Abstract
(+)-Sesamin, a furofuran class lignan, is widespread in vascular plants and represented by Sesamum spp. (+)-Sesamin has been of rapidly growing interest because of its beneficial biological effects in mammals, but its biosynthesis and physiological roles in plants remain to be clarified. It is speculated to be synthesized from (+)-pinoresinol by means of (+)-piperitol by formation of two methylenedioxy bridges mediated by two distinct Sesamum indicum cytochrome P450 (SiP450) proteins. Here, we report an SiP450, CYP81Q1, that alone catalyzes (+)-sesamin biosynthesis from (+)-pinoresinol by means of (+)-piperitol by forming two methylenedioxy bridges. The CYP81Q1 gene expression profile was temporally consistent with the accumulation pattern of (+)-sesamin during seed development. The CYP81Q1-GFP chimera protein was colocalized with an endoplasmic reticulum (ER)-targeting chimera protein, indicating that (+)-sesamin biosynthesis occurs on the ER cytoplasmic surface. Moreover, we isolated two CYP81Q1 homologs from other Sesamum spp. Sesamum radiatum CYP81Q2 showed dual (+)-piperitol/(+)-sesamin synthetic activity. CYP81Q2, as well as CYP81Q1, therefore, corresponds to a (+)-piperitol/(+)-sesamin synthase in lignan biosynthesis. In contrast, Sesamum alatum CYP81Q3 showed no activity, in accord with (+)-sesamin being deficient in S. alatum. Our findings not only provide insight into lignan biosynthesis but also unravel a unique mode of cytochrome P450 action.
Keywords: P450, Sesamum indicum, pinoresinol, piperitol
Sesame (Pedaliaceae, Sesamum indicum L.), a major oil seed crop of high nutritional value, is widely cultivated throughout the world. Sesame oil, ≈50% of seed weight, contains large amounts of natural antioxidants (1, 2). The potent antioxidant properties of sesame seed extract mainly are attributable to the presence of lignans (3). Lignans are phenylpropanoid dimers in which phenylpropane units are linked by the central carbons of the side chains, and they constitute an abundant class of phenylpropanoids (4). They are widely distributed in vascular plants. (+)-Sesamin, first isolated in 1890, is a major furofuran lignan in sesame seeds and is distinguished by two characteristic methylenedioxy bridges (2) (Fig. 1A). It has a growth-retarding effect on some plants, as well as a synergistic effect with pyretherum insecticides (5, 6). (+)-Sesamin has been shown to have a wide range of biological effects: protection against ethanol- and carbon tetrachloride-induced liver damage (7, 8), improvement of fatty acid metabolism (9–11), antiinflammatory effects (12), increased vitamin E activities (13, 14), inhibition of vascular superoxide production (15, 16), and induction of nitric oxide (17). Therefore, it is an extremely interesting natural compound from the viewpoint of health care (18).
Fig. 1.
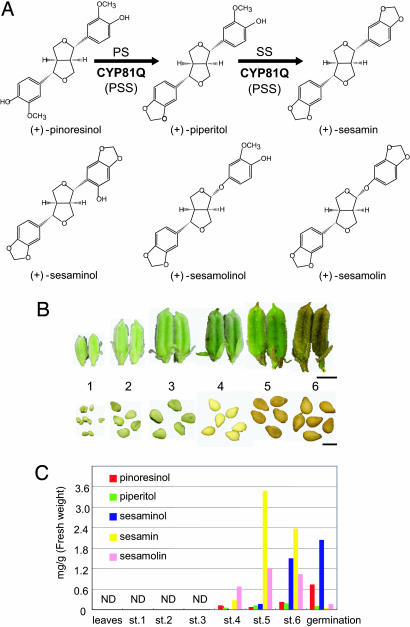
Developmental regulation of lignan biosynthesis. (A) Lignan structures in Sesamum spp. (B) Sesame seed development. Numbers correspond to developmental seed stages. (Scale bars: Upper, 1 cm; Lower, 2 mm.) (C) Developmental accumulation changes in five sesame lignans determined by HPLC after treatment with β-glucosidase. ND, not detected.
The (+)-sesamin biosynthetic pathway has been deduced from results of radioisotope tracer and enzymatic experiments (19). (+)-Pinoresinol, the first lignan in the lignan pathway, is synthesized by stereoselective phenoxy radical coupling of two units of achiral E-coniferyl alcohol (Fig. 1A) (19–21). (+)-Piperitol is synthesized from it by formation of a methylenedioxy bridge (22), and (+)-sesamin also is synthesized from (+)-piperitol by formation of such a bridge (23). These two stepwise methylenedioxy bridge formations were postulated to be catalyzed by two consecutively acting S. indicum cytochrome P450 (SiP450) enzymes, piperitol synthase (PS) and sesamin synthase (SS), because only PS activity was observed in sesame seed microsomes, and the methylenedioxy bridge formation was inhibited by various cytochrome P450 enzyme inhibitors (22) (Fig. 1A).
In alkaloid biosynthesis, methylenedioxy bridge formations have for the first time been known to be cytochrome P450-dependent (24, 25). Eventually, CYP719A1 was identified from Coptis japonica as the first methylenedioxy bridge-forming P450 involved in isoquinoline alkaloid biosynthesis (26). This finding suggested that SiP450s, which belong to the CYP719A subfamily, are involved in (+)-sesamin biosynthetic methylenedioxy bridge formations. Since then, SiP450 genes have been identified by comparative EST analysis of sesame seeds, but their functions have yet to be examined (27).
