Abstract
Multiresistant Klebsiella pneumoniae caused a nosocomial outbreak. Resistance patterns of the presumed outbreak isolates varied among and within patients. In order to control the outbreak, screening for extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae was commenced. A number of susceptible K. pneumoniae strains were stored to serve as controls in genetic strain typing. Typing by pulsed-field gel electrophoresis proved the clonality of the strains in the recognized outbreak patients. Typing of the control strains by pulsed-field gel electrophoresis showed that at least one patient had been missed by the ESBL screening procedure. Further genetic typing confirmed the presence of the SHV-5 ESBL gene in all but one of the outbreak strains. Variable presence of integrons that carried the aminoglycoside resistance genes aadB and aadA2 was found. A gyrA mutation in codon 83 was present in all outbreak strains tested, despite considerable differences in ciprofloxacin MICs. The MICs of ciprofloxacin and the chemically unrelated drug cefoxitin were correlated (r = 0.86, P < 0.01) and were compatible with the overexpression of an efflux pump in a subset of the outbreak strains. We conclude that outbreak strains that express an ESBL gene only at a low level may pass unnoticed in a screening procedure, when the laboratory is unaware of variable ESBL expression. In this particular outbreak, screening for strains for which ciprofloxacin MICs were ≥0.25 μg/ml would in retrospect have been the most sensitive method for detection of the K. pneumoniae outbreak strain.
Nosocomial outbreaks in reference centers due to multiresistant Klebsiella pneumoniae isolates have been described frequently (6, 19, 21). In The Netherlands the incidence of extended-spectrum β-lactamase (ESBL)-producing bacteria is low (22). Outbreaks appear to be rare in The Netherlands, especially in general acute-care hospitals. Many factors need to be addressed simultaneously in bringing an outbreak under control: appropriate isolation measures need to be taken, hand hygiene procedures may need reinforcement, possible environmental reservoirs need elimination, and antibiotics policies may need reconsideration. Screening in search of colonized patients needs to be instituted, and decisions on how to perform such screening have to be made (10). The aim of this study was to elucidate resistance mechanisms in the outbreak strain that are relevant to the description of the observed course of the outbreak. This was done in order to retrospectively evaluate the appropriateness of the decisions made at the beginning of the outbreak.
(Part of this work was presented at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 2001.)
MATERIALS AND METHODS
Hospital and patients.
The Zuiderzee Hospital Lelystad is a 250-bed facility providing care for a population of 70,000. The Intensive Care Unit/Coronary Care Unit (ICU) has eight beds and is able to care for three ventilated patients. The general surgical nursing ward has two units of 14 beds each. Samples for bacteriology are routinely sent to the Municipal Public Health Laboratory in Amsterdam. Patients involved in the outbreak were detected between March 1997 and June 1999. All patients with one or more cultures positive for ESBL-producing K. pneumoniae isolates were included as cases. Nosocomial infections (bloodstream infection, urinary tract infection, wound infection, or respiratory tract infection [RTI]) were classified according to Centers for Disease Control and Prevention criteria (7). If ESBL-producing Klebsiella was isolated from a patient but these criteria were not met, it was concluded that the patient was colonized. While patients positive by culture for ESBL-producing Klebsiella stayed in the ICU, these and all other patients were screened once a week or before they left the ward. In the surgical ward, patients were screened weekly if they were in the ward longer than 2 weeks, received antibiotics for reasons other than surgical prophylaxis, or had diabetes or other medical conditions predisposing to infection.
Microbiological methods.
Clinical samples (blood culture, urine, sputum, wound swab, etc.) were processed by conventional methods. Screening cultures included urine if the patient was catheterized, wound swabs if wounds were present, a sputum or throat swab, and a feces or rectal swab. Samples used to screen for epidemic multidrug-resistant (MDR) K. pneumoniae isolates were inoculated directly on MacConkey agar and into nutrient broth containing gentamicin (10 μg/ml). Fecal samples were diluted 1:1 in 0.9% NaCl prior to inoculation. After overnight incubation, broth was subcultured onto MacConkey agar. The epidemic strain was suspected if large pink colonies were present. Suspected colonies were tested for both β-glucosidase production (on CPS ID2 agar [Biomerieux, Lyon, France]) and aztreonam resistance. Resistance to aztreonam was tested by disk diffusion (Rosco, Taastrup, Denmark), by using the inhibition zone (26 mm) recommended by the manufacturer, in screening for ESBL production. Strains meeting both criteria were typed biochemically; double-disk diffusion and the E test combination of ceftazidime and ceftazidime-clavulanic acid were used for initial confirmation of ESBL production. Strains were stored at −70°C in a medium containing glycerol. Once a patient was identified as either colonized or infected, subsequent isolates with a different resistance pattern were stored as well. A number of susceptible Klebsiella strains from screened patients not colonized with resistant strains were also stored as control strains. Susceptibility testing of β-lactams and cephalosporins was repeated by the agar dilution method, performed on Mueller-Hinton agar with an inoculum of 5 × 104 CFU per spot. NCCLS breakpoint criteria were applied (14). MICs of ceftazidime, aztreonam, ciprofloxacin, cefoxitin, and gentamicin were determined.
