Abstract
In Escherichia coli, a family of cold shock proteins (CSPs) function as transcription antiterminators or translational enhancers at low temperature by destabilizing RNA secondary structure. A wheat nucleic acid-binding protein (WCSP1) was found to contain a cold shock domain (CSD) bearing high similarity to E. coli cold shock proteins. In the present study, a series of mutations were introduced into WCSP1, and its functionality was investigated by using in vivo and in vitro assays in the context of functional conservation with E. coli CSPs. Constitutive expression of WT WCSP1 in an E. coli cspA, cspB, cspE, cspG quadruple deletion mutant complemented its cold-sensitive phenotype, suggesting that WCSP1 shares a function with E. coli CSPs for cold adaptation. In addition, transcription antitermination activity was demonstrated for WCSP1 by using an E. coli strain that has a hairpin loop upstream of a chloramphenicol resistance gene. In vitro dsDNA melting assays clearly demonstrated that WCSP1 melts dsDNA, an activity that was positively correlated to the ability to bind ssDNA. When mutations were introduced at critical residues within the consensus RNA binding motifs (RNP1 and RNP2) of WCSP1, it failed to melt dsDNA. Studies with WCSP1-GFP fusion proteins documented patterns that are consistent with ER and nuclear localization. In vivo and in vitro functional analyses, coupled with subcellular localization data, suggest that WCSP1 may function as a RNA chaperone to destabilize secondary structure and is involved in the regulation of translation under low temperature.
Keywords: cold acclimation, cold shock protein, RNA binding, RNA chaperone, wheat
The response of prokaryotes to low temperature stress has been extensively studied in E. coli and is characterized by the accumulation of cold shock proteins (CSPs). Bacterial CSPs are small proteins that consist of a single nucleic acid-binding domain, which is termed the cold shock domain (CSD). The CSD is proposed to be an ancient molecule that was present before the origin of single-cell life (1) and is the most evolutionary conserved nucleic acid-binding domain within prokaryotes and eukaryotes (1–3). CSPs have been extensively described in >50 Gram-negative and Gram-positive bacterial species (1). E. coli contains nine CSPs (4, 5); however, its individual members are differentially regulated in response to low temperature stress (4). CspA, the most predominant CSP, may accumulate up to 10% of total proteins subsequent to low temperature exposure (6).
The structure of E. coli CspA contains five stranded β-barrel sheets with two juxtaposed consensus RNA-binding domains (RNP1 and RNP2) that reside on separate β-sheets (7, 8). A similar structure also was observed for a Bacillus subtilis CSP, CspB (9). Mutational analysis of critical aromatic core residues within RNP1 and RNP2 of CspB confirmed their essentiality for ssDNA-binding activity (10). Base stacking between critical aromatic residues within RNP1 and RNP2 enables CSPs to bind DNA (1) with apparent nonspecificity (11–13). The functional significance of bacterial CSPs to low temperature stress is directly related to RNA behavior at low temperature. RNA molecules typically form stable secondary structures in response to low temperature (14). Because of the design of prokaryotic transcriptional machinery, cold-induced RNA secondary structure may impose premature transcription termination. CspA, CspC, and CspE were confirmed to possess in vivo and in vitro transcription antitermination activity (15). CspA is also thought to enhance translation at low temperature through the elimination of stabilized RNA secondary structures (16).
CSPs are not usually found in eukaryotes, but exist as a nucleic acid-binding domain within multidomain proteins, called CSD proteins. It is believed that RNA-binding proteins evolved from small molecules such as the CSD and contain combinations of various nucleic acid binding modules (17). Eukaryotic CSD proteins are characterized by variable N-terminal sequences (18), a CSD, and diverse auxiliary C-terminal domains such as basic/aromatic islands, RG repeats and retroviral-type CCHC zinc fingers (1, 17). This feature contrasts with bacterial CSPs, which are solely comprised of a single CSD (17). Among the most widely studied eukaryotic CSD proteins is the Y-box protein family. Y-box proteins were initially named for their ability to preferentially bind the Y-box sequence (CTGATTGGYYAA) of MHC class II promoters through their CSD (19). All vertebrate Y-box proteins contain a variable N-terminal domain, a CSD, and a C-terminal auxiliary domain. As reviewed by ref. 17, although many Y-box proteins function to regulate transcription and some demonstrate RNA specificity (20), others have an mRNA stabilization role in the cytoplasm. In Xenopus oocytes, FRGY2 complexes with RNA and functions as a core component of messenger ribonucleoprotein (mRNP) complexes (21–23). FRGY2 possesses RNA-binding sequence specificity through its CSD (20, 24) and functions to control translation by masking RNA (24, 25). All of these functions are not directly related to cold shock response or cold adaptation.