To clarify the underlying mechanism of (+)-sesamin biosynthesis, we comprehensively investigated the SiP450 genes that are responsible for SS. Interestingly, we identified a previously uncharacterized SiP450, CYP81Q1, which serves both (+)-sesamin and (+)-piperitol by formation of methylenedioxy bridges on (+)-piperitol and (+)-pinoresinol, respectively. Namely, CYP81Q1 corresponds to a (+)-piperitol/(+)-sesamin synthase (PSS). Unexpectedly, CYP81Q1 has only 24% amino acid sequence identity to CYP719A1. We also identified a functional homolog of CYP81Q1 in Sesamum radiatum, evidence that this methylenedioxy bridge-forming enzyme is structurally and functionally conserved in the Sesamum genus. Our findings provide the biochemical and genetic tools to produce a beneficial natural product, (+)-sesamin.
Results
Sesame Lignans Are Synthesized Specifically in the Later Phase of Seed Development.
Lignan mixtures extracted from S. indicum seeds at six developmental stages, germinating seeds, and leaves were analyzed by HPLC (Fig. 1B). (+)-Sesamin, detected first at stage 4, reached a maximum at stage 5 and then was slightly decreased at stage 6 in fully matured seeds (Fig. 1C). On germination, (+)-sesamin content markedly was decreased, indicating that (+)-sesamin biosynthesis is activated at least as early as at stage 4 but not at germination. (+)-Pinoresinol and (+)-piperitol, precursors of (+)-sesamin, were not detected before (+)-sesamin accumulation. During germination, a substantial amount of (+)-pinoresinol, but not (+)-piperitol, was accumulated, evidence that (+)-pinoresinol biosynthesis is regulated separately from (+)-piperitol and (+)-sesamin biosynthesis. In contrast, (+)-piperitol biosynthesis was tightly coupled with (+)-sesamin biosynthesis, because there was no substantial accumulation of (+)-piperitol at any stage.
(+)-Sesaminol, a 2′ hydroxylated derivative of (+)-sesamin (28), apparently was accumulated in fully matured seeds (stage 6) and increased at germination (Fig. 1 A and C). (+)-Sesamolin, also a major sesame lignan (22, 23), had an accumulation pattern similar to that of (+)-sesamin (Fig. 1 A and C). None of the lignans analyzed in this study was detected in seeds from stage 1 to 3 or in leaves (Fig. 1C), indicating that sesame lignan biosynthesis is specifically activated in the later phases of seed development. The lignan metabolite ratio changes markedly at germination.
Identification of CYP81Q1.
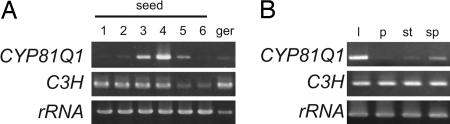
The (+)-sesamin accumulation profile predicted that the gene that encodes SS is expressed substantially in the later phases of seed development (Fig. 1C). Consequently, a cDNA library was constructed from seeds in stages 4–5 and screened with probe mixtures of 13 Arabidopsis cytochrome P450 genes. To obtain various SiP450 genes, these probes were selected from different P450 subfamilies. One hundred ninety-two positive clones were obtained after a second screening and clustered finally in 17 independent molecular species of SiP450. To further screen these candidate SiP450 genes, their expression patterns were examined by RT-PCR. One candidate SiP450, SiP189, had an expression profile consistent with the developmental accumulation pattern of (+)-sesamin. Its expression began at stage 3, reached a maximum at stage 4, and then decreased before seed maturation was complete (Figs. 1C and 2A). It also was expressed in leaves but not in germinating seeds, petals, or stems, whereas the S. indicum p-coumarate 3-hydroxylase (C3H, CYP98A20) gene, which is involved in phenylpropanoid pathway, was expressed ubiquitously (Fig. 2B) (29).
Fig. 2.
Gene expression analyses by RT-PCR. Temporal CYP81Q1 expression patterns during seed development (A) and in aerial organs (B). C3H is S. indicum p-coumarate 3-hydroxylase (CYP98A20) expressed in phenylpropanoid biosynthesis (29). ger, germinating seeds treated with H2O for 2 days; l, leaves; p, petals; st, stems; sp, seed pods.
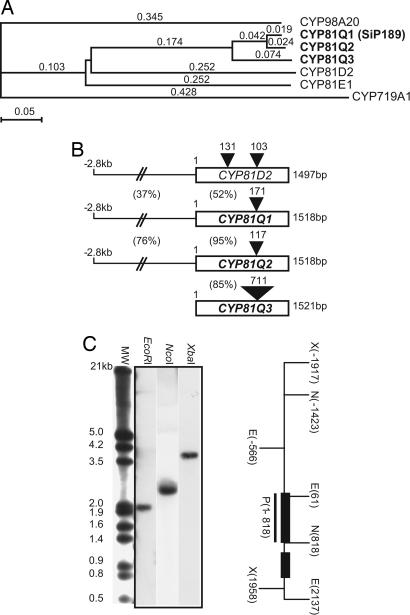
SiP189 bears an ORF of 1,518 bp for 506 aa (Fig. 3B). It has 52%, 49%, and 24% amino acid sequence identity, respectively, to a functionally unknown Arabidopsis P450, CYP81D2 (NM 119899); Glycyrrhiza CYP81E1, catalyzing a 2′ hydroxylation of isoflavone (30); and Coptis CYP719A1 (26) (Fig. 3 A and C). The SiP189 gene has a single 171-bp intron 898 bp downstream from the start codon, whereas the CYP81D2 gene has two introns in its genomic structure (Fig. 3B). Southern blot analysis showed that the SiP189 gene exists as a single gene in the S. indicum genome (Fig. 3C). SiP189 has been designated CYP81Q1 by the P450 nomenclature committee. It has conserved regions in cytochrome P450 proteins (Fig. 6, which is published as supporting information on the PNAS web site), a helix K region, an aromatic region, and a heme-binding region that includes the proximal Cys residue at the COOH-terminal region. The distal Thr residue that corresponds to Thr-252 in the I helix of P450cam is conserved in many P450 proteins (31); however, CYP81Q1 does not have the conserved Thr residue but does have Ala-308 (Fig. 6).
