Fig. 3A). Also, DNA sequencing of the amplified product indicated that both alleles were expressed in the patient’s blood cells (Fig. 3B).
Fig. 3.
Transcription of both CLC alleles in the patient blood cells. (A) CLC expression was analyzed by RT-PCR in peripheral blood cells. GAPDH transcripts were used as controls. (B) cDNA of patient CLC was sequenced. (Left) Coexpression of both alleles encoding either a stop and a WT codon at cDNA position 321. (Right) Coexpression of both WT and 590 G>T alleles.
Production and Analysis of the Proteins Encoded by the Two Mutated CLC Alleles.
To analyze the contribution of the identified mutations to the pathology, they were introduced into CLC WT cDNA by site-directed mutagenesis, and the mutated forms of the cytokine were analyzed for expression and biological activity.
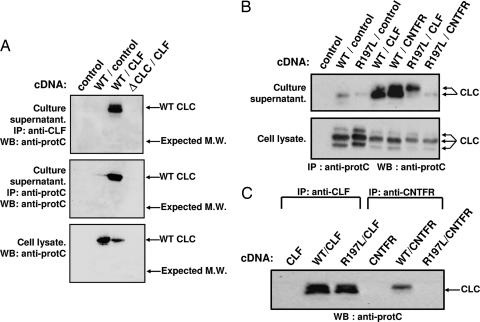
A CLC cDNA interrupted at position 107 (ΔCLC) was generated and coexpressed with CLF in transfected Cos-7 mammalian cells. Culture supernatants and cell lysates were analyzed for the presence of the putative truncated ΔCLC–CLF composite cytokine (Fig. 4A). ΔCLC–CLF was enriched by immunoprecipitation using mAbs directed against CLF or the protein C epitope tag fused to the different CLC forms. The purified fractions were analyzed by Western blotting. Whereas synthesis and secretion of WT CLC were clearly observed, we failed to detect any ΔCLC protein in culture supernatants or Cos-7 cell lysates (Fig. 4A). This result indicates that truncated CLC containing a stop codon at position 107 could not be correctly processed and expressed in mammalian cells.
Fig. 4.
Secretion and interaction of WT and mutant CLC proteins with soluble CNTFRα and CLF. Cos-7 cells were transfected with cDNAs encoding WT or mutated forms of proteins. Immunoprecipitations were performed on cell lysates or culture supernatants by using anti-CNTFRα or anti-CLF mAbs, or an anti-protein C epitope fused to CLC or ΔCLC, and proteins were analyzed by Western blotting (WB). WT and mutant CLC were detected by using a biotinylated anti-protein C tag Ab. (A) Absence of detectable expression of the truncated form of CLC (ΔCLC). WT and ΔCLC were expressed with CLF, and proteins were detected by immunoprecipitation and Western blot analysis. No protein band could be observed at the position corresponding to the expected ΔCLC molecular weight (indicated by arrows). (B) Whereas both WT and R197L CLC form secreted complexes with CLF, only WT CLC is released when coexpressed with soluble CNTFRα. The complexes were immunoprecipitated (IP) from cell culture supernatants or cell lysates, as indicated. Slight variations in CLC molecular weight represent different glycosylation states of the protein. (C) Interaction of R197L CLC and WT CLC with CLF and soluble CNTFRα were compared. Transfected cell culture supernatants were immunoprecipitated with the indicated Abs before CLC detection using an anti-protein C tag Ab.