Macrorestriction analysis of DNA by PFGE.
A representative set of the presumed K. pneumoniae outbreak isolates was characterized by pulsed-field gel electrophoresis (PFGE). Two sets of nine Klebsiella control strains were characterized by PFGE as well. The first set consisted of Klebsiella strains isolated from the ICU and surgical wards. The other control strains were identified from the internal medicine and other wards. From each isolate grown on blood agar plates, 1 colony was picked and suspended in 100 μl of EET buffer (100 mM EDTA, 10 mM EGTA, 10 mM Tris-HCl). Bacterial suspensions were embedded in agarose plugs by mixing with equal volumes of 1% agarose solution; 200-μl plugs were prepared. Plugs were incubated overnight with proteinase K (1 mg/ml) and sodium dodecyl sulfate (1%) and were subsequently washed with a buffer (10 mM Tris-1 mM EDTA) six times for 30 min each time. Subsequently the plugs were stabilized twice, for 30 min each time, in 120 μl of a buffer (SuRe cut H buffer; Boehringer, Mannheim, Germany) and were digested with 40 U of XbaI during overnight incubation at 37°C. Plugs were washed four times, for 30 min each time, with 0.5× Tris-borate-EDTA. The DNA present in the agarose plugs was analyzed on a 1% agarose gel by PFGE (CHEF DR III) at 14°C and 6 V/cm in 0.5× Tris-borate-EDTA by using pulse times of 5 to 35 s at an angle of 120° (−60° to +60°) for 20 h. The agarose gel was stained afterwards in ethidium bromide (5 mg/liter) and photographed under UV illumination. Identical patterns were assigned a letter to designate the type.
Characterization of integrons.
PCR for the integron variable region, containing one or more genes inserted as cassettes, was carried out as described by Levesque et al. (9). The sequence for the forward primer in the 5′ conserved segment was 5′-GGC ATC CAA GCA GCA AG-3′, and that for the reverse primer in the 3′ conserved segment was 5′-AAG CAG ACT TGA CCT GA-3′. DNA for PCR was isolated by suspending bacteria in Aqua dest and subsequently boiling the suspension for 10 min. The debris was spun down at 10,000 × g for 2 min. A 10-μl volume of the supernatant was added to the PCR mix and amplified under the following conditions: predenaturation at 94°C for 10 min, followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 5 min at 74°C. Amplification products were analyzed by agarose gel electrophoresis in the presence of a 100-bp DNA ladder for size assessment (Gibco/BRL Life Technologies, Breda, The Netherlands). Besides length assessment, sequencing of the inserted genes was performed by cloning of the excised amplicon. Sequencing was performed on an ABI 373 DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.) with the Thermo Sequenase cycle sequencing ready reaction kit (Amersham, Little Chalfont, Buckinghamshire, England).
SHV-PCR.