Fig. 3.
Identification and characterization of CYP81Q genes. (A) P450 protein phylogenic tree. CYP719A1 is the Coptis methylenedioxy bridge-forming P450 (BAB68769), CYP81D2 is a functionally unknown Arabidopsis P450 (NM 119899), CYP98A20 is Sesamum p-coumarate 3-hydroxylase (AAL47545), and CYP91E1 is Glycyrrhiza isoflavone 2′-hydroxylase (AB001379). The phylogenic tree was constructed with clustalw macvector 7.2.2 software (53). Branch length is proportional to the estimated divergence of each protein. (Scale bar: 5% change.) (B) Genomic structures of three CYP81Q genes and the CYP81D2 gene. Black triangles with numbers indicate intron position and size. The percentage shown between the 5′ noncoding region and ORF indicates the DNA sequence identity of the 5′ noncoding regions and amino acid sequence identity of the ORFs. (C) Genomic Southern blot analysis. S. indicum genomic DNA was digested with the restriction enzyme shown at the top of each lane. The hybridization signal was shown as a single band in each digestion. Underlining indicates the probe region (P) corresponding to exon 1. Black boxes denote exons of the CYP81Q1 gene. E, EcoRI; N, NcoI; X, XbaI.
CYP81Q1 Is a PSS.
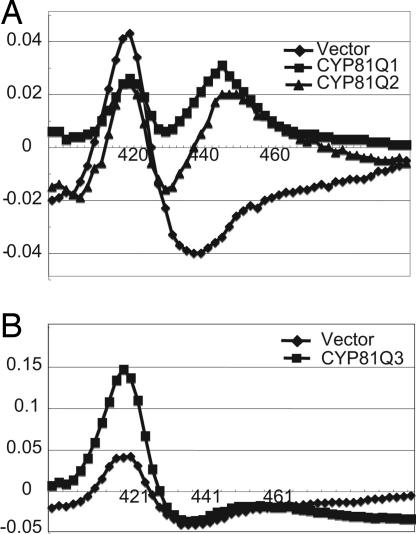
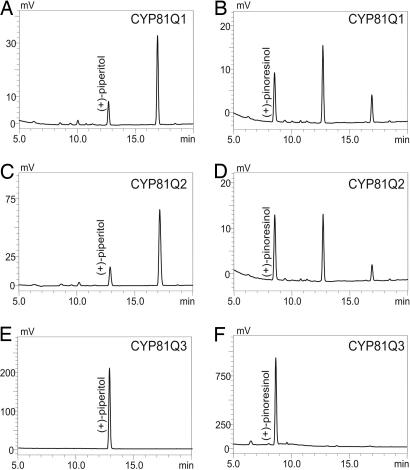
The recombinant CYP81Q1 protein was expressed heterologously in yeast to determine its enzymatic activity. A microsomal fraction was extracted, and its reduced CO-difference spectra were measured (Fig. 4A). Compared with the microsomal fraction of the control yeast carrying an empty vector, the fraction expressing CYP81Q1 gene showed significant absorption at 449 nm, which indicated that the recombinant CYP81Q1 protein was expressed as an active P450. The microsomal fraction was incubated with (+)-piperitol and NADPH, and the reaction mixture was analyzed by HPLC. A product with a furofran lignan-specific spectrum represented by two absorptions at 230 and 280 nm was eluted at a retention time (RT) of 16.9 min, which is the same as the RT of authentic (+)-sesamin (Fig. 5A). Interestingly, the microsomal fraction also catalyzed the transformation of (+)-pinoresinol (RT of 8.6 min) into two lignan products eluted at RTs of 12.7 and 16.9 min, identical to the RTs of authentic (+)-piperitol and (+)-sesamin, respectively (Fig. 5B). Liquid chromatography-MS analysis gave the respective molecular weights of these two products: RT 12.7, m/z = 374 (M+NH4+); RT 16.9, m/z = 372 (M+NH4+) (Fig. 7, which is published as supporting information on the PNAS web site). In contrast, no lignan conversion occurred in the control reactions. These findings show that CYP81Q1 has activity that is responsible for both PS and SS. Its dual methylenedioxy bridge-forming activity required NADPH, because the absence of NADPH abolished CYP81Q1 activity toward (+)-pinoresinol and (+)-piperitol (Fig. 8, which is published as supporting information on the PNAS web site). Apparent Km values toward (+)-pinoresinol and (+)-piperitol were 10.2 ± 1.4 and 11.7 ± 1.7 μM, respectively.
Fig. 4.
Reduced CO-difference spectra of microsomal preparations from different recombinant yeasts. These spectra were recorded by using microsomal fractions of recombinant yeast (1 mg of protein/ml). (A) Diamonds, pYE22m; squares, CYP81Q1; triangles, CYP81Q2. (B) Diamonds, pYE22m; squares, CYP81Q3.
Fig. 5.
Chromatograms showing absorption at 280 nm in the in vitro enzyme assays of microsomal preparations. (A and B) CYP81Q1 reactions with (+)-piperitol (A) and (+)-pinoresinol (B). (C and D) CYP81Q2 reactions with (+)-piperitol (C) and (+)-pinoresinol (D). (E and F) CYP81Q3 reactions with (+)-piperitol (E) and (+)-pinoresinol (F). The standards (+)-pinoresinol, (+)-piperitol, and (+)-sesamin were eluted at RTs of 8.6, 12.8, and 16.9 min, respectively.