We next generated an epitope tagged R197L CLC mutant corresponding to the protein encoded by the second allele of the patient. Besides CLF, CLC could also associate to soluble forms of CNTFRα to generate a second secreted composite cytokine capable of recruiting the LIFR and gp130 receptor components (20). Therefore, we expressed the R197L mutant either alone or in combination with CLF or soluble CNTFRα, and we analyzed cytokine production by Western blotting (Fig. 4B). As reported (20, 25), CLC was released in the cell culture medium when coexpressed with CLF or soluble CNTFRα. Interestingly, R197L mutated CLC was secreted only when coexpressed with CLF (Fig. 4B). In the presence of soluble CNTFRα, R197L CLC remained trapped within the cells (Fig. 4 B and C). This result corroborates with the structural localization of residue 197 in the site 1 region of CLC contacting CNTFRα, whereas CLC binding to CLF (involving site 3) was not affected (25). To test whether the R197L mutant and CLF were released as a stable composite cytokine, transfectant culture media were immunoprecipitated with an Ab directed against CLF. The purified fraction was subjected to Western blot analysis using Ab directed against the CLC epitope tag. Results indicated the formation of a stable secreted R197L CLC–CLF complex (Fig. 4C).
The R197L CLC of the Patient Does Not Bind to or Activate CNTFRα.
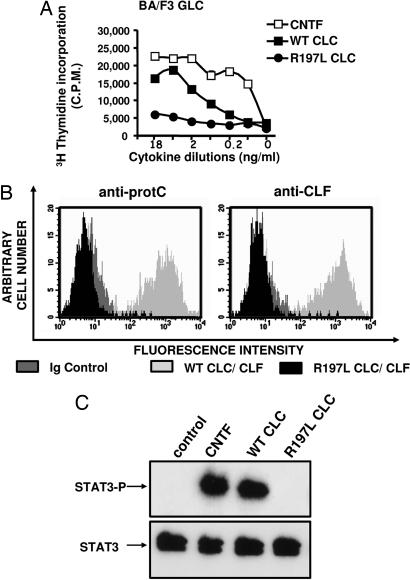
To investigate the functional activity of R197L CLC, we used derivatives of the IL-3-dependent Ba/F3 cell line rendered responsive to CNTF by transfection with gp130, LIFR, and CNTFRα (referred to here as “BAF GLC”). Although this cell line proliferated in response to WT CLC as observed (14, 20), it failed to respond to R197L CLC (but a slight residual activity was observed for the highest tested concentrations) (Fig. 5A). By using the same cell line, we analyzed the capacity of the secreted mutant composite cytokine to bind to the tripartite CNTF receptor (Fig. 5B). Cytokine binding was monitored by flow cytometry using Abs directed against CLF or the CLC epitope tag, as described in ref. 14. No interaction between the R197L CLC–CLF composite cytokine and CNTFRα could be detected (Fig. 5B). To further analyze the properties of the R197L CLC, we used the SK-N-GP neuroblastoma cell line spontaneously expressing the tripartite CNTF receptor. We studied the induction of STAT3 Tyr phosphorylation, a signaling pathway recruited in response to CNTF receptor activation by CLC (Fig. 5C). No STAT3 Tyr phosphorylation could be evidenced in response to R197L CLC, whereas a clear signal was detected when the SK-N-GP cells were activated with WT form of cytokine. Together, these results indicate that the R197L mutated form of CLC failed to recognize and activate the tripartite CNTF receptor.
Fig. 5.
Biological activities of WT and R197L CLC. (A) Proliferation analysis of BA/F3 GLC cell line in response to serial dilutions of purified WT (■) or R197L (●) CLC. CNTF was used as a positive control (□) (most of the standard error deviation triplicate values were inferior to the symbol size). C.P.M., counts per minute. (B) Flow cytometry analysis of the binding of WT (light gray histogram) or R197L (black histogram) CLC–CLF complexes to BAF GLC cells. Bound cytokine was revealed by using either an anti-protein C tag or an anti-CLF mAb. (C) Analysis of STAT3 Tyr phosphorylation induced by WT or the R197L CLC in the SK-N-GP human neuroblastoma cell line.
Arg-197 Is Crucial for CLC Binding to CNTFRα.