The presence of an ESBL SHV type was confirmed by PCR as described by Nuesch-Inderbinen et al. (17). Briefly, 1 bacterial colony was suspended in 200 μl of H2O, boiled for 10 min, and centrifuged for 2 min. A 10-μl volume of the supernatant was added to 40μl of a reaction buffer containing 10 mM Tris-HCl buffer (pH 9.0), 50 mM KCl, 0.01% gelatin, 0.1% Triton X-100, 1.5 mM MgCl2, 0.2 μM each deoxynucleoside triphosphate, and 0.25 U of Taq DNA polymerase (Sphaero Q, Leiden, The Netherlands) for SHV-PCR. The sequences for the forward and reverse primer were 5′-GCC CGG GTT ATT CTT ATT TGT CGC3′ and 5′-TCT TTC CGA TGC CGC CGC CAG TCA-3′, respectively. Primers were used at a concentration of 0.5 μM. Amplification involved an initial denaturation of 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C. A final extension of 10 min at 72°C was performed. DNA was amplified with a model 60 thermocycler (Biomed, Theres, Germany). SHV-PCR products were subjected to restriction enzyme digestion with NheI for 2 h at 37°C as previously described (17). A 5-μl volume of the enzyme product was mixed with 5 μl of a buffer containing 20 mM Tris acetate (pH 7.5), 20 mM magnesium acetate, and 100 mM potassium acetate and with 4 U of NheI (Pharmacia Biotech, Uppsala, Sweden). Fragments were electrophoresed in 1% agarose. For sequencing of the SHV product, the forward primer OS-1 (5′-TCG GGC CGC GTA GGC ATG AT-3′) and the reverse primer OS-2 (5′-AGC AGG GCG ACA ATC CCG CG-3′) (1) were used. The 625-bp fragment was excised, cloned, and sequenced as described above.
Quinolone resistance.
The quinolone resistance-determining regions of the gyrA and parC genes were amplified and sequenced by using PCR and the ABI Prism Big Dye cycle sequencing ready reaction kit on an ABI Prism 377 DNA sequencer (both from Perkin-Elmer Applied Biosystems) (4). The gyrA forward primer (5′-GGA TGT CCG AGA TGG CCT GAA GC-3′) and reverse primer (5′-CGC CAG ACA GCC GTT AAT CAC TT-3′) and the parC forward primer (5′-AAT GCC AGC GCC AAA TTC AAA AAG-3′) and reverse primer (5′-CCC CCA GTT TCC CTG ACC ATC C-3′) were obtained from GIBCO BRL Life Technologies (Rockville, Md.).
IEF.
The β-lactamases were provisionally characterized by isoelectric focusing (IEF) according to the method of Matthew et al. (13). Briefly, after dilution of an overnight culture in Trypticase soy broth and a further 4-h incubation at 37°C, the cells were harvested and washed once with 1 M phosphate buffer (pH 7.0). The enzyme was released by ultrasonic treatment. After centrifugation for 15 min at 12,000 × g, the supernatant was spotted onto commercially prepared polyacrylamide gel plates (pH 3 to 9) (Pharmacia LKB) and electrophoresed by using a Pharmacia gel system. Enzymes were visualized by staining with a 0.05% (500-μg/ml) solution of nitrocefin (Becton Dickinson Microbiology Systems, Cockeysville, Md.) following IEF. The isoelectric point (pI) of SHV-5 was estimated by comparison with reference strains with known pIs (TEM-1 and SHV-1).
Statistics.
Spearman's nonparametric correlations of MIC data were calculated by using SPSS for Windows software.
RESULTS
Description of the outbreak.
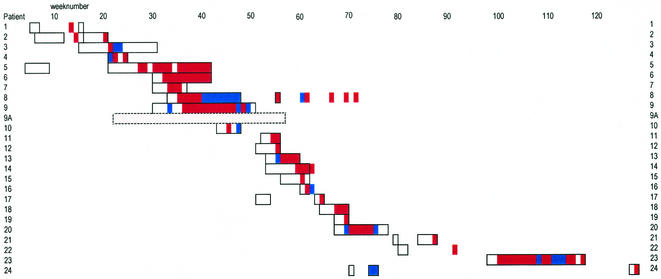
The course of the outbreak is depicted in Fig. 1, and patient characteristics are summarized in Table 1. Figure 1 includes a short “pre-outbreak” episode which shows simultaneous admission of some outbreak patients in the surgical ward. The first patient recognized as having an MDR Klebsiella infection while in the hospital was patient 2. This patient developed a lower RTI while in the ICU. The RTI was complicated with sepsis, and the patient died, despite treatment with imipenem and amikacin. Six weeks earlier, a wound swab of this patient had been taken at the surgical outpatient department. The sample had grown an ESBL-producing Klebsiella strain, but that strain was susceptible to gentamicin and ciprofloxacin. Seven weeks before patient 2 died, patient 1, then an outpatient, had MDR Klebsiella in a urine sample. Both outpatient department isolates were stored in the laboratory, but no link between these patients was immediately apparent. As soon as nosocomial spread in the ICU was noticed (patients 3 and 4), screening was commenced. No subsequent spread was found in the ICU. Patient 5 stayed at the surgical ward and had MDR Klebsiella detected while patient 3 was still in the ICU. Patient 5 perhaps contracted the MDR Klebsiella on a previous admission, while in the surgical ward with patients 1 and 2. Patients 6, 8, and 9 required prolonged isolated care. Only patient 6 was nursed in an internal ward.