As shown in Fig. 1, (+)-sesamolin has developmental and structural features similar to (+)-sesamin (19, 22). Because (+)-sesamolinol, a Sesamum lignan, has a structural potential to form a methylenedioxy bridge, giving (+)-sesamolin (32) (Fig. 1A), we hypothesized that (+)-sesamolin might be produced by methylenedioxy bridge formation from (+)-sesamolinol. To examine this hypothesis, the microsomal fraction was incubated with (+)-sesamolinol and NADPH. However, no product of (+)-sesamolinol was observed, suggesting that CYP81Q1 does not catalyze (+)-sesamolin biosynthesis from (+)-sesamolinol (Fig. 8). Besides, other potential furofran class lignans, (+)-phillygenin and (+)-epipinoresinol, (+)-pinoresinol 4′-O-glucoside, which are found in Forsythia species, were examined (Fig. 1A) (33, 34). None of these lignans served as a substrate of CYP81Q1 (Fig. 8 and data not shown). Taken together, we concluded CYP81Q1 to be the P450 that is responsible for PSS in furofuran lignan biosynthesis.
To determine the subcellular compartment in which (+)-piperitol and (+)-sesamin biosyntheses occur, a CYP81Q1-GFP chimera protein was expressed in onion (Allium cepa) epidermal cells by means of a transient expression system (35–37). The CYP81Q1-GFP protein had an endoplasmic reticulum (ER)-like network pattern, whereas GFP was localized solely in the cytoplasm and nucleus (Fig. 9, which is published as supporting information on the PNAS web site). The CYP81Q1-GFP protein was colocalized with SP-mRFP-HDEL chimera protein (in which mRFP is monomeric red fluorescent protein), an ER marker (37–39), demonstrating that (+)-piperitol and (+)-sesamin biosyntheses occur on the ER cytoplasmic surface (Fig. 9).
Functional Characterization of CYP81Q2 and CYP81Q3 from Two Sesamum Species.
The African/Indian Sesamum spp. S. radiatum produces (+)-sesamin in its seeds (40), indicating that the dual methylenedioxy bridge-forming enzyme is present in this species. We isolated a CYP81Q1 homolog (designated CYP81Q2) from S. radiatum seeds by RT-PCR. The CYP81Q2 gene was expressed in the seeds but not in the leaves, stems, seed pods, or flowers (Fig. 10, which is published as supporting information on the PNAS web site). We also isolated a CYP81Q1 homolog (designated CYP81Q3) from an African Sesamum spp., Sesamum alatum, which is reported to lack (+)-sesamin (40). The CYP81Q3 gene was expressed substantially in immature seeds (Fig. 10). CYP81Q2 and CYP81Q3 showed a significant structural similarity to CYP81Q1 (Fig. 3 A and B).
Recombinant proteins of the two CYP81Q1 homologs were expressed in yeast, and we evaluated their enzymatic activities. Yeast microsomes expressing the CYP81Q2 gene had NADPH-dependent PSS activity (Fig. 5 C and D). Moreover, neither (+)-sesamolinol nor (+)-phillygenin was used, evidence that CYP81Q2 is a functional homolog of CYP81Q1 (Fig. 8). In contrast, microsomes of yeast expressing the CYP81Q3 cDNA had neither PS nor SS activity (Fig. 5 E and F). They showed significant absorption at 421 nm but not at 450 nm, whereas CYP81Q2 showed absorption at 449 nm, indicating that CYP81Q3 is not correctly folded (Fig. 4). Therefore, CYP81Q3 seems to be a nonfunctional P450. These results coincide with the absence of (+)-sesamin in S. alatum (40).
Discussion
Developmental Regulation of Lignan Metabolism.
In this study, we identified a Sesamum P450, CYP81Q1, that catalyzes dual methylendioxy bridge formation on (+)-pinoresinol, yielding a (+)-sesamin by means of (+)-piperitol. Thus, CYP81Q1 is characterized as a cytochrome P450 involved in lignan biosynthesis.
(+)-Sesamin rapidly decreases on germination, whereas (+)-pinoresinol and (+)-sesaminol increase (Fig. 1C). This metabolism suggests two possible regulations: (+)-sesamin biosynthesis is inactivated and/or (+)-sesamin metabolism is activated on germination. Because CYP81Q1 gene expression is absent in germinating seeds, (+)-pinoresinol accumulation should be the consequence of (+)-sesamin biosynthesis inactivation. On the other hand, the increasing (+)-sesaminol suggests that it is a major metabolite of (+)-sesamin, although the (+)-sesaminol biosynthetic mechanism is still unknown. Moreover, (+)-sesaminol and (+)-pinoresinol were undetectable in germinating seeds unless the extracted lignan mixtures were treated with β-glycosidase (data not shown), indicating that (+)-sesaminol and (+)-pinoresinol exist as their glucosides (41, 42) and suggesting that glycosylation, as well as hydroxylation, is involved in furofuran lignan metabolism.
Because CYP81Q1 is a single gene (Fig. 3C), functional redundant P450s are unlikely to exist in the S. indicum genome. Considering that CYP81Q1 did not use (+)-sesamolinol as a substrate (Fig. 8), other classes of P450s, such as CYP719A or non-P450 enzymes, may catalyze the (+)-sesamolin biosynthetic methylenedioxy bridge formation. As previously proposed in ref. 19, another potential (+)-sesamolin biosynthetic pathway from (+)-sesamin by means of (+)-epi-sesaminone also cannot be ruled out, because (+)-sesamolin conversion from (+)-sesamolinol has never been observed experimentally. Recently, we identified a recessive mutant that lacks (+)-sesamolin but has (+)-sesamin (unpublished data). The mutant will provide a clue to elucidate this issue.
Dual Catalytic Mode of CYP81Q Proteins.