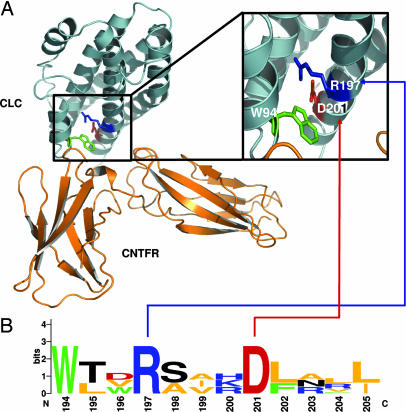
To further investigate the role of the R197L mutation in the binding of CLC to CNTFRα, a structural analysis of both proteins was carried out. Models were built by homology molecular modeling based on protein secondary structures. Ligand–receptor interactions were then simulated by using a rigid-body-docking algorithm and constraints based on surface complementarities of both proteins. A ribbon representation of the energy-minimized complex of CLC–CNTFRα is shown in Fig. 6A. The region of CLC involved in the interaction with CNTFRα (site 1) is formed by the N-terminal part of the a D-helix of CLC associated with a second short α-helix (named the αA′-helix) and located between the αA- and αB-helices (Fig. 2C). The complementary CNTFRα region formed an elbow-shaped motif, with a binding site composed of loops from D2 and D3 receptor domains that interacts with αA′- and αD-helices (26). Molecular docking indicated that the W94 side chain of CLC, which is conserved with the other CNTFRα ligands (CNTF and NP; Fig. 2C), dominated the binding interface with CNTFRα and contributed to hydrophobic interactions. Side chains of R197 and D201 residues in CLC were not involved directly in the contact point but they formed hydrogen bonds with the W94 residue. The position of these residues contributed to the stability of site 1 by packing the αA′-helix against the four-helix-bundle core and maintaining the correct orientation of W94. Structural analysis of all known CNTFRα ligands underscored the importance of these two residues for receptor binding (Fig. 6B). Multiple sequence alignment shows that R197 and D201 are part of a motif that is strictly conserved among the CNTFRα ligands (13, 25). These structural prediction data highlight the key role of R197 in the formation of the CLC binding site 1 to CNTFRα, as confirmed by the biochemical analysis of the R197L CLC mutant.
Fig. 6.
Molecular modeling and docking of CLC and CNTFRα. Aromatic residues are shown in green, positive residues are shown in blue, negative residues are shown in red, hydrophobic residues are shown in orange, and polar residues are shown in black. (A) CLC and CNTFRα are represented by blue and orange ribbons, respectively, by using pymol. (Inset) Magnified view of the boxed area of CLC-interacting site 1. (B) Motif conservation of residues implicated in site 1 of known orthologs of CLC, CNTF, and NP shown with logo.
Discussion
The CNTF receptor pathway is important for both development and maintenance of the nervous system and muscles (10). Two CNTFRα ligands have been identified in man, CNTF and CLC–CLF (1, 14). We recently identified mutations inactivating the synthesis of CLF in CISS patients (23). These patients have a complex clinical picture, which includes defects in thermosensation and pain sensitivity, lack of appetite, kyphoscoliosis, and muscular atrophy. Although only a few CISS patients with mutations in the CLF gene have been studied, some variation in the phenotype is observed with more pronounced symptoms in the patients presenting CLF stop mutations (23).
In this article, we identified a different form of the disease (CISS2) linked to a CLC deficiency, with clinical features very similar to mildly affected CISS1 patients. The CISS2 patient also had signs of a mild and predominantly sensory neuropathy. Whether the neuropathy is acquired or forms part of the syndrome would have to be determined by the study of additional CISS2 patients. To our knowledge, none of the CISS1 patients that have been reported have shown any sign of neuropathy. However, studies in rodents established that decrease or absence in CNTF synthesis in aging contribute to a reduction in muscle strength, possibly indicating a similar function for CLC (6, 11). High concentrations of R197L CLC led to a residual activity on BAF GLC cells, despite an absence of detectable binding to CNTFRα, as shown in Fig. 5. This residual effect might be explained by a direct recruitment of gp130 and LIFR through sites 2 and site 3, with high CLC concentrations as shown before for CNTF or IL-6 (27, 28). Potential site 2- and 3-mediated residual activity might account for the observed differences between the CISS2 patient and the CISS1 patients with the most severe symptoms.