In plants, a distinctive class of CSD proteins was discovered. Plant CSD homologues typically contain two distinct nucleic acid-binding modules (a single N-terminal CSD and variable quantities of C-terminal retroviral-like CCHC zinc fingers), which are interspersed by glycine-rich regions, with a few exceptions (26, 27). Our initial study identified plant CSD orthologues within 19 different genera from lower plants, monocots, dicots, and higher plants. Arabidopsis contains four unique CSD proteins and displayed differential regulation in response to low temperature stress (27). In our previous study, a CSD protein from wheat (WCSP1) was isolated and characterized (29). WCSP1 mRNA is up-regulated during cold acclimation and WCSP1 protein accumulated to high levels during cold acclimation and binds ssDNA, dsDNA, and RNA. C-terminal zinc fingers are necessary for dsDNA-binding activity and an intact CSD is sufficient for binding to both ssDNA and RNA (29). We also recently confirmed that intact WCSP1 is boiling stable and capable of binding DNA/mRNA subsequent to boiling treatments (30).
In the present study, collective observations gained from in vitro DNA binding and melting assays, in vivo functional complementation of bacterial mutants, and subcellular localization allowed us to conclude that WCSP1 is capable of functioning in a similar manner as bacterial CSPs.
Results
In Vitro ssDNA-Binding Activity of Mutant WCSP1 Constructs.
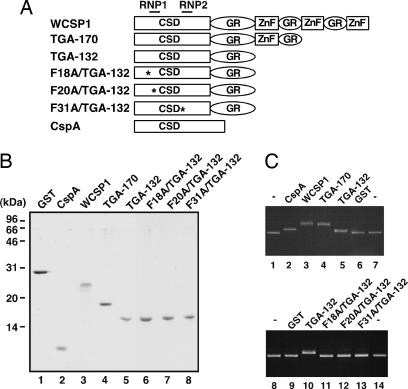
WCSP1 contains an N-terminal CSD, glycine-rich region and three C-terminal retroviral-like CCHC zinc fingers interspersed within glycine-rich domains. C-terminal zinc fingers were sequentially eliminated by adding a premature stop codon at amino acid 170 (TGA-170) and 132 (TGA-132), respectively. In addition, F to A substitutions were introduced within RNA binding domains (RNP1 and RNP2), a strategy which targeted amino acids that were found to be critical for CspB (ref. 10; Fig. 1A). All constructs were subsequently assessed for ssDNA-binding activity.
Fig. 1.
Recombinant protein purification and ssDNA-binding activity of WCSP1 and mutant WCSP1 constructs. (A) Design of WCSP1 mutant constructs. C-terminal zinc fingers were selectively eliminated from WT WCSP1 by the addition of premature stop codons and three critical residues within RNP1 and RNP2 (∗) were point mutated from F to A (F18A, F20A, and F31A). (B) Recombinant protein purification of WCSP1 and its mutant constructs. GST protein and E. coli CspA protein were used as a negative and positive control, respectively. Approximately 1 μg of each construct was separated by SDS/PAGE for visualization of recombinant proteins. (C) DNA gel-shift assay with mutant WCSP1 constructs. Approximately 500 pmol of purified recombinant fusion proteins were added to ssDNA-binding reactions and separated by agarose-gel electrophoresis. Gel shifts were subsequently visualized by EtBr staining.
Recombinant bacterial CspA, WT, and mutant WCSP1 proteins were produced as C-terminal fusions to a GST-affinity tag. Highly purified single bands of eluted intact proteins (without a GST tag) were visualized with SDS/PAGE (Fig. 1B), quantified with a Bradford protein assay and equally applied (500 pmol) to a ssDNA gel-shift assay (Fig. 1C). WT WCSP1 and TGA-170 exhibited gel shifts that were higher than the positive control CspA, an effect that is most likely attributed their difference in molecular weight. Although TGA-132 shifted ssDNA slightly in a similar level to that of CspA, constructs that were identical to TGA-132, which differed only in mutations of critical core amino acid residues (F to A substitutions within RNP1 or RNP2), completely lost ssDNA-binding abilities. Purified GST served as a negative control and confirmed that ssDNA-binding activities were specific to CspA and WCSP1 constructs. Collectively, these data indicated that RNP1 and RNP2 within the CSD are critical for the ssDNA-binding activity of WCSP1.
WCSP1 Complements a Cold Sensitive E. coli Mutant.
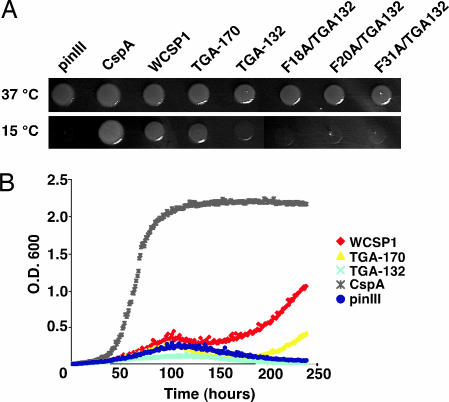
A quadruple deletion E. coli mutant (BX04), which lacks four CSPs was used to confirm the essential roles of CspA, B, E, and G for growth at low temperature (15°C) (31). We cloned WT and mutant WCSP1 constructs into an inducible pINIII expression vector (31) and used this mutant to test the functionality of WCSP1. In comparison with the positive control CspA, WT WCSP1 and TGA-170 clearly complemented the mutant. However, TGA-132, which lacked all C-terminal zinc fingers, did not recover growth of the mutant (Fig. 2A). Similarly, derivatives of TGA-132 which contained individual site-directed mutations within RNP-1 and RNP-2, failed to grow at low temperatures (Fig. 2A). Although growth recovery was delayed in comparison to CspA, WCSP1 and TGA-170 complemented the mutant in liquid culture (Fig. 2B). Collectively, these data indicated that WCSP1 shares a function with bacterial CSPs.