Of particular interest is CYP81Q’s unique dual-catalyzing mode, because all known P450 proteins catalyze only single methylenedioxy bridge formation (26, 43). Dual catalysis might be a reasonable consequence, because (+)-pinoresinol is a symmetric lignan, and its two aromatic rings are equivalent. This dual catalysis agrees well with the fact that only small amounts of (+)-piperitol and its possible 4′-O-methylated product, (+)-kobusin, were present in the six developmental seed stages examined (Fig. 1C) (22, 23). However, it contradicts the previous study in which only (+)-piperitol biosynthetic activity in the seed microsomal fraction was detected (22). These inconsistent results might come from instability of the native CYP81Q1 protein in the microsomal fraction prepared from sesame seeds. As shown in Figs. 4 and 5, no absorption at 450 nm or enzymatic activity was observed in the microsomal fraction expressing CYP81Q3, although no obvious mutation likely to impair P450 activity was observed (Fig. 6). These results imply that CYP81Q3 has unknown mutations that result in an inactive P450 conformation. However, it is possible that this expression system is unsuitable to express CYP81Q3.
P450 proteins have a single active site coupled with heme in their folded structures (44); therefore, the dual methylenedioxy bridge formation occurs stepwise. Considering that methylenedioxy bridge formations occur on different aromatic rings located on both ends of (+)-pinoresinol, the dual catalysis is distinguished from other known P450-mediated multiple catalyses, such as dual hydroxylation of flavonoid by CYP75B and multiple oxidation in gibberellin biosynthesis by CYP88A (45, 46). (+)-Piperitol is as accessible to the substrate pocket of CYP81Q proteins as (+)-pinoresinol (Fig. 5); therefore, two alternative models for the mode of CYP81Q action could be proposed. One is that the CYP81Q proteins have a wide active site in which sequential methylenedioxy bridge formations occur. In this model, (+)-piperitol probably reverses in the active site after the first catalysis because the second methylenedioxy bridge is formed at the opposite side. Another possible model is that the (+)-piperitol produced by the first catalysis is released from the CYP81Q proteins. Most of the (+)-piperitol is recaptured in the reverse direction for the second catalysis, and a small part of the (+)-piperitol is metabolized to its derivatives, such as a (+)-kobusin.
Conserved Thr in the distal I helix is considered to be the residue that is important for O–O bond scission in the P450 reaction cycle (31, 47). Interestingly, all CYP81Q proteins have an Ala instead of a conserved Thr in the distal I helix (Fig. 6). CYP719A1 also lacks the conserved Thr but does have the Ser (26), showing that conserved Thr is not indispensable for methylenedioxy bridge-forming P450s. Notably, the conserved Thr is also replaced by Ser in isoflavone synthases (CYP93C), and the Ser residue plays an important role for the isoflavone synthetic aryl migration (48).
CYP81Q Gene Evolution.
Pedaliaceae Sesamum and Ranunculaceae Coptis are distantly related species (49). Sesamum CYP81Q1 and Coptis CYP719A1 have only 24% sequence identity to each other, although both of them are methylenedioxy bridge-forming P450s. Besides, CYP719A1 does not catalyze methylenedioxy bridge formation on (+)-pinoresinol (unpublished data). These differences suggest that these methylenedioxy bridge-forming P450s evolved independently.
(+)-Sesamin is widely found in vascular plants, in the Asterales, Scropholariales, Lamiales, Solanales, Apiales, Sapindales, Aristolochiales, Piperales, Laurales, and Magnoliales orders in the Magnoliophyta division. It even is present in Ginkgoales and Coniferales in the Gymnospermophyta division (49, 50). Its wide distribution suggests that an ortholog of PSS occurred in an ancestral plant common to both angiosperms and gymnosperms and has been widespread in vascular plants.
(+)-Pinoresinol is a common early precursor not only for furofuran lignans but also for other classes of lignans, including furan with 9(9′)-oxygen, 9,9′-dihydroxydibenzylbutane, 9,9′-dihyroxyaryltetralin, dibenzylbutyrolactone, and aryltetralinlactone (4, 19). Forsythia intermedia pinoresinol/lariciresinol reductase is known to be a branch-point enzyme for converting (+)-pinoresinol to (+)-lariciresinol and (−)-secoisolariciresinol, a furan and a 9,9′-dihydroxydibenzylbutane lignan, respectively (51). In contrast, PSS is also a pivotal branch-point enzyme that directs to the furofuran lignan biosynthetic pathway from (+)-pinoresinol. Despite the wide distribution of lignans in vascular plants, their physiological roles remain totally unknown. These roles may be elucidated by further study of lignans’ bioactivities and biosyntheses.
Materials and Methods
Plant Materials.
S. indicum cv. Masekin, the standard plant used (11), was grown in an experimental field in Tsukuba City, Ibaragi Prefecture, Japan. S. radiatum and S. alatum were grown in a greenhouse. Six developmental seed stages were defined by developmental features (Fig. 1B).
Seed stages were as follows: stage 1, seed pods no longer than 1.5 cm; stage 2, seed pods between 1.5 and 2.5 cm; stage 3, light-green seed pods longer than 2.5 cm; stage 4, deep-green seed pods longer than 2.5 cm with white seeds; stage 5, deep-green seed pods longer than 2.5 cm with brown sepals and seeds; stage 6, brown seed pods longer than 2.5 cm with fully matured seeds. Stage 6 seeds that had been soaked in water at 25°C for 1 week provided the germinating seeds.
Chemicals.
Lignans used in this study were extracted and purified from S. indicum seeds and Forsythia leaves as described in refs. 3, 33, 34, 41, and 42.
HPLC.
