Acid soils are widespread throughout the world, being particularly prevalent in tropical and subtropical countries where food production is a major concern. The low pH of these soils solubilizes ionic forms of aluminum (Al) into the soil solution, and these Al species are toxic to plants, dramatically inhibiting root growth and function, which in turn results in severe crop-yield losses.
The severity of this Al toxicity constraint to crop production was placed into perspective by da Silva (1), who described the early breeding efforts aimed at developing wheat cultivars adapted to soil acidity in areas of southern Brazil. Accordingly, in the early 1900s, these acid, Al-toxic soils imposed such a dramatic selection pressure that tolerant varieties yielding only ≈800–1,000 kg per hectare were considered satisfactory, whereas completely susceptible germplasm died before giving any yield. Fortunately, breeding for Al tolerance not only in wheat but also in several other crops has been extremely successful in expanding agriculture onto large areas of acid soils throughout the world, greatly contributing to food security worldwide, particularly in the poorest regions of the globe. One of the major Al tolerance genes that has played a vital role in this success is the focus of the work by Hoekenga et al. (2) in a recent issue of PNAS. By investigating Al tolerance in the model species Arabidopsis thaliana, the authors have made a significant contribution to the understanding of some important evolutionary aspects of plant Al tolerance, which can be used to foster a new molecular perspective for Al tolerance breeding in the grasses.
Al tolerance in the tolerant wheat cultivar BH1146 is conditioned by a single major locus that controls nearly 85% of the phenotypic variation in a cross with the Al-sensitive cultivar Anahuac; this locus, designated AltBH, was genetically mapped to the long arm of chromosome 4D (3). Parallel to that, a number of physiological studies have strongly indicated that Al tolerance in wheat is mainly achieved through an exclusion mechanism that acts to keep Al away from sensitive sites in the root apex. This is achieved by the chelation of Al by malate, which is released from the wheat root apex upon exposure to Al (4, 5). The molecular basis for the AltBH gene has recently been elucidated with the cloning of the ALMT1 (aluminum activated malate transporter) gene, which was found by Sasaki et al. (6) to encode for an Al-activated malate transporter that is more highly expressed in the root apices of a tolerant wheat near-isogenic line (NIL) than in its sensitive counterpart. However, because the tolerance source for this NIL was not BH1146, evidence that the ALMT1 gene underlies AltBH was provided in a later study focusing on the genetic and physical mapping of ALMT1 in wheat. Raman et al. (7) showed that ALMT1 colocalizes with AltBH in the long arm of wheat chromosome 4D, strongly suggesting that the malate transporter corresponds to AltBH. The conserved position of the barley Al tolerance gene, Alp, on the long arm of the chromosome 4 (8) and that of Alt3 on the long arm of rye 4R (9, 10) reinforced the proposition made by Garvin and Carver (11) that Al tolerance in the Triticeae tribe (wheat, barley, and rye) is controlled by parallel mutations in orthologous loci. However, this apparent conservation appears to persist across a wider evolutionary continuum, as a major Al tolerance QTL on rice chromosome 3 is likely orthologous to the Al tolerance loci in the Triticeae (12).
Hoekenga et al. (2) found that an Arabidopsis homolog of the wheat ALMT1 gene, designated AtALMT1, plays a pivotal but not exclusive role in Arabidopsis Al tolerance by encoding an Al-activated root malate efflux transporter. However, this work also showed that an effective use of comparative genomics based on fully sequenced genomes of model plant species will require a much more sophisticated framework than that provided solely by marker-based identification of colinear regions, particularly when gene families are involved. Sequence similarity analysis to the complete sequence of the Arabidopsis genome uncovered a much more complex pattern than what was previously understood based on work in the cereal species. ALMT1 in Arabidopsis is part of a complex gene family whose members appear to differ not only in their sequence but also in their expression patterns. Interestingly, AtALMT8, which is most similar in sequence to wheat ALMT1, is not expressed in Arabidopsis roots, whereas AtALMT1 is the functional homolog expressed in root cells. In addition, whereas wheat ALMT1 is constitutively expressed, expression of Arabidopsis AtALMT1 is Al-inducible. This striking functional divergence raises an important question: when using comparative genomics to model species for gene isolation, what is the likelihood that the “best” match of a gene family actually corresponds to the functional homolog? At least for AtALMT1, it was clear that this was not the case.