Fig. 2.
Complementation of a low temperature sensitivity of an E. coli cspA, cspB, cspE, cspG quadruple mutant (BX04) with WCSP1 and mutant constructs. (A) Overnight liquid cultures were adjusted for OD, uniformly spotted onto LB-carbenicillin plates, and grown at either 15 or 37°C. Growth after 5 days at 15°C clearly revealed bacterial growth for the positive control CspA, WCSP1, and TGA-170. (B) Overnight liquid cultures were adjusted for OD and added to LB-carbenicillin liquid media for a time-course growth analysis at 15°C.
WCSP1 Exhibits in Vivo Transcription Antitermination Activity.
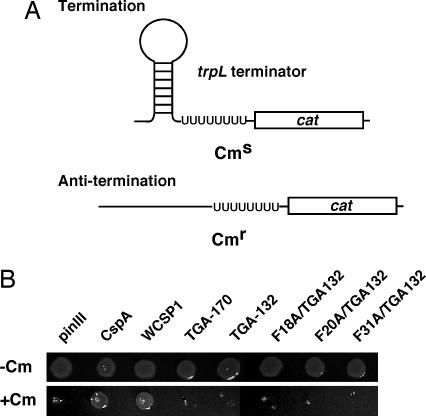
One of the evident in vivo functions of E. coli CSPs is transcription antitermination activity (15). We therefore questioned whether plant CSD proteins also are capable of functioning as transcription antiterminators; similar to E. coli CspA. An E. coli strain developed by Landick et al. (32) contains an incorporated chloramphenicol resistance gene downstream from a trpL terminator (Fig. 3A) and serves as an efficient system to assess transcription antitermination activity of CSPs (15, 33, 34). Identical pINIII constructs that were used to assess low temperature complementation were transformed into RL211 cells and were used for an in vivo determination of transcription antitermination activity (Fig. 3B). RL211 cultures were grown in liquid medium and spotted onto LB-carbenicillin plates with or without chloramphenicol. Similar to the previous report (15), RL211 cells expressing cspA grew on chloramphenicol plates, thereby confirming that our experimental approach was functioning properly. Unlike TGA-170 and TGA-132, we confirmed that WT WCSP1 exhibited transcription antitermination activity. In other words, WCSP1 has an activity that is capable of melting RNA secondary structure in vivo, and full-length WCSP1 is necessary for this activity.
Fig. 3.
Transcription anti-termination assay with RL211 E. coli strain for in vivo assessment of transcription antitermination activity. (A) RL211 contains a hairpin loop upstream from a chloramphenicol resistance gene. Expression of proteins that are capable of relaxing the hairpin loop will result in chloramphenicol resistance. (B) Fresh liquid cultures were adjusted for uniform OD, spotted onto LB-carbenicillin agar (with or without) chloramphenicol, and grown at 37°C. Colony growth was clearly observed with WCSP1, an indication that WT WCSP1 is capable of showing an in vivo RNA chaperone activity.
Nucleic Acid Melting Activity of WCSP1.
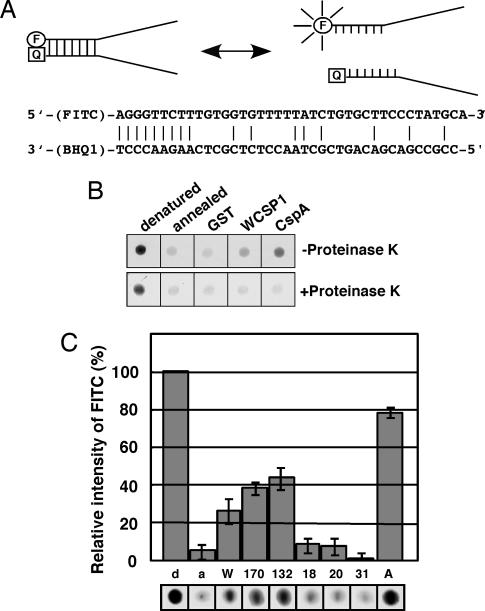
Because in vivo studies demonstrated that WCSP1 has a function common to CSPs in E. coli, we examined its nucleic acid melting activity by using an in vitro molecular beacon system as a means to quantitatively analyze molecular functions of the WCSP1 protein. Two partially complementing oligonucleotides, one of which was FITC-labeled at 5′ terminus and the other was labeled with a quencher (BHQ1) at the 3′ terminus, were used as a substrate (Fig. 4A). Because of the close proximity of the fluorescent tag and quencher, fluorescence is efficiently quenched when the two substrates remain annealed to each other. When a protein melts the substrate, the fluorescent tag and quencher spatially separate and cause fluorescence. When compared with the denatured sample, the annealed molecular beacon only emitted an ≈4% relative FITC intensity (Fig. 4B). Addition of GST resulted in no significant change, but when purified E. coli CspA was added to the reaction, fluorescence was stably increased to ≈78% of the denatured beacons (Fig. 4B). Through the use of positive (CspA) and negative (GST) controls, these data collectively corroborated our experimental approach. When purified WCSP1 was added to the reaction, fluorescence intensity increased in comparison with the negative controls and, thereby, demonstrated that WCSP1 is capable of melting the double-stranded nucleic acid. However, its ability to melt DNA was not as efficient as E. coli CspA in these in vitro assays (Fig. 4B). Addition of proteinase K to the reaction mixture after the maximum fluorescence resulted in reduced fluorescence as the degradation of WCSP1 and E. coli CspA protein occurred (Fig. 4B). These data confirmed the role of proteins in the DNA-melting activity because the quencher was able to maintain a close proximity to the fluorescence tag and suppressed the emittance of light (Fig. 4B). Taken together, we conclude that increased fluorescence of the beacon is due to nucleic acid melting that is exhibited by WCSP1.