One gram of each sesame tissue sample was pulverized in liquid N2, and lignans were extracted with 3 ml of 80% ethanol containing 1% DMSO (vol/vol). The extracts were digested at 40°C for 5 h with 6 units/ml almond β-glucosidase (Sigma) in 0.1 M sodium phosphate buffer (pH 4.6). The resulting samples were adjusted to 50% acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid (TFA) and then centrifuged at 15,000 × g for 5 min. The supernatant (100 μl) was passed through a Millex-LH filter (0.45 μm/4 mm; Millipore) and subjected to HPLC analysis with an LC-2010A HT system (Shimadzu) equipped with a photodiode array system (SPD-M10A VP, 200–400 nm; Shimadzu) and a Develosil C30-UG-5 column (4.6 × 150 mm; Nomura Chemical, Aichi, Japan). Each sample was eluted with a linear gradient of 35–90% solvent B [90% acetonitrile containing 0.1% (vol/vol) TFA] in solvent A [H2O containing 0.1% (vol/vol) TFA] for 20 min at a flow rate of 0.6 ml/min and then was eluted with 90% solvent B for 7 min. Lignans were monitored at 280 nm. The liquid chromatography-MS/MS procedure is described in Supporting Methods, which is published as supporting information on the PNAS web site.
cDNA Library Screening.
An RNeasy Plant Mini Kit (Qiagen, Valencia, CA) was used to extract total RNA from Sesamum seeds. Poly(A)+ RNA was obtained from total RNAs by use of an oligotex-MAG mRNA purification kit (Takara Bio, Tokyo). A cDNA library was constructed with 5 μg of poly(A)+ RNA by means of a ZAP Express cDNA Synthesis Kit and ZAP Express cDNA Gigapack3 Gold Cloning Kit (Stratagene). This library had a titer of 1 × 107 plaque-forming units/ml. For the screening probes, mixtures of 13 Arabidopsis P450 genes (1, CYP90A; 2, CYP72B; 3, CYP71B; 4, CYP84A; 5, CYP96A; 6, CYP710A; 7, CYP86A; 8, CYP74; 9, CYP75B; 10, CYP79F; 11, CYP81D; 12, CYP705A; 13, CYP83A) were amplified with the gene-specific primers described in Table 1, which is published as supporting information on the PNAS web site.
PCR-based digoxigenin (DIG) labeling was performed as described in ref. 52. Approximately 300,000 plaque-forming units of the cDNA library were screened with a mixture of the 13 DIG-labeled Arabidopsis P450 probes. Screening and detection of positive clones were performed with a DIG-DNA labeling and detection kit (Roche, Mannheim, Germany). Positive clones were detected under low stringent hybridization conditions described in ref. 41, and 192 positive clones were obtained after the second screening. These clones were excised and subcloned into the pBK-CMV plasmid (Stratagene), and the nucleotide sequences were determined. Sequencing reactions were performed with a BigDye-terminator version 3.1 cycle sequencing kit (Applied Biosystems) and analyzed in a 3100 Genetic analyzer (Applied Biosystems). The resulting sequences were clustered with known P450 proteins by means of clustalw macvector 7.2.2 software (Accelrys, San Diego) (53), resulting in 17 molecular species of the SiP450 gene.
RT-PCR.
cDNAs were synthesized from the total RNA of each Sesamum plant tissue with SuperScript III (Invitrogen). The PCR with ExTaq DNA polymerase (Takara) was run at 94°C for 3 min followed by 28 cycles at 94°C for 1 min, at 54°C for 1 min, and at 72°C for 2 min. CYP81Q2 and CYP81Q3 genes were amplified with primer set 15 shown in Table 1. Sesamum ribosomal 18SRNA (AJ236041) and Sesamum p-coumarate 3-hydroxylase (AAL47545) were amplified with the primer sets 16 and 17, respectively, shown in Table 1.
Cloning.
Because SiP189 had an incomplete ORF, its 5′ end was amplified with a GeneRacer kit (Invitrogen), and primer set 14 then was determined by cycle sequencing as described above. Complete cDNA was amplified with primer set 15 and then subcloned into a pCR-Blunt II-TOPO vector (Invitrogen), resulting in CYP81Q1/pCR-Blunt-TOPOII. CYP81Q2 and CYP81Q3 genes were amplified from the cDNAs of S. radiatum and S. alatum, respectively, by primer set 15 and then subcloned into a pCR-Blunt II-TOPO vector (Invitrogen), resulting in CYP81Q2/pCR-Blunt-TOPOII and CYP81Q3/pCR-Blunt-TOPOII.
Genomic Southern Blotting Analysis.
See Supporting Methods.
Genomic Library Screening and Shotgun Sequencing of a Genomic CYP81Q1 Clone.
See Supporting Methods.
Enzymatic Assay.
Full-length cDNA of CYP81Q1 was excised from the CYP81Q1/pCR-Blunt-TOPOII plasmid with BamHI and XhoI and inserted into the BamHI and SalI sites of the pYE22m yeast expression vector, whose multiple cloning site is flanked by the GAPDH gene promoter and the GAPDH terminator (45). Yeast INVsc strain (Invitrogen) was transformed by the conventional method with pYE22m carrying CYP81Q1 cDNA (45). cDNAs of CYP81Q2 and CYP81Q3 were also introduced in pYE22m by the same procedure. Expression of the three CYP81Q genes in yeast was confirmed by RT-PCRs (data not shown). The yeasts were incubated for 36 h at 30°C in 400 ml of YNBDglc medium [0.67% (wt/vol) yeast nitrogen base/2% (wt/vol) glucose/20 mg/liter amino acids, except tryptophan]. Microsome fractions were collected by ultracentrifugation from the yeast transformant culture (26, 45, 54), and the sediments were suspended in 1 ml of suspension buffer [0.1 M potassium phosphate buffer, pH 7.4/20% (vol/vol) glycerol/0.3 μl/ml mercaptoethanol]. Microsomal absorption was measured from 400 to 500 nm with a spectrophotometer (U-3000P; Hitachi, Tokyo) as reported in refs. 45 and 54. The reaction mixture (300 μl) consisted of 250 μl of microsome/30 μl of 1 M potassium phosphate buffer, pH 7.4/10 μl of 50 mM NADPH/10 μl of lignans (1 mg/ml). It was incubated for 30 min at 30°C, and the reaction was terminated by the addition of an equal amount of 100% acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid. This mixture was subjected to HPLC and liquid chromatography-MS analyses as described above. Km values and standard errors were estimated by fitting the initial velocity data to the Michaelis–Menten equation by nonlinear regression methods (55).