Part of the symptoms of the CISS2 patient, anhidrosis in hot weather, displays similarity to the syndrome of congenital insensitivity to pain with anhidrosis, which is due to inactivating mutations in the nerve growth factor receptor TRKA (29). Thermosensation and nociception are mediated by sensory neurons of the dorsal root ganglia that terminate as free nerve endings in the skin and involve the activation of specific transient receptor potential channels (30). Interestingly, it was reported recently that exposure of rodent nociceptive neurons to the IL-6–soluble IL-6 receptor complex lead to the potentiation of heat-activated inward currents, suggesting that these neurons might be CLC target cells (31). Preliminary observations indicate that nociceptive and thermosensitive mouse dorsal root ganglia neurons express the CNTFRα components and are CLC-responsive.
Sweat glands release soluble factors inducing differentiation steps from a noradrenergic to a cholinergic transmitter phenotype required for correct sweat gland secretory responsiveness (32). It was established that most of these effects were mediated through gp130 receptor-acting cytokines (33, 34). Our studies suggest that the CLC–CLF composite cytokine may contribute to this process. The possibility for CLC to modulate the thermoregulation of central nervous system origin should also be considered. The tripartite CNTF receptor complex is expressed by both central and peripheral neurons opening the possibility for CLC to also act at both levels, similarly to that recently demonstrated for CNTF in energy balance (35).
We could not find additional R197L and Y107X mutations among 140 CLC gene copies, indicating a low prevalence of these variants in the human population. Also, the autosomal recessive transmission of CLC-mutated alleles is likely to account for the very low incidence of the CISS2 syndrome. In humans, two receptors belonging to the IL-6/gp130 family of mediators have been associated with disease. In addition to the CLF soluble receptor in CISS, mutations in LIFR have been reported in Stuve–Wiedemann syndrome, a skeletal disorder with feeding and swallowing difficulties, paradoxical sweating at low temperature, muscle hypotonia, and respiratory insufficiency (36). Part of these symptoms overlap between both diseases in agreement with the LIFR role in CLC signaling. In contrast, homozygous inactivation of CNTF, present in 2.3% of examined human populations, is clinically silent, indicating functional redundancy among the CNTFRα ligands (12). This situation is very similar to that observed for a number of type I cytokine receptors for which mutations associated with diseases have been identified, whereas no deleterious mutations have been reported for the corresponding cytokines. For example, inactivating mutations for IL-2Rγ, IL-7R, IL-12Rβ1, and IL-12/p40 were associated with congenital immune deficiencies (37). Development of a CISS phenotype with mutations in the CLC gene suggests a specific, nonredundant function for this cytokine in humans.
CLC gene mutations were identified in a CISS2 patient with skeletal anomalies, a mild sensorimotor neuropathy, and alteration in thermoregulation. Analysis and structural modeling of these mutations implicate a lack of interaction between the cytokine and CNTFRα in the pathology. The prevalent role of CLC, as well as CLF, in the control of sweating can provide valuable information for prospective investigations of less dramatic sweating disorders or hyperhidrosis.
Materials and Methods
Subject and Clinical Evaluation.
In the CISS2, patient sweating in response to various ambient temperatures was examined and documented. Comprehensive studies of autonomic cardiovascular reflexes and postganglionic sympathetic efferent functions were performed and were normal. Hematological, biochemical, and serological tests were carried out and were normal.
DNA Sequencing and Site-Directed Mutagenesis.