Fig. 4.
Nucleic acid-melting activity of WCSP1. (A) Schematic representation for the model of the DNA-melting assay with molecular beacons. F, FITC; Q, black hole quencher. Annealing of two substrates quenches fluorescence (Left) and spatial separation results in fluorescence (Right). (B) Effects of GST, WT WCSP1, and CspA on melting of molecular beacons was described in Methods. Proteinase K was added to the reaction mixtures to confirm protein-mediated melting activity of WCSP1 and CspA (Lower). (C) Ability of WCSP1 and its mutant constructs to melt DNA. Relative fluorescence with each protein is shown in comparison to completely heat-denatured molecular beacons (100% relative intensity equals fluorescence from denatured beacons). d, denatured; a, annealed; W, WCSP1; 170, TGA-170; 132, TGA-132; 18, F18A/TGA-132; 20, F20A/TGA-132; 31, F31A/TGA-132; A, CspA.
Through the use of purified recombinant WCSP1 mutant proteins, it was clear that the elimination of C-terminal zinc fingers did not effect the ability of WCSP1 to melt DNA (Fig. 4C). It therefore was concluded that this ability resided within the CSD. We previously demonstrated that site-directed mutagenesis of putatively critical core amino acids within RNP-1 and RNP-2 eliminated the ssDNA-binding ability of WCSP1 (Fig. 1C). It is of particular interest to note that mutation of these core amino acids nearly eliminated any ability of WCSP1 to melt DNA (Fig. 4C). These data allow us to conclude that WCSP1 is capable of function in a similar manner to its prokaryotic counterpart.
Subcellular Localization of WCSP1.
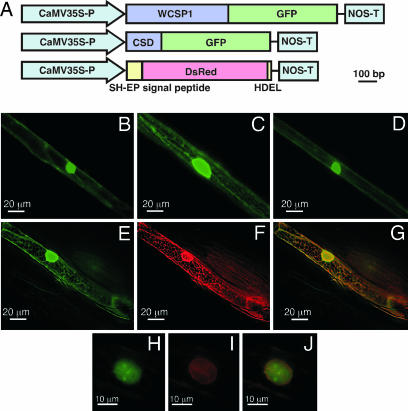
We constructed WCSP1::GFP fusion proteins to gain a better understanding for the putative function of WCSP1 in planta (Fig. 5A). Although the GFP-only positive control showed signals of cytosolic and nuclear localization (Fig. 5B), WCSP1-GFP exhibited signals within the nucleus and cortical ER network-like structures (Fig. 5C). When only the CSD region of WCSP1 was fused to GFP, the putative ER-associated labeling pattern disappeared (Fig. 5D). These data suggested that the C-terminal region of WCSP1 is a determinant for its putative ER-like localization. To confirm the ER localization of WCSP1-GFP, we cobombarded a marker gene for ER localization (DsRed-HDEL) (35). Within the identical cell, a clear GFP (Fig. 5E) and DsRed image (Fig. 5F) showed similar patterns, and the merged image showed colocalization at the ER network and nuclear envelope (Fig. 5G). These observations clearly support the notion that WCSP1 is ER localized. Nuclei of WCSP1-GFP-expressing cells exhibited a spot-like labeling pattern, bearing similarity to the pattern of Cajal bodies localization (Fig. 5H; ref. 36). Similar spots were not detected with DsRed for the ER marker protein (Fig. 5I) and did not c-localize within the merged image (Fig. 5J). From these data, it was concluded that WCSP1 localizes to ER and nuclear regions in wheat cells and requires its C-terminal region for ER localization.
Fig. 5.
Transient GFP localization in wheat epidermal cells. (A) Constructs used in the transient expression experiments. (B–D) Fluorescent images of the wheat leaf sheath epidermal cells transformed with GFP vector controls (B), WCSP1::GFP (C), and CSD::GFP (D). (E–J) Deconvolution images of the cells cobombarded with WCSP1::GFP (E and H) and an ER marker SH-EP::DsRed::HDEL (F and I). (G) A merged image of E and F. (J) A merged image of H and I.