Subcellular Localization.
See Supporting Methods.
Supplementary Material
Acknowledgments
We thank Dr. R. Y. Tsien (University of California, San Diego) for providing the monomeric red fluorescent protein (mRFP) vector; Dr. Y. Niwa (University of Shizuoka, Shizuoka, Japan) for the GFP vector; Drs. I. Hara-Nishimura and K. Tamura (both of Kyoto University) for the SP-mRFP-HDEL vector and confocal microscope system; Drs. H. Inui and H. Ohkawa (both of Kobe University, Kobe, Japan) for use of a spectrophotometer; Dr. F. Sato (Kyoto University) for providing the CYP719A1 plasmid; Dr. M. Kawase (National Institute of Agrobiological Science, Tsukuba, Japan) for the S. alatum and S. radiatum seeds; Y. Takeuchi, Y. Kobayashi, and H. Toyonaga for experimental support; and Drs. Y. Yamada, T. Osawa, M. Yamaguchi, T. Kusumi, W. Miki, T. Ashikari, T. Kawasaki, M. Mizutani, M. Nakao, H. Okuhara, Y. Moriguchi-Ueyama, and S. Shirato-Yasumoto for helpful discussions.
Abbreviations
- SiP450
Sesamum indicum cytochrome P450
- RT
retention time
- PSS
(+)-piperitol/(+)-sesamin synthase
- PS
piperitol synthase
- SS
sesamin synthase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the DNA Databank of Japan database [accession nos. AB194714 (CYP81Q1), AB194715 (CYP81Q2), and AB194716 (CYP81Q3)].
References
- 1.Budowski P., Markley K. S. Chem. Rev. 1951;48:125–151. doi: 10.1021/cr60149a005. [DOI] [PubMed] [Google Scholar]
- 2.Hearon W. M., MacGregor W. S. Chem. Rev. 1955;55:957–1067. [Google Scholar]
- 3.Fukuda Y., Osawa T., Namiki M., Ozaki T. Agric. Biol. Chem. 1985;49:301–306. [Google Scholar]
- 4.Umezawa T. Phytochem. Rev. 2003;2:371–390. [Google Scholar]
- 5.Bhiravamurty P. V., Kanakara R. D., Rao E. V., Sastry K. V. Curr. Sci. 1979;48:949–950. [Google Scholar]
- 6.Haller H. L., MacGovran E. R., Goodhue L. D., Sullivan W. N. J. Org. Chem. 1942;7:183–184. [Google Scholar]
- 7.Akimoto K., Kitagawa Y., Akamatsu T., Hirose N., Sugano M., Shimizu S., Yamada H. Ann. Nutr. Metab. 1993;37:218–224. doi: 10.1159/000177771. [DOI] [PubMed] [Google Scholar]
- 8.Nakai M., Harada M., Nakahara K., Akimoto K., Shibata H., Miki W., Kiso Y. J. Agric. Food Chem. 2003;51:1666–1670. doi: 10.1021/jf0258961. [DOI] [PubMed] [Google Scholar]
- 9.Umeda-Sawada R., Fujiwara Y., Abe H., Seyama Y. J. Nutr. Sci. Vitaminol. 2003;49:442–446. doi: 10.3177/jnsv.49.442. [DOI] [PubMed] [Google Scholar]
- 10.Tsuruoka N., Kidokoro A., Matsumoto I., Abe K., Kiso Y. Biosci. Biotech. Biochem. 2005;69:179–188. doi: 10.1271/bbb.69.179. [DOI] [PubMed] [Google Scholar]
- 11.Sirato-Yasumoto S., Katsuta M., Okuyama Y., Takahashi Y., Ide T. J. Agric. Food Chem. 2001;49:2647–2651. doi: 10.1021/jf001362t. [DOI] [PubMed] [Google Scholar]
- 12.Utsunomiya T., Chavali S. R., Zhong W. W., Forse R. A. Am. J. Clin. Nutr. 2000;72:804–808. doi: 10.1093/ajcn/72.3.804. [DOI] [PubMed] [Google Scholar]
- 13.Kamal-Eldin A., Pettersson D., Appelqvist L-A. Lipids. 1995;30:499–505. doi: 10.1007/BF02537023. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda S., Tohyama T., Yamashita K. J. Nutr. 2002;132:961–966. doi: 10.1093/jn/132.5.961. [DOI] [PubMed] [Google Scholar]
- 15.Nakano D., Itoh C., Takaoka M., Kiso Y., Tanaka T., Matsumura Y. Biol. Pharm. Bull. 2002;25:1247–1249. doi: 10.1248/bpb.25.1247. [DOI] [PubMed] [Google Scholar]
- 16.Nakano D., Itoh C., Ishii F., Kawanishi H., Takaoka M., Kiso Y., Tsuruoka N., Tanaka T., Matsumura Y. Biol. Pharm. Bull. 2003;26:1701–1705. doi: 10.1248/bpb.26.1701. [DOI] [PubMed] [Google Scholar]
- 17.Lee C. C., Chen P. R., Lin S., Tsai S. C., Wang B. W., Chen W. W., Tsai C. E., Shyu K. G. J. Hypertens. 2004;22:2329–2338. doi: 10.1097/00004872-200412000-00015. [DOI] [PubMed] [Google Scholar]
- 18.MacRae W. D., Tower G. H. N. Phytochemistry. 1984;23:1207–1220. [Google Scholar]
- 19.Davin L. B., Lewis N. G. Phytochem. Rev. 2003;2:257–288. [Google Scholar]
- 20.Davin L. B., Wang H.-B., Crowell A. L., Bedgar D. L., Martin D. M., Sarkanen S., Lewis N. G. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 21.Halls S. C., Davin L. B., Kramer D. M., Lewis N. G. Biochemistry. 2004;43:2587–2595. doi: 10.1021/bi035959o. [DOI] [PubMed] [Google Scholar]
- 22.Jiao Y., Davin L. B., Lewis N. G. Phytochemistry. 1998;49:387–394. [Google Scholar]