FIG. 1.
Time course of an outbreak of MDR K. pneumoniae. The smallest unit in the figure is a week; boxes represent the time from admission to the end of the patient's hospital stay. If a patient was admitted for at least 1 day, the patient week is boxed. Similarly, weeks have been colored either red (for MDR Klebsiella positive) or blue (for MDR Klebsiella negative). Color without a box indicates an outpatient. No color in a box or bar means that no (screening) cultures were performed on that patient in that week. One patient (patient 9A) was detected only retrospectively and is represented by a dashed box. This patient had been screened several times, but ESBL production was not detected in K. pneumoniae isolates. PFGE typing, however, indicated that this patient carried a strain of the epidemic type.
TABLE 1.
Patient characteristics
| Patient | Sexa | Age (yr) | Historyb | Fecal carriage | Infectionc |
|---|---|---|---|---|---|
| 1 | M | 67 | Bladder carcinoma | UTI | |
| 2 | F | 64 | Vascular complications of DB | RTI + BI | |
| 3 | M | 57 | Stomach carcinoma, DB | RTI | |
| 4 | M | 81 | Cardiac failure, valve replacement | RTI + BI | |
| 5 | F | 85 | Wasting after hip fracture | + | UTI |
| 6 | M | 74 | Pancreatitis, COPD | + | RTI |
| 7 | F | 67 | Renal carcinoma, DB | + | UTI |
| 8 | M | 63 | Vascular complications of DB | + | UTI |
| 9 | F | 88 | Hip fracture | + | UTI |
| 9A | F | 84 | Fracture of pubic bone, DB | − | COL |
| 10 | F | 80 | Diabetic foot | − | COL |
| 11 | F | 74 | Colon carcinoma, DB | − | RTI |
| 12 | M | 43 | Rupture of the cecum | + | COL |
| 13 | F | 80 | Bacteremia | + | RTI |
| 14 | F | 55 | Vascular surgery | − | UTI |
| 15 | M | 71 | Hip fracture | − | COL |
| 16 | F | 42 | Breast cancer | + | COL |
| 17 | F | 76 | Rupture of the cecum | − | UTI |
| 18 | F | 77 | Hip fracture | + | UTI |
| 19 | F | 62 | Hemicolectomy | + | COL |
| 20 | F | 84 | Diabetic foot | − | COL |
| 21 | M | 61 | Bladder carcinoma | + | UTI |
| 22 | M | 78 | Urinary retention | UTI | |
| 23 | M | 74 | Femur fracture | + | UTI |
| 24 | M | 72 | Cardiac failure, DB | RTI |
M, male; F, female.
DB, diabetes; COPD, chronic obstructive pulmonary disease.
UTI, urinary tract infection; BI, bloodstream infection; COL, colonization.
The outbreak seemed to have ended with the discharge of patient 9. One month later, however, an RTI with MDR Klebsiella was found in patient 11 (a 74-year-old woman). This patient had been transferred from the surgical ward to the ICU. By the end of this subsequent episode, nine more patients had been found to be infected or colonized. In the following year, four more isolated cases occurred. Not a single case was found in the 2 years that passed since the discharge of patient 24.
In summary, MDR K. pneumoniae isolates were demonstrated in diagnostic and screening cultures of 24 patients. Ten out of 16 patients were initially found by screening of feces or rectal swab specimens. Of the 24 patients involved, 17 met criteria for a nosocomial infection and 7 were colonized. Seven patients had RTIs, two of which were complicated with bacteremia; all others had urinary tract infections. PFGE typing of control strains (see below) identified isolate 575 as part of the outbreak. The patient infected with this strain was included in Table 1 and Fig. 1 as patient 9A. During the outbreak 136 patients tested negative for ESBL-producing Klebsiella on one or more occasions.
Infection control measures.