Discussion
In prokaryotes, CSPs have been extensively characterized and known to play a critical role for the cold adaptation of cells. However, at the present time, proteins that play a similar role in the adaptation to low temperature have not yet been identified in eukaryotic cells. Wheat WCSP1 is considered as a candidate protein for the eukaryotic counterpart, because it shares CSDs with the bacterial CSPs and accumulates to higher levels in response to cold. In this study, we demonstrated that WCSP1 shares in vitro and in vivo functions with bacterial CSPs. This report sheds light on unexplored aspects of common cold adaptation mechanisms within prokaryotic and eukaryotic cells.
WCSP1 protein consists of three distinct domains, which are the CSD, CCHC zinc finger, and glycine-rich domain. CCHC zinc fingers are recognized as ssDNA nucleic acid-binding modules, and similarly, the bacterial and animal CSDs exhibit preference for ssDNA and dsDNA, respectively, as a substrate (3). In our first line of investigation, we were interested to understand the contribution of individual domains to the known ssDNA-binding activity of WCSP1 (29). In comparison with the positive control CspA, it was clearly evident that WCSP1 still maintained an ability to bind ssDNA when C-terminal zinc fingers were eliminated from WCSP1 (Fig. 1C). These data corroborate the notion that C-terminal zinc fingers are not essential for the ssDNA-binding activity of WCSP1. Because it was apparent that the ssDNA-binding activity originated from the CSD, we mutated critical core residues that were found to be essential for ssDNA binding in bacterial CspB (10). Mutation of these conserved residues within the CSD of WCSP1 resulted in a complete loss of ssDNA affinity (Fig. 1C). These data are consistent with the notion that plant CSDs may function similarly in their interaction with DNA/RNA as their prokaryotic counterparts, possibly through side-chain interactions and hydrophobic stacking.
One of the clear functions of E. coli CSPs is the suppression of transcriptional termination in the metY-rpsO operon (15). In this system, several rho-independent transcription terminators that reside within the metY-rpsO operon are destabilized by cold-induced CSPs and, as a result, downstream genes of the operon can be transcribed. Therefore, CSPs are involved in cold-induced transcriptional derepression of the metY-rpsO operon genes, including nusA, infB, rbpA, and pnp. It is interesting to note that CspE also was identified as a major nascent RNA-binding protein within the transcription complex of E. coli (37). Because of the difference in the process of transcription termination between prokaryotic and eukaryotic systems, it seems unlikely that WCSP1 functions as a transcription antiterminator in wheat; however, at this time, we cannot completely rule out its role in other processes involving transcription. Instead, the exhibited RNA chaperone activity and putative ER localization of WCSP1 suggest that WCSP1 is involved in the translation process. Under low temperature conditions, translation may be hampered by the stabilization of mRNA secondary structure (16). It therefore is reasonable to consider that the RNA chaperone activity of WCSP1 can destabilize the secondary structure and allow efficient translation under low temperature. Along similar lines, it is important to note that E. coli CspA is capable of enhancing translation under low temperature conditions (38).
Homologues of bacterial CSPs can be found as functional domain within eukaryotic multidomain proteins. Eukaryotic homologues typically contain variable N-terminal sequences (18), a CSD, and diverse auxiliary C-terminal domains such as basic/aromatic islands, RG repeats, and retroviral-like CCHC zinc fingers (1, 17). Previous studies have focused on the correlation of domain structure-to-function and found that C-terminal domains may mediate protein–protein interactions and increase the specificity of nucleic acid-binding activity (18). Unlike the majority of eukaryotic CSD proteins, plant homologues typically contain retroviral-like CCHC zinc finger domains (26, 27, 29). However, a CSD protein from C. elegans (LIN-28), which was found to be critical for developmental timing, also contains a similar modular structure with both an N-terminal CSD and CCHC zinc fingers (39). In cyanobacteria and plants, RNA recognition motif-type RNA-binding proteins have been described to accumulate in response to low temperature (40–43); however, their direct functional relation to this stress has not been clearly elucidated. Very recently, it was demonstrated that a RNA recognition motif-type RNA-binding protein is related to freeze tolerance in Arabidopsis (44).
E. coli CspE was functionally analyzed for its ability to bind ssDNA, to complement a low temperature-sensitive mutant and to exhibit transcription antitermination activity (33, 34). Unlike RNA-binding activity, DNA-melting activity and transcription antitermination were found to be directly correlated in E. coli CspE (34). DNA-melting activity also was required for the ability to complement the low temperature sensitive cspA, cspB, cspE, cspG quadruple deletion mutant (34). We have confirmed that a eukaryotic CSD protein is capable of functioning similar to E. coli counterparts. However, in our current study, we found a direct relation between the ability of WCSP1 to bind ssDNA (Fig. 1C), complement a low temperature-sensitive mutant (Fig. 2), exhibit transcription antitermination (Fig. 3), and melt DNA (Fig. 4). In all of these lines of investigation, full-length WCSP1 was found to be essential for all in vitro and in vivo activities, and site-directed mutagenesis of core amino acids with RNP1 and RNP2 determined that they were critically important. Similar to E. coli CSPs, our data collectively support the supposition that plant CSD homologues are involved in regulating translation at low temperature.