- 23.Kato M. J., Chu A. C., Davin L. B., Lewis N. G. Phytochemistry. 1998;47:583–591. [Google Scholar]
- 24.Bauer W., Zenk M. H. Phytochemisty. 1991;30:2953–2961. [Google Scholar]
- 25.Rueffer M., Zenk M. H. Phytochemisty. 1994;36:1219–1223. [Google Scholar]
- 26.Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inoue K., Sato F. J. Biol. Chem. 2003;278:38557–38565. doi: 10.1074/jbc.M302470200. [DOI] [PubMed] [Google Scholar]
- 27.Suh M. C., Kim M. J., Hur C.-G., Bae J. M., Park Y. I., Chung C.-H., Kang C.-W., Ohlrogge J. B. Plant Mol. Biol. 2003;52:1107–1123. doi: 10.1023/b:plan.0000004304.22770.e9. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda Y., Nagata M., Osawa T., Namiki M. J. Am. Oil Chem. Soc. 1986;63:1027–1031. [Google Scholar]
- 29.Anterola A. M., Jeon J.-H., Davin L. B., Lewis N. G. J. Biol. Chem. 2002;277:18272–18280. doi: 10.1074/jbc.M112051200. [DOI] [PubMed] [Google Scholar]
- 30.Akashi T., Ayabe S. Biochem. Biophys. Res. Commun. 1998;251:67–70. doi: 10.1006/bbrc.1998.9414. [DOI] [PubMed] [Google Scholar]
- 31.Imai M., Watanabe Y., Matsushima-Hibiya Y., Makino R., Koga H., Horiuchi T., Ishimura Y. Proc. Natl. Acad. Sci. USA. 1989;86:7823–7827. doi: 10.1073/pnas.86.20.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osawa T., Nagata M., Namiki M., Fukuda Y. Agric. Biol. Chem. 1985;49:3351–3352. [Google Scholar]
- 33.Kitagawa S., Hisada S., Nishibe S. Phytochemistry. 1984;23:1635–1636. [Google Scholar]
- 34.Chiba M., Hisada S., Nishibe S., Thieme H. Phytochemistry. 1980;19:335–336. [Google Scholar]
- 35.Noji M., Inoue K., Kimura N., Gouda A., Saito K. J. Biol. Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- 36.Chiu W.-L., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. Curr. Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S., Watanabe M., Nishimura M., Hara-Nishimura I. Plant J. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K., Yamada K., Shimada T., Hara-Nishimura I. Plant J. 2004;39:393–402. doi: 10.1111/j.1365-313X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- 39.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bedigian D., Seigler D. S., Harlan J. R. Biochem. Syst. Ecol. 1985;13:133–139. [Google Scholar]
- 41.Katsuzaki H., Kawasumi M., Kawakishi S., Osawa T. Biosci. Biotech. Biochem. 1992;56:2087–2088. [Google Scholar]
- 42.Katsuzaki H., Kawakishi S., Osawa T. Phytochemistry. 1994;35:773–776. doi: 10.1016/s0031-9422(00)90603-4. [DOI] [PubMed] [Google Scholar]
- 43.Clemens S., Barz W. Phytochemistry. 1996;41:457–460. [Google Scholar]
- 44.Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 45.Holton T. A., Brugliera F., Lester D. R., Tanaka Y., Hyland C. D., Menting J. G., Lu C. Y., Farcy E., Stevenson T. W., Cornish E. C. Nature. 1993;366:276–279. doi: 10.1038/366276a0. [DOI] [PubMed] [Google Scholar]
- 46.Helliwell C. A., Chandler P. M., Poole A., Dennis E. S., Peacock W. J. Proc. Natl. Acad. Sci. USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeom H., Sligar S. G., Li H., Poulos T. L., Fulco A. J. Biochemistry. 1995;14:14733–14740. doi: 10.1021/bi00045a014. [DOI] [PubMed] [Google Scholar]
- 48.Sawada Y., Kinoshita K., Akashi T., Aoki T., Ayabe S. Plant J. 2002;31:555–564. doi: 10.1046/j.1365-313x.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- 49.Soltis P. S., Soltis D. E., Chase M. W. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- 50.Umezawa T. Wood Res. 2003;90:27–110. [Google Scholar]
- 51.Dinkova-Kostova A. T., Gang D. R., Davin L. B., Bedgar D. L., Chu A., Lewis N. G. J. Biol. Chem. 1996;271:29473–29482. doi: 10.1074/jbc.271.46.29473. [DOI] [PubMed] [Google Scholar]
- 52.Yonekura-Sakakibara K., Tanaka Y., Fukuchi-Mizutani M., Fujiwara H., Fukui Y., Ashikari T., Murakami Y., Yamaguchi M., Kusumi T. Plant Cell Physiol. 2000;41:495–502. doi: 10.1093/pcp/41.4.495. [DOI] [PubMed] [Google Scholar]
- 53.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akashi T., Fukuchi-Mizutani M., Aoki T., Ueyama Y., Yonekura-Sakakibara Y., Tanaka Y., Ayabe S. Plant Cell Physiol. 1999;40:1182–1186. doi: 10.1093/oxfordjournals.pcp.a029505. [DOI] [PubMed] [Google Scholar]
- 55.Leatherbarrow R. J. Trends Biochem. Sci. 1990;15:455–458. doi: 10.1016/0968-0004(90)90295-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.