PCR primers for amplification of exons and flanking introns for CLF, CLC, CNTF, CNTFRα, LIFR, and gp130 were designed by using oligo 6.3 software (Molecular Biology Insights, Cascade, CO). PCR and RT-PCR amplifications were performed under standard conditions. The PCR products were sequenced by using the ABI Prism BigDye terminator sequencing kit version 1 and the ABI 3100 genetic analyzer (Applied Biosystems). DNA sequences were analyzed by using the staden software package with the program seqed (Applied Biosystems). The following primers surrounding an intron region were chosen to amplify CLC cDNA: 5′-CTCCATCCAGAAAACCTATG-3′ and 5′-CTGTAGGCCTCGTAGTTCTG-3′. The pcDNA3 vector containing the cDNA encoding the human CLC-tagged protein C epitope was subjected to site-directed mutagenesis by using the QuikChange site-directed mutagenesis kit (Stratagene). Mutation was performed on R197 of CLC by replacing it with a leucin. The cDNA encoding the first 107 residues of CLC (ΔCLC) was PCR-amplified and fused to a protein C coding epitope. The fragment was then inserted in a pcDNA3 vector containing an internal ribosomal entry site and the CLF cDNA.
Sequence Alignments and Molecular Modeling.
Orthologs of CLC (human, GenBank accession no. NP_037378.1; and mouse, NP_064336), CNTF (human, NP_000605.1; mouse, Q02011; rat, NP_037298.1; pig, O02732; chicken, P51642; and rabbit, P14188), and NP (chimp, AAS66750; mouse, AAR17733; and rat, AAS66749) were retrieved from the GenBank database. The multiple sequence alignment was obtained by using the programs t-coffee and genedoc. For simplicity, only human CLC, human CNTF, and mouse NP are shown in Fig. 2. CLC was modeled by using modeller, based on multiple sequence alignment and secondary-structure predictions (Network Prediction Server @nalysis, Pole Bio-Informatique Lyonnais). Structural coordinates of human CNTF [Protein Data Bank (PDB) entry 1CNT] and LIF (PDB entry 1PVH) were selected as molecular templates; the coordinates of an additional α-helix predicted in the AB loop were computed with the Biopolymer module of insight ii (Accelrys, San Diego) and integrated during the structure-calculation process. The D3 domain of CNTFRα was obtained from the PDB (entry 1UC6); the D2 domain was modeled based on the structure of IL-6Rα (PDB entry 1P9M) and of the IL-12/p40 subunit for the DE loop (PDB entry 1F45); the angle between D2 and D3 and their assembly was based on the overall structure of IL-6Rα. All of the models were energy-minimized and quality-verified by using the programs PROFILES_3D and procheck.
Reagents, Cells, Transfections, and Protein Purification.
Anti-CNTFRα (AN-C2; IgG2a) and anti-human CLF (AN-F-C6; IgG1) mAbs were generated in the laboratory. The anti-protein C (HPC4) Ab was purchased from Roche Diagnostics. The anti-STAT-3 polyclonal Ab and the phospho-705-STAT-3 mAb were from Santa Cruz Biotechnology and New England Biolabs, respectively. The Cos-7 fibroblast and SK-N-GP human neuroblastoma cell lines were grown in RPMI medium 1640 supplemented with 10% FCS. The culture medium of Ba/F3 cells modified to express the functional receptor for CNTF (BAF GLC) was supplemented with 5 ng/ml human CNTF (R & D Systems). Peripheral blood cells from the patient were activated for 24 h in culture with 10 nM phorbol myristate acetate and 3 nM phytohemagglutinin before RNA extraction and reverse transcription. cDNAs encoding WT CLC, mutant forms of CLC, CLF, or soluble CNTFRα were transfected in Cos-7 cells by using ExGen reagent (Euromedex, Souffelweyersheim, France) (14). WT and R197 CLC were purified from Cos-7 cell lysates by affinity chromatography with an anti-protein C-affinity column (Roche Diagnostics) (20). Protein purity and concentrations were determined by SDS/PAGE and silver staining.