Screening for ESBL-producing K. pneumoniae was immediately started when nosocomial spread in the ICU was noted. Patients with positive screening cultures were isolated in separate rooms or were placed together and nursed in cohort. Nurses had to put on gowns and gloves before entering the isolation room. In the first episode, most of the outbreak patients in the surgical ward were bedridden. In the second episode, most patients were mobile and were discharged as quickly as possible. However, some of those who had to stay did not adhere to their confinement in isolation and may have contributed to the spread of the strain. In addition, hand-disinfecting procedures were reinforced. The use of an alcohol-based hand-disinfecting agent was introduced first in the surgical ward and later throughout the hospital. Wet environments within the ward were checked for possible modes of transmission. Plastic washing bowls were not always properly cleaned and dried, especially during the weekend. However, cultures did not grow MDR K. pneumoniae. We decided to replace the plastic washing bowls with steel bowls (8).
Molecular typing results.
All recognized ESBL-producing strains were of PFGE type A. Some strains showed minor differences in their PFGE patterns and were assigned subtypes. Non-ESBL-producing strains isolated from recognized patients were of other PFGE types, except isolates 510 and 521. Table 2 summarizes the results. The two sets of nine Klebsiella strains that were characterized by PFGE, serving as controls, all showed different types except strain 575. This strain had apparently been missed by the screening procedure for ESBL production.
TABLE 2.
Characteristics of epidemic isolates
| Patient | Isolate | PFGE type | Integron size (bp) | SHV-PCR | GyrA mutation | Inferred efflux pump |
|---|---|---|---|---|---|---|
| 1 | 407 | A | 800, 1,000 | + | + | + |
| 2 | 406 | A1a | —,b1,000 | + | + | − |
| 409 | 800, 1,000 | + | + | + | ||
| 3 | 412 | A1 | 800, 1,000 | + | + | + |
| 4 | 413 | A1 | 800, 1,000 | + | + | − |
| 5 | 419 | A2 | 800, 1,000 | + | + | + |
| 422 | —, — | + | + | + | ||
| 498 | 800, 1,000 | + | + | − | ||
| 6 | 421 | A3 | 800,— | + | + | + |
| 480 | —, — | + | + | − | ||
| 7 | 482 | A2 | 800, 1,000 | + | + | + |
| 8 | 424 | A4 | —, — | + | + | + |
| 9 | 474 | A3 | 800,— | + | + | − |
| 510 | 800,— | + | + | − | ||
| 521 | A4′ | —, — | − | + | − | |
| 9A | 575 | A4 | 800,— | + | + | − |
| 10 | 523 | A3 | 800,— | + | + | + |
| 11 | 561 | 800,— | + | + | − | |
| 12 | 562 | A3 | 800,— | + | + | − |
| 13 | 563 | A3 | 800,— | + | + | + |
| 15 | 565 | A3 | 800,— | + | − | |
| 16 | 568 | A3 | 800,— | + | − | |
| 17 | 567 | A5 | 800,— | + | − | |
| 18 | 625 | A3 | 800,— | + | − | |
| 19 | 626 | A3 | 800,— | + | − | |
| 20 | 627 | A3 | —, — | + | − | |
| 21 | 628 | 800,— | + | |||
| 22 | 629 | —, — | + | |||
| 23 | 630 | 800,— | + | |||
| 24 | 23304 | —, — | + |
PFGE types showing close relatedness are indicated by extension numbers (subtypes).
—, gene fragment, mutation, or mechanism not detected or not present.
Results of antibiotic susceptibility testing.
Patterns of resistance to antimicrobial agents differed considerably among the isolates in testing by disk diffusion. These initial results were confirmed by quantitative MICs as shown in Table 3. All strains of the dominant PFGE type were resistant to ceftazidime, except for isolates 510, 521, and 575.
TABLE 3.