One curious aspect of the work by Hoekenga et al. (2) was the detection of a major (or possibly two) QTL on Arabidopsis chromosome 1 that conditions Al tolerance and malate release in a recombinant inbred line population derived from Landsberg erecta × Columbia. This QTL lies in close physical proximity to AtALMT1, yet the data presented showed that these two loci are distinct. The authors hypothesized that the QTL could underlie an AtALMT1 activator, which could act upstream of the pathway leading to malate release by the transporter. Interestingly, a decrease in both Al tolerance and root malate release in ditelosomic lines of the wheat cultivar Chinese Spring, in which chromosome arms other than 4DL (where AltBH is located) were missing, was attributed to the loss of different genes independently influencing malate release (13). These observations suggest that Al tolerance, as conditioned by malate release, might indeed be originally a quantitative trait, yet exhibits simple inheritance patterns in crosses such as BH1146 × Anahuac because of the presence of nonlimiting alleles at accessory loci. Finally, an elegant genetic complementation test performed by Hoekenga et al. clearly reinforced the importance of background effects on the genetic control of Al tolerance. The Landsberg ecotype is more sensitive to Al than Columbia and shows a much smaller root malate release in the presence of Al. Landsberg was crossed to a T-DNA knockout mutant for AtALMT1 in the Columbia background, and the resulting F1 hybrid was analyzed for Al tolerance and malate release. The F1 hybrid showed Al tolerance and malate release similar to that seen for wild-type Columbia, which was significantly greater than in Landsberg. This finding indicates that a single copy of the Landsberg allele of AtALMT1, when present in the F1 genetic background, mediates the same level of malate transport as two copies of the Columbia allele in its native environment. Genetic background effects have been previously implicated in wheat Al tolerance (14–16). For instance, Johnson et al. (15) reported incomplete transfer of Al tolerance from Atlas to a derived NIL, which could be due either to the loss of one or more additional genes present in Atlas or to a reduced expression of tolerance in the hard red winter-type genetic background. Even more striking was the result reported by Aniol and Gustafson (17), showing that the expression of Al tolerance genes located on rye chromosomes (including 4RL where Alt3 resides) incorporated into sensitive wheat was suppressed in the wheat genetic background. Such a dramatic effect based on a single gene copy as that observed by Hoekenga et al. offered a molecular verification of this phenomenon, showing the importance of this level of regulation on the expression of the Al tolerance phenotype. This result also emphasizes the importance of choosing the more appropriate recurrent parent on backcross breeding programs, in addition to the Al tolerance donors, so that these background effects can act to the breeder’s advantage for the development of more highly Al tolerant crops.
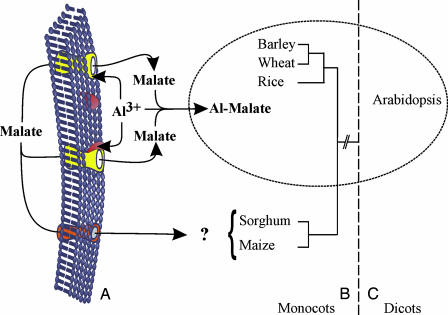
The work by Hoekenga et al. (2) was the first report of Al tolerance gene conservation between moncot and dicot species (Fig. 1). Another such example involves the orthologous genes responsible for gibberellin insensitivity in Arabidopsis, wheat, and maize (18) that condition short plant height. The conservation uncovered by Hoekenga et al. is interesting in that Al tolerance is not an inherent characteristic of wheat, but rather represents a derived state (11). The conservation of such a gene over a broad evolutionary spectrum raises the possibility that a deeper search for homologs in species such as sorghum and maize may identify yet unknown sources of Al tolerance gene diversity in these crops. Should these functional homologs not be found, would there be additional factors that were lost in the course of the evolutionary changes that gave rise to these species, which are required for their functionality? These questions provoked by the work of Hoekenga et al. (2) are of paramount importance for crop Al tolerance, and the answers may conceivably contribute to the enhancement of the current levels of Al tolerance in economically important grass species.