Immunoprecipitation, Western Blotting, and Tyr Phosphorylation Analysis.
Cell supernatants were harvested 48 h after transfection with the appropriate cDNAs, and cells were lysed in SDS/PAGE sample buffer. Cell supernatants were then immunoprecipitated overnight by using an anti-protein C tag, an anti-CLF, or an anti-CNTFRα mAb at 10 μg/ml. Complexes were isolated by using beads coupled to protein A and subjected to Western blot analysis by using a biotinylated anti-protein C mAb and streptavidin peroxidase. For Tyr phosphorylation analysis, SK-N-GP cells were stimulated for 10 min with the indicated cytokine. Cells lysates were subjected to SDS/PAGE and Western blot analysis by using a mAb specific for a phosphorylated form of STAT-3 (14).
Biological Assays and Flow Cytometry Analysis.
BAF GLC cells were seeded in 96-well plates at a concentration of 5 × 103 cells per well and incubated for 72 h. Serial dilutions of tested cytokines were performed in triplicate. Thymidine incorporation was measured as described in ref. 14. For flow cytometry, BAF GLC cells were successively incubated for 30 min at 4°C with indicated cytokines, the appropriate primary Ab directed against the cytokine (10 μg/ml), or an isotype control Ab, before using a phycoerythrin-conjugated anti-mouse Ab. Fluorescence was subsequently analyzed on a FACScalibur flow cytometer (Beckton Dickinson).
Acknowledgments
We thank J. S. Bringsli and I. B. Tjelflaat for genomic DNA sequencing work, J. M. Mathys for help in preparing blood cells and cDNAs, L. Preisser and E. Ravon for RT-PCR analyses and cDNA sequencing, G. Elson for careful review of the manuscript, and K.-J. Kallen (Christian Albrechts Universität zu Kiel, Kiel, Germany) for providing BAF GLC cells. This work was supported by Canadian Institute of Health Research Grant MOP-57832, Association Française Contre les Myopathies Grant 9884, and a grant from the Post Genome Program of the Contrat Etat Région. J.-F.G. is a Canada Research Chair recipient, and F.R. is supported by the Angers Agglomeration.
Abbreviations
- CNTF
ciliary neurotrophic factor
- CLC
cardiotrophin-like cytokine
- CLF
cytokine-like factor
- CNTFRα
CNTF α-component receptor
- NP
neuropoietin
- CISS
cold-induced sweating syndrome
- LIFR
leukemia inhibitory factor receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ip N. Y., Yancopoulos G. D. Annu. Rev. Neurosci. 1996;19:491–515. doi: 10.1146/annurev.ne.19.030196.002423. [DOI] [PubMed] [Google Scholar]
- 2.Ip N. Y., McClain J., Barrezueta N. X., Aldrich T. H., Pan L., Li Y., Wiegand S. J., Friedman B., Davis S., Yancopoulos G. D. Neuron. 1993;10:89–102. doi: 10.1016/0896-6273(93)90245-m. [DOI] [PubMed] [Google Scholar]
- 3.Sendtner M., Schmalbruch H., Stockli K. A., Carroll P., Kreutzberg G. W., Thoenen H. Nature. 1992;358:502–504. doi: 10.1038/358502a0. [DOI] [PubMed] [Google Scholar]
- 4.Mitsumoto H., Ikeda K., Klinkosz B., Cedarbaum J. M., Wong V., Lindsay R. M. Science. 1994;265:1107–1110. doi: 10.1126/science.8066451. [DOI] [PubMed] [Google Scholar]