MICs of several antibiotics for the outbreak isolates
| Patient | Isolatea | MIC (μg/ml)b
|
||||
|---|---|---|---|---|---|---|
| CAZ | ATM | GEN | CIP | FOX | ||
| 1 | 407 | >256 | >256 | 64 | 0.5 | 16 |
| 2 | 406 | 256 | 256 | <0.25 | 0.25 | 4 |
| 409 | >256 | >256 | 32 | 2 | 64 | |
| 3 | 412 | >256 | >256 | 32 | 2 | 64 |
| 4 | 413 | 128 | 128 | 32 | 0.25 | 4 |
| 5 | 419 | >256 | >256 | 16 | 2 | 64 |
| 422 | 256 | 256 | <0.25 | 2 | 64 | |
| 498 | >256 | >256 | 32 | 0.25 | 2 | |
| 6 | 421 | >256 | >256 | 16 | 2 | 32 |
| 480 | 128 | 128 | 0.5 | 0.25 | 2 | |
| 7 | 482 | 256 | >256 | 16 | 2 | 64 |
| 8 | 424 | >256 | >256 | <0.25 | 2 | 64 |
| 9 | 474 | 256 | 128 | 32 | 0.25 | 4 |
| 510 | 2 | 1 | 32 | 0.25 | 4 | |
| 521 | 0.5 | <0.25 | <0.25 | 0.25 | 2 | |
| 9A | 575 | 4 | 2 | 0.5 | 0.25 | 2 |
| 10 | 523 | 256 | >256 | <0.25 | 2 | 32 |
| 11 | 561 | 256 | 128 | 32 | 0.25 | 2 |
| 12 | 562 | >256 | >256 | 32 | 0.25 | 4 |
| 13 | 563 | 256 | 256 | 32 | 4 | 16 |
| 15 | 565 | 256 | 256 | 32 | 0.25 | 4 |
| 16 | 568 | 256 | 32 | 32 | 0.25 | 4 |
| 17 | 567 | 256 | 128 | 32 | 0.25 | 4 |
| 18 | 625 | >256 | >256 | 32 | 0.25 | 4 |
| 19 | 626 | 256 | 256 | 32 | 0.25 | 4 |
| 20 | 627 | 128 | 64 | 0.5 | 0.25 | 8 |
Isolates are listed by the patients from whom they were isolated, and only the isolates differing from previous isolates from the same patient are shown. The MDR Klebsiella strain of patient 14 was lost.
ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; FOX, cefoxitin; GEN, gentamicin.
Characterization of resistance mechanisms.
To identify which β-lactamase enzyme was responsible, β-lactamase enzymes were extracted and characterized by IEF. In fact, two β-lactamases with pIs of 7.6 and 8.2 were demonstrated. The latter was suspect for SHV-5. This was confirmed by cloning and sequencing. Furthermore, strains were tested for the presence of the ESBL gene by SHV-PCR and subsequent NheI digestion of the amplicons (Table 2). Two isolates for which the ceftazidime MIC was low (isolates 510 and 575) were SHV-5 positive by this molecular technique. One isolate of the predominant PFGE type A (isolate 521) did not contain the SHV-5 gene and was fully susceptible to the antibiotics tested.
Integrons.
Integron amplicons of 800 and 1,000 bp were detected in 23 out of 30 outbreak strains (Table 2). Strains containing both fragments were found early in the outbreak, while later in the outbreak only the 800-bp fragment was detected. The 800-bp fragment contained the aadB gene. The presence of this fragment corresponded with gentamicin MICs above the breakpoint. The 1,000-bp fragment contained the aadA2 gene, coding for resistance to streptomycin.
Sequencing of gyrA and parC genes.
A TCC-to-TAC mutation of codon 83 of the gyrA gene was found in all tested PFGE type A strains, resulting in a Ser-to-Tyr amino acid change. No mutations were found in the quinolone resistance-determining region of the parC gene.
Correlation.
Spearman's nonparametric correlation coefficients of MIC data were calculated for all possible combinations of the antibiotics in Table 3. The highest correlation coefficient (0.88; P < 0.01) was found for ceftazidime and aztreonam. The next highest correlation coefficient (0.86; P < 0.01) was found for cefoxitin and ciprofloxacin. These substances are chemically unrelated but do share lipophilic properties. No correlation was found between gentamicin and any of the other antibiotics.