Knowledge of CSD protein subcellular localization would provide valuable information in regards to predicting putative in vivo function. In the present study, we performed transient expression analysis of WCSP1-GFP fusion proteins and found GFP localization in both the ER and nucleus. The pattern of nuclear localization was similar to that of a maize RNA recognition motif-type RNA-binding protein MA16 (45). Because WCSP1 lacks transactivation or repressor domains, the nuclear localization suggests that WCSP1 may be involved in nuclear RNA-processing events. Concerning the association to ER, it is important to note that WCSP1 lacks putative signal sequences and an ER retention signal. Furthermore, we confirmed that the C-terminal glycine-rich and zinc finger regions are necessary for ER localization. These data suggest that WCSP1 may be anchored to the ER through a protein–protein interaction. A putative association to ER networks supports the notion that WCSP1, in fact, may have an in vivo functional role that is related to translation.
It is important to note that translation at low temperature was recently determined to be functionally significant for the acquisition of freeze tolerance in Arabidopsis (46). A mutant (los1-1), which is dysfunctional in an elongation factor, has reduced freeze tolerance and has an impaired ability to translate proteins at 0°C. Furthermore, a DEAD-Box RNA helicase (LOS4) also was found to be critically involved in chilling sensitivity of Arabidopsis and, thereby, provides evidence that RNA metabolism is directly involved in plant’s response to low temperature (47, 48). Taken together with our collective functional data, these important studies indirectly support the hypothesis that CSD proteins may play a similar role and function in a similar manner to prokaryotic counterparts in planta and augment translation of a set of proteins at low temperatures and, thereby, have a positive effect on the low temperature survival of plants.
Materials and Methods
Site-Directed Mutagenesis and Plasmid Cloning.
Site-directed mutagenesis was performed with the Gene Editor kit according to suppliers instructions (Promega). A full-length WCSP1 cDNA with an in-frame N-terminal BamHI restriction site (29) was subcloned into the pBluescript S/K- vector and used as the template for mutations. Premature stop codons were created by using the following: TGA-170, 5′-CCGCGAGTGATACAAGTGC-3′; TGA-132, 5′-CCGTGGATGATACAAGTGC-′3 primers. Phenylalanine to alanine point mutations were created within the cold shock domain of the TGA-132 template by using the following mutagenic primers: (F18A, 5′-CGTCACCAAGGGGCCGGCTTCATCTCC-3′; F20A, 5′-CAAGGGGTTCGGCGCCATCTCCCCGGAG-3′; F31A, 5′-CAGCGAGGACCTCGCCGTCCACCAGTCC-3′.
For bacterial complementation studies, constructs were cloned into the pINIII vector by adding an in-frame N-terminal NdeI site with the primer 5′-TCCCATATGGGGGAGAGGGTCAAG-3′ and a C-terminal BamHI site by using 5′-TGTGGATCCTAGTGGGCCTTGTTGGG-3′. Constructs for purification of recombinant proteins were cloned into the pGEX-6P3 vector (Amersham Pharmacia Biosciences). WCSP1 and all mutant constructs contained an in-frame BamHI site, which created an N-terminal fusion to GST. A C-terminal XhoI site was added to WCSP1 and mutant constructs via PCR with the following reverse primer: 5′-TCTCTCGAGGTGGGCCTTGTTGGGGCAC-3′. E. coli CspA was amplified from the pINIII-CspA template for cloning into pGEX-6P3 with the following primers: 5′-TCTGGATCCATGTCCGGTAAAATGACTGG-3′ and 5′-TCTCTCGAGTTACAGGCTGGTTACGTTAC-3′. Amplified PCR products were digested with BamHI-NdeI and BamHI-XhoI and ligated into predigested pINIII or pGEX-6p3 vector, respectively.
The complete ORF of WCSP1 was cloned into the sGFP(s65T) (49) vector for a C-terminal fusion to GFP. The GeneEditor mutagenesis kit was used to incorporate an in-frame N-terminal NcoI site and a C-terminal SalI site with the following mutagenic primers: 5′-GCTAGGGTTTTGGTCGACCGGTTTCGGCGGCGC-3′ and 5′-CCCAACAAGGCCCACTCCATGGCACAGCTCTTCAG-3′, respectively. The CSD::GFP construct was created by PCR amplifying WCSP1::GFP template with the previously described SalI primer and the following forward NcoI primer: 5′-TCCCATGGGTGCGGTGACGTCGGA-3′. The Vigna mungo signal peptide construct (SH-EP::DsRed::HDEL), which was used for a positive control for ER localization (35), was a kind gift from Takashi Okamoto (Tokyo Metropolitan University, Tokyo).