- 5.Helgren M. E., Squinto S. P., Davis H. L., Parry D. J., Boulton T. G., Heck C. S., Zhu Y., Yancopoulos G. D., Lindsay R. M., DiStefano P. S. Cell. 1994;76:493–504. doi: 10.1016/0092-8674(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 6.Guillet C., Auguste P., Mayo W., Kreher P., Gascan H. J. Neurosci. 1999;19:1257–1262. doi: 10.1523/JNEUROSCI.19-04-01257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis S., Aldrich T. H., Stahl N., Pan L., Taga T., Kishimoto T., Ip N. Y., Yancopoulos G. D. Science. 1993;260:1805–1808. doi: 10.1126/science.8390097. [DOI] [PubMed] [Google Scholar]
- 8.Grotzinger J., Kernebeck T., Kallen K.-J., Rose-John S. Biol. Chem. 1999;380:803–813. doi: 10.1515/BC.1999.100. [DOI] [PubMed] [Google Scholar]
- 9.Stahl N., Farruggella T. J., Boulton T. G., Zhong Z., Darnell J. E., Jr., Yancopoulos G. D. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 10.DeChiara T. M., Vejsada R., Poueymirou W. T., Acheson A., Suri C., Conover J. C., Friedman B., McClain J., Pan L., Stahl N., et al. Cell. 1995;83:313–322. doi: 10.1016/0092-8674(95)90172-8. [DOI] [PubMed] [Google Scholar]
- 11.Masu Y., Wolf E., Holtmann B., Sendtner M., Brem G., Thoenen H. Nature. 1993;365:27–32. doi: 10.1038/365027a0. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi R., Yokoji H., Misawa H., Hayashi M., Hu J., Deguchi T. Nat. Genet. 1994;7:79–84. doi: 10.1038/ng0594-79. [DOI] [PubMed] [Google Scholar]
- 13.Derouet D., Rousseau F., Alfonsi F., Froger J., Hermann J., Barbier F., Perret D., Diveu C., Guillet C., Preisser L., et al. Proc. Natl. Acad. Sci. USA. 2004;101:4827–4832. doi: 10.1073/pnas.0306178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elson G. C., Lelievre E., Guillet C., Chevalier S., Plun-Favreau H., Froger J., Suard I., de Coignac A. B., Delneste Y., Bonnefoy J. Y., et al. Nat. Neurosci. 2000;3:867–872. doi: 10.1038/78765. [DOI] [PubMed] [Google Scholar]
- 15.Senaldi G., Varnum B. C., Sarmiento U., Starnes C., Lile J., Scully S., Guo J., Elliott G., McNinch J., Shaklee C. L., et al. Proc. Natl. Acad. Sci. USA. 1999;96:11458–11463. doi: 10.1073/pnas.96.20.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y., Wang W., Yourey P. A., Gohari S., Zukauskas D., Zhang J., Ruben S., Alderson R. F. Biochem. Biophys. Res. Commun. 1999;262:132–138. doi: 10.1006/bbrc.1999.1181. [DOI] [PubMed] [Google Scholar]
- 17.Bazan J. F. Neuron. 1991;7:197–208. doi: 10.1016/0896-6273(91)90258-2. [DOI] [PubMed] [Google Scholar]
- 18.Forger N. G., Prevette D., de Lapeyriere O., de Bovis B., Wang S., Bartlett P., Oppenheim R. W. J. Neurosci. 2003;23:8854–8858. doi: 10.1523/JNEUROSCI.23-26-08854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elson G. C., Graber P., Losberger C., Herren S., Gretener D., Menoud L. N., Wells T. N., Kosco-Vilbois M. H., Gauchat J. F. J. Immunol. 1998;161:1371–1379. [PubMed] [Google Scholar]
- 20.Plun-Favreau H., Elson G., Chabbert M., Froger J., deLapeyriere O., Lelievre E., Guillet C., Hermann J., Gauchat J. F., Gascan H., Chevalier S. EMBO J. 2001;20:1692–1703. doi: 10.1093/emboj/20.7.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trinchieri G., Pflanz S., Kastelein R. A. Immunity. 2003;19:641–644. doi: 10.1016/s1074-7613(03)00296-6. [DOI] [PubMed] [Google Scholar]