DISCUSSION
This paper has been focused on the striking variability of resistance patterns of the K. pneumoniae outbreak strain. The resistance pattern is the key to designing a screening procedure in order to detect and isolate patients colonized with an outbreak strain (10). ESBL production, gentamicin resistance, and ciprofloxacin resistance were noted as significant problems in the presumed outbreak strain. The screening procedure focused on ESBL production by suspected Klebsiella isolates, by testing for both β-glucosidase production and aztreonam resistance. Parallel incubation of samples in gentamicin-containing broth allowed detection of strains with gentamicin resistance as an additional feature. PFGE typing demonstrated that all the outbreak strains belonged to a single clone, despite their differences in resistance patterns. PFGE typing of the control strains identified an additional patient (patient 9A) as part of the outbreak, by detection of a strain (isolate 575) that had been missed in the screening procedure. This patient had been tested on several occasions. The differences found in susceptibility to aminoglycosides could be explained by the presence of two integron fragments, one coding for gentamicin (aadB) resistance and one for streptomycin [aadA(2)] resistance. Strains isolated early in this nosocomial outbreak mostly contained both fragments, while strains isolated later in the outbreak contained only the 800-bp fragment. Integrons have been implicated in nosocomial infections before, but we do not know of previous studies reporting the variable presence of integrons among clonal nosocomial outbreak strains (M. E. Jones, E. Peters, A. M. Weersink, A. Fluit, and J. Verhoef, Letter, Lancet 349:1742-1743, 1997). Martinez-Freijo et al. found integron fragments of similar length with identical genes in Enterobacteriaceae from different regions in Europe (11). In nine strains containing a 1,000-bp fragment, they detected the aadA gene, as we did in our K. pneumoniae strains. They also found four integron fragments of 800 bp containing the aacA4 gene; we found the aadB gene in our 800-bp fragment.
Resistance to ciprofloxacin was investigated by sequencing the gyrA and parC genes. All outbreak strains tested had the same gyrA mutation. Considerable differences in the MICs of ciprofloxacin were noted among outbreak isolates with identical topoisomerase genes. The presence or lack of an overexpressed efflux pump is a possible explanation for these MIC differences (20). A role for an efflux pump is supported by the finding of a correlation between the MICs of the unrelated antibiotics ciprofloxacin and cefoxitin. Both ciprofloxacin and cefoxitin have lipophilic properties and are thus substrates for efflux pumps of the RND type (15). In addition, cefoxitin is a poor substrate for ESBLs. The role of energy-dependent efflux in quinolone resistance has been described for K. pneumoniae isolates (5, 12). Also, the interplay between efflux pumps and topoisomerase mutations has been studied for Escherichia coli and shows a multiplicative effect on the MICs of fluoroquinolones (18).
Difficulties in the recognition of ESBL-producing bacteria have been noted before (3). In the present study, two isolates (isolates 510 and 575) belonging to the epidemic type were not recognized as ESBL producers despite the presence of the SHV-5 gene. The ceftazidime MICs for these isolates were 2 to 4 μg/ml, whereas those for recognized outbreak isolates were 128 μg/ml or higher. In retrospect, these strains ought to have been recognized as ESBL producers in our aztreonam screening procedure. These MICs are above 1 μg/ml, corresponding to the 26-mm inhibition zone recommended by the NCCLS. During the outbreak we were insufficiently aware that outbreak strains might show inhibition zones almost as wide as the threshold zone, whereas the first recognized outbreak strains did not show a zone to aztreonam at all.
Such differences in MICs may be explained, at least in part, by different levels of ESBL production. Xiang et al. reported that clonal SHV-5-producing K. pneumoniae strains differed fivefold in the level of β-lactamase production (23). Still, ceftazidime MICs for low-level β-lactamase producers were sufficiently high, 16 μg/ml or higher, for easy detection. We noted that both our missed strains lacked evidence for overexpression of an efflux pump. Resistance to β-lactams due to the multidrug efflux pump ArcAB in Salmonella enterica serovar Typhimurium has been studied by Nikaido et al. (16). A β-lactam that has more lipophilic side chains is pumped out more efficiently by the efflux pump. For instance, for nafcillin the overexpressed efflux pump raises the MIC by a factor of 128. The cefoxitin MIC for the wild-type Salmonella strain was 4 μg/ml, which is similar to those for our “low-level” Klebsiella strains (2 to 8 μg/ml). The MIC for the efflux pump-overexpressing Salmonella strain was 16 μg/ml, similar to those for our “high-level” Klebsiella strains (16 to 64 μg/ml). The study by Nikaido et al. did not include ceftazidime, but MICs of cefotaxime and ceftriaxone were increased four- and twofold, respectively. For ceftazidime the contribution of an overexpressed efflux pump would probably be in the same range. Overexpression of an efflux pump is thus unlikely to increase MICs of ceftazidime to the same extent as MICs of ciprofloxacin. However, a strain that carries the SHV-5 gene but produces this enzyme only at a low level and at the same time does not overexpress an efflux pump may pass unnoticed in a screening procedure aimed at ESBL production. Nosocomial spread may remain undetected, and an outbreak with a prolonged course may result.