Fig. 1.
Range of Al tolerance conservation in plants as conferred by Al-activated malate release encoded by ALMT1. (A) The ALMT1 protein is an Al-activated malate transporter located in the root cell plasma membrane of the tolerant genotype (6). (B) Genetic mapping indicates that functional ALMT1 homologs are present in Triticeae species (3, 8, 9, 10) and in rice (12). (C) Hoekenga et al. showed that functional conservation of ALMT1 extends to the dicot, Arabidopsis. The question mark on B indicates that a putative functional ALMT1 homolog (protein in red on the plasma membrane) has yet to be found in maize and sorghum. [Image in A reprinted with permission from ref. 19 (copyright 2004 by Annual Reviews, www.annualreviews.org).]
Footnotes
See companion article on page 9738 in issue 25 of volume 103.
Conflict of interest statement: No conflicts declared.
References
- 1.da Silva A. R. Proceedings of the Workshop on Plant Adaptation to Mineral Stress in Problem Soils, Beltsville, MD; Ithaca, NY: Cornell University Agricultural Experiment Station; 1976. pp. 223–231. [Google Scholar]
- 2.Hoekenga O. A., Maron L. G., Piñeros M. A., Cançado G. M. A., Shaff J., Kobayashi Y., Ryan P. R., Dong B., Delhaize E., Sasaki T., et al. Proc. Natl. Acad. Sci. USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riede C. R., Anderson J. A. Crop Sci. 1996;36:905–909. [Google Scholar]
- 4.Delhaize E., Craig S., Beaton C. D., Bennet R. J., Jagadish V. C., Randall P. J. Plant Physiol. 1993;103:685–693. doi: 10.1104/pp.103.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delhaize E., Ryan P. R., Randall P. J. Plant Physiol. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S. J., Ryan P. R., Delhaize E., Matsumoto H. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 7.Raman H., Zhang K., Cakir M., Appels R., Garvin D. F., Maron L. G., Kochian L. V., Moroni J. S., Raman R., Imtiaz M., et al. Genome. 2005;48:781–791. doi: 10.1139/g05-054. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y., Sorrells M. E., Kochian L. V., Garvin D. F. Crop Sci. 2000;40:778–782. [Google Scholar]
- 9.Miftahudin , Scoles G. J., Gustafson J. P. Theor. Appl. Genet. 2002;104:626–631. [Google Scholar]
- 10.Miftahudin, Chikmawati T., Ross K., Scoles G. J., Gustafson J. P. Theor. Appl. Genet. 2005;110:906–913. doi: 10.1007/s00122-004-1909-0. [DOI] [PubMed] [Google Scholar]
- 11.Garvin D. F., Carver B. F. Handbook of Soil Acidity. New York: Dekker; 2003. pp. 387–406. [Google Scholar]
- 12.Nguyen B. D., Brar D. S., Bui B. C., Nguyen T. V., Pham L. N., Nguyen H. T. Theor. Appl. Genet. 2003;106:583–593. doi: 10.1007/s00122-002-1072-4. [DOI] [PubMed] [Google Scholar]
- 13.Papernik L. A., Bethea A. S., Singleton T. E., Magalhaes J. V., Garvin D. F., Kochian L. V. Planta. 2001;212:829–834. doi: 10.1007/s004250000444. [DOI] [PubMed] [Google Scholar]
- 14.Carver B. F., Ownby J. D. Adv. Agron. 1995;54:117–172. [Google Scholar]
- 15.Johnson J. P., Carver B. F., Baligar V. C. Crop Sci. 1997;37:103–108. [Google Scholar]
- 16.Tang Y., Garvin D. F., Kochian L. V., Sorrells M. E., Carver B. F. Crop Sci. 2002;42:1541–1546. [Google Scholar]
- 17.Aniol A., Gustafson J. P. Can. J. Genet. Cytol. 1984;26:701–705. [Google Scholar]
- 18.Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., Flintham J. E., Beales J., Fish L. J., Worland A. J., Pelica F., et al. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 19.Kochian L. V., Hoekenga O. A., Piñeros M. A. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]