- 22.Alexander W. S., Rakar S., Robb L., Farley A., Willson T. A., Zhang J. G., Hartley L., Kikuchi Y., Kojima T., Nomura H., et al. Curr. Biol. 1999;9:605–608. doi: 10.1016/s0960-9822(99)80266-8. [DOI] [PubMed] [Google Scholar]
- 23.Knappskog P. M., Majewski J., Livneh A., Nilsen P. T., Bringsli J. S., Ott J., Boman H. Am. J. Hum. Genet. 2003;72:375–383. doi: 10.1086/346120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohar E., Shoenfeld Y., Udassin R., Magazanik A., Revach M. Lancet. 1978;2:1073–1074. doi: 10.1016/s0140-6736(78)91805-6. [DOI] [PubMed] [Google Scholar]
- 25.Perret D., Guillet C., Elson G., Froger J., Plun-Favreau H., Rousseau F., Chabbert M., Gauchat J. F., Gascan H. J. Biol. Chem. 2004;279:43961–43970. doi: 10.1074/jbc.M407686200. [DOI] [PubMed] [Google Scholar]
- 26.Man D., He W., Sze K. H., Gong K., Smith D. K., Zhu G., Ip N. Y. J. Biol. Chem. 2003;278:23285–23294. doi: 10.1074/jbc.M301976200. [DOI] [PubMed] [Google Scholar]
- 27.Gearing D. P., Ziegler S. F., Comeau M. R., Friend D., Thoma B., Cosman D., Park L., Mosley B. Proc. Natl. Acad. Sci. USA. 1994;91:1119–1123. doi: 10.1073/pnas.91.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroers A., Hecht O., Kallen K.-J., Pachta M., Rose-John S., Grotzinger J. Protein Sci. 2005;14:783–790. doi: 10.1110/ps.041117105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indo Y., Tsuruta M., Hayashida Y., Karim M. A., Ohta K., Kawano T., Mitsubuchi H., Tonoki H., Awaya Y., Matsuda I. Nat. Genet. 1996;13:485–488. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- 30.Tominaga M., Caterina M. J. J. Neurobiol. 2004;61:3–12. doi: 10.1002/neu.20079. [DOI] [PubMed] [Google Scholar]
- 31.Obreja O., Biasio W., Andratsch M., Lips K. S., Rathee P. K., Ludwig A., Rose-John S., Kress M. Brain. 2005;128:1634–1641. doi: 10.1093/brain/awh490. [DOI] [PubMed] [Google Scholar]
- 32.Grant M. P., Francis N. J., Landis S. C. Mol. Cell. Neurosci. 1995;6:32–42. doi: 10.1006/mcne.1995.1004. [DOI] [PubMed] [Google Scholar]
- 33.Yamamori T., Fukada K., Aebersold R., Korsching S., Fann M. J., Patterson P. H. Science. 1989;246:1412–1416. doi: 10.1126/science.2512641. [DOI] [PubMed] [Google Scholar]
- 34.Stanke M., Duong C. V., Pape M., Geissen M., Burbach G., Deller T., Gascan H., Parlato R., Schutz G., Rohrer H. Development (Cambridge, U.K.) 2006;133:141–150. doi: 10.1242/dev.02189. [DOI] [PubMed] [Google Scholar]
- 35.Kokoeva M. V., Yin H., Flier J. S. Science. 2005;310:679–683. doi: 10.1126/science.1115360. [DOI] [PubMed] [Google Scholar]
- 36.Dagoneau N., Scheffer D., Huber C., Al-Gazali L. I., Di Rocco M., Godard A., Martinovic J., Raas-Rothschild A., Sigaudy S., Unger S., et al. Am. J. Hum. Genet. 2004;74:298–305. doi: 10.1086/381715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picard C., Casanova J. L. Curr. Opin. Pediatr. 2004;16:648–658. doi: 10.1097/01.mop.0000145919.92477.5f. [DOI] [PubMed] [Google Scholar]