Despite the frequency of multiresistance in nosocomial outbreak strains, the role of an overexpressed efflux pump in nosocomial outbreaks has not often been mentioned. In a large outbreak with Pseudomonas aeruginosa, all isolates were found to overexpress an efflux pump (2). The K. pneumoniae isolates of the outbreak reported here could retrospectively be separated into a group that did overexpress an inferred efflux pump and a group that did not. Recently developed efflux pump inhibitors for RND type pumps may facilitate studies of the epidemiology of overexpressed efflux pumps in nosocomial bacteria.(J. Blais, D. Cho, K. Tangen, C. Ford, A. Lee, O. Lomovskaya, and S. Chamberlain, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1266, 1999). Retrospectively, a screening method aimed at detecting ciprofloxacin MICs above the wild-type level, 0.25 μg/ml instead of 0.03 μg/ml, would have been a more sensitive method for detection of K. pneumoniae outbreak strains. We do not know how many more colonized patients would have been detected, or whether such an approach would have hampered the specificity of the screening procedure. The results presented here should be useful to those encountering similar problems.
REFERENCES
- 1.Arlet, G., and A. Philippon. 1991. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable beta-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 66:19-25. (Erratum, 68:125.) [DOI] [PubMed] [Google Scholar]
- 2.Bertrand, X., P. Bailly, G. Blasco, P. Balvay, A. Boillot, and D. Talon. 2000. Large outbreak in a surgical intensive care unit of colonization or infection with Pseudomonas aeruginosa that overexpressed an active efflux pump. Clin. Infect. Dis. 31:E9-E14. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deguchi, T., A. Fukuoka, M. Yasuda, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, S. Ishihara, Y. Ban, and Y. Kawada. 1997. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 41:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deguchi, T., T. Kawamura, M. Yasuda, M. Nakano, H. Fukuda, H. Kato, N. Kato, Y. Okano, and Y. Kawada. 1997. In vivo selection of Klebsiella pneumoniae strains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob. Agents Chemother. 41:1609-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French, G. L., K. P. Shannon, and N. Simmons. 1996. Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and beta-lactam-beta-lactamase inhibitor combinations by hyperproduction of SHV-5 beta-lactamase. J. Clin. Microbiol. 34:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1998. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128-140. (Erratum, 16:177.) [DOI] [PubMed]
- 8.Joynson, D. H. 1978. Bowls and bacteria. J. Hyg. (London) 80:423-425. [DOI] [PMC free article] [PubMed]
- 9.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacKenzie, F. M., and I. M. Gould. 1998. Extended spectrum beta-lactamases. J. Infect. 36:255-258. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, J. Verhoef, and M. E. Jones. 1999. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Martinez, L., I. Garcia, S. Ballesta, V. J. Benedi, S. Hernandez-Alles, and A. Pascual. 1998. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 42:1850-1852. (Erratum, 42:2772.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing: twelfth informational supplement, M100-S9, 19 (1). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuesch-Inderbinen, M. T., H. Hachler, and F. H. Kayser. 1996. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398-402. [DOI] [PubMed] [Google Scholar]
- 18.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pena, C., M. Pujol, C. Ardanuy, A. Ricart, R. Pallares, J. Linares, J. Ariza, and F. Gudiol. 1998. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 42:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prodinger, W. M., M. Fille, A. Bauernfeind, I. Stemplinger, S. Amann, B. Pfausler, C. Lass-Florl, and M. P. Dierich. 1996. Molecular epidemiology of Klebsiella pneumoniae producing SHV-5 beta-lactamase: parallel outbreaks due to multiple plasmid transfer. J. Clin. Microbiol. 34:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stobberingh, E. E., J. Arends, J. A. Hoogkamp-Korstanje, W. H. Goessens, M. R. Visser, A. G. Buiting, Y. J. Debets-Ossenkopp, R. J. van Ketel, M. L. van Ogtrop, L. J. Sabbe, G. P. Voorn, H. L. Winter, and J. H. van Zeijl. 1999. Occurrence of extended-spectrum beta-lactamases (ESBL) in Dutch hospitals. Infection 27:348-354. [DOI] [PubMed] [Google Scholar]
- 23.Xiang, X., K. Shannon, and G. French. 1997. Mechanism and stability of hyperproduction of the extended-spectrum beta-lactamase SHV-5 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 40:525-532. [DOI] [PubMed] [Google Scholar]