Recombinant Protein Purification, Quantification, and SDS/PAGE Analysis.
pGEX-6P3 constructs were transformed into BL21 DE3 competent cells (Promega). Transformants were cultured in 250 ml of 2× YT-carbenicillin (100 μg/ml) medium until a 0.6 OD at 600 nm was reached and IPTG (0.5 mM) was added for a 3-h induction period. Bacterial pellets were resuspended in 5 ml of PBS and sonicated as described in ref. 29. Disrupted cells were centrifuged at 12,000 × g for 10 min at 4°C, and supernatant was applied to a glutathione-Sepharose column for purification and PreScission Protease digestion according to the supplier’s instructions (Amersham Pharmacia Biosciences). Eluate was concentrated and washed with three PBS buffer exchanges in Centricon spin columns (Millipore). Concentrated samples were spectrophotometrically quantified with the RC DC protein assay system (Bio-Rad) and equally loaded (1 μg per lane) for SDS/PAGE analysis by following standard procedures.
ssDNA-Binding Analysis.
DNA-binding analyses were performed similarly as described in ref. 29. Briefly, purified 500 pmol of individual recombinant protein constructs were added to ice cold-binding buffer (20 mM Tris·HCl, pH 7.5/2 mM EDTA/4 mM KCl/5% glycerol/50 μg/ml BSA) and M13 phage ssDNA (150 ng). Subsequent to a 10-min incubation period on ice, samples were separated on a 1% agarose gel and stained with ethidium bromide for visualization of gel shifts.
In Vitro DNA-Melting Assay.
A fluorescent molecular beacon system developed in a study in ref. 34 was modified and used for an in vitro dsDNA-melting assay. Two partially complimenting oligonucleotides were labeled with FITC and BHQ1 (black hole quencher), respectively: 5′-FITC-AGGGTTCTTTGTGGTGTTTTTATCTGTGCTTCCCTATGCA-3′ and 5′-GGCGGCTGCTGTCAGCGATTGGAGAGCGAGTTCTTGGGA-BHQ1-3′ (Hokkaido System Science, Hokkaido, Japan). The nucleotide substrates were mixed (FITC: BHQ1 = 1:20) and denatured at 95°C for 2 min and incubated on ice for annealing. Annealed DNA was used as a beacon (substrate) for the assay. One picomole molecular beacon was incubated on ice with GST, WCSP1, mutant WCSP1, or CspA proteins (300 pmol) in 15 μl of 10 mM Tris·HCl, pH 7.5, solution. Fluorescence measurements were visualized on an LAS-3000 image analyzer (Fujifilm, Japan) with excitation and emission wavelengths of 460 and 515 nm, respectively. Fluorescence intensity was calculated with Image Gauge software (Fujifilm). Proteinase K (1.4 units) was added to the reactions to check the contribution of proteins to DNA-melting activity.
Transient Expression of GFP Fusion Constructs in Wheat Leaves.
Plasmid DNA was bound to 1.0-μm gold particles (Bio-Rad) according to the manufacturer’s instruction. The DNA-covered gold particles were bombarded into 2 week-old wheat seedling leaf sheath tissue with a PDS-1000 biolistic delivery system (Bio-Rad) with a rupture setting of 1,100 psi. Bombarded leaves were maintained in the dark for 16 h at 25°C and viewed on glass slides with a FW4000 fluorescence imaging workstation (Leica, Cambridge, UK). Deconvolution images were obtained with Leica deblur software.
Acknowledgments
We thank Drs. Masayori Inouye (Robert Wood Johnson Medical School, Piscataway, NJ), Robert Landick (University of Wisconsin, Madison), Takashi Okamoto, and Yasuo Niwa (University of Shizuoka, Shizuoka, Japan) for graciously providing the E. coli strains and plasmids and Drs. Sangita Phadtare and Keita Sutoh for helpful discussions. This work was supported by grants from Japan Society for the Promotion of Science (KAKENHI Scientific Research B 15380231) and Akiyama Foundation. D.T.K. was supported by a Science and Technology Agency of Japan Postdoctoral Fellowship.
Abbreviations
- CSD
cold shock domain
- CSP
cold shock protein.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Graumann P. L., Marahiel M. A. Trends Biochem. Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe A. P., Tafuri S., Ranjan M., Familari M. New Biol. 1992;4:290–298. [PubMed] [Google Scholar]
- 3.Wolffe A. P. BioEssays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka K., Fang L., Inouye M. Mol. Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang N., Yamanaka K., Inouye M. J. Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein J., Pollitt N. S., Inouye M. Proc. Natl. Acad. Sci. USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newkirk K., Feng W., Jiang W., Tejero R., Emerson S. D., Inouye M., Montelione G. T. Proc. Natl. Acad. Sci. USA. 1994;91:5114–5118. doi: 10.1073/pnas.91.11.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schindelin H., Jiang W., Inouye M., Heinemann U. Proc. Natl. Acad. Sci. USA. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindelin H., Marahiel M. A., Heinemann U. Nature. 1993;364:164–168. doi: 10.1038/364164a0. [DOI] [PubMed] [Google Scholar]
- 10.Schroder K., Graumann P., Schnuchel A., Holak T. A., Marahiel M. A. Mol. Microbiol. 1995;16:699–708. doi: 10.1111/j.1365-2958.1995.tb02431.x. [DOI] [PubMed] [Google Scholar]
- 11.Lopez M. M., Yutani K., Makhatadze G. I. J. Biol. Chem. 1999;274:33601–33608. doi: 10.1074/jbc.274.47.33601. [DOI] [PubMed] [Google Scholar]
- 12.Lopez M. M., Makhatadze G. I. Biochim. Biophys. Acta. 2000;1479:196–202. doi: 10.1016/s0167-4838(00)00048-0. [DOI] [PubMed] [Google Scholar]
- 13.Lopez M. M., Yutani K., Makhatadze G. I. J. Biol. Chem. 2001;276:15511–15518. doi: 10.1074/jbc.M010474200. [DOI] [PubMed] [Google Scholar]
- 14.Polissi A., De Laurentis W., Zangrossi S., Briani F., Longhi V., Pesole G., Deho G. Res. Microbiol. 2003;154:573–580. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- 15.Bae W., Xia B., Inouye M., Severinov K. Proc. Natl. Acad. Sci. USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W., Hou Y., Inouye M. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 17.Sommerville J. BioEssays. 1999;21:319–325. doi: 10.1002/(SICI)1521-1878(199904)21:4<319::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. BioEssays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 19.Didier D. K., Schiffenbauer J., Woulfe S. L., Zacheis M., Schwartz B. D. Proc. Natl. Acad. Sci. USA. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouvet P., Matsumoto K., Wolffe A. P. J. Biol. Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 21.Deschamps S., Viel A., Garrigos M., Denis H., le Maire M. J. Biol. Chem. 1992;267:13799–13802. [PubMed] [Google Scholar]
- 22.Murray M. T., Schiller D. L., Franke W. W. Proc. Natl. Acad. Sci. USA. 1992;89:11–15. doi: 10.1073/pnas.89.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto K., Tanaka K. J., Aoki K., Sameshima M., Tsujimoto M. Biochem. Biophys. Res. Commun. 2003;306:53–58. doi: 10.1016/s0006-291x(03)00909-4. [DOI] [PubMed] [Google Scholar]
- 24.Bouvet P., Wolffe A. P. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 25.Ranjan M., Tafuri S. R., Wolffe A. P. Genes Dev. 1993;7:1725–1736. doi: 10.1101/gad.7.9.1725. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley P. D., Palis J. Plant Cell. 1994;6:1522–1523. doi: 10.1105/tpc.6.11.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlson D., Imai R. Plant Physiol. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mussgnug J. H., Wobbe L., Elles I., Claus C., Hamilton M., Fink A., Kahmann U., Kapazoglou A., Mullineaux C. W., Hippler M., et al. Plant Cell. 2005;17:3409–3421. doi: 10.1105/tpc.105.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlson D., Nakaminami K., Toyomasu T., Imai R. J. Biol. Chem. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- 30.Nakaminami K., Sasaki K., Kajita S., Takeda H., Karlson D., Ohgi K., Imai R. FEBS Lett. 2005;579:4887–4891. doi: 10.1016/j.febslet.2005.07.074. [DOI] [PubMed] [Google Scholar]
- 31.Xia B., Ke H., Inouye M. Mol. Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 32.Landick R., Stewart J., Lee D. N. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 33.Phadtare S., Tyagi S., Inouye M., Severinov K. J. Biol. Chem. 2002;277:46706–46711. doi: 10.1074/jbc.M208118200. [DOI] [PubMed] [Google Scholar]
- 34.Phadtare S., Inouye M., Severinov K. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto T., Shimada T., Hara-Nishimura I., Nishimura M., Minamikawa T. Plant Physiol. 2003;132:1892–1900. doi: 10.1104/pp.103.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw P. J., Brown J. W. Curr. Opin. Plant Biol. 2004;7:614–620. doi: 10.1016/j.pbi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Hanna M. M., Liu K. J. Mol. Biol. 1998;282:227–239. doi: 10.1006/jmbi.1998.2005. [DOI] [PubMed] [Google Scholar]
- 38.Brandi A., Pietroni P., Gualerzi C. O., Pon C. L. Mol. Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 39.Moss E. G., Lee R. C., Ambros V. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 40.Sato N. Nucleic Acids Res. 1995;23:2161–2167. doi: 10.1093/nar/23.12.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baudo M., Meza-Zepeda L., Palva E., Heino P. J. Exp. Bot. 1999;50:1867–1868. [Google Scholar]
- 42.Carpenter C. D., Kreps J. A., Simon A. E. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn M. A., Brown K., Lightowlers R., Hughes M. A. Plant Mol. Biol. 1996;30:947–959. doi: 10.1007/BF00020806. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y. O., Kim J. S., Kang H. Plant J. 2005;42:890–900. doi: 10.1111/j.1365-313X.2005.02420.x. [DOI] [PubMed] [Google Scholar]
- 45.Gendra E., Moreno A., Alba M. M., Pages M. Plant J. 2004;38:875–886. doi: 10.1111/j.1365-313X.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- 46.Guo Y., Xiong L., Ishitani M., Zhu J.-K. Proc. Natl. Acad. Sci. USA. 2002;99:7786–7791. doi: 10.1073/pnas.112040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong Z., Lee H., Xiong L., Jagendorf A., Stevenson B., Zhu J.-K. Proc. Natl. Acad. Sci. USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., Zhu J.-K. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]