Abstract
Her2/neu (Her2) is a tyrosine kinase belonging to the EGF receptor (EGFR)/ErbB family and is overexpressed in 20–30% of human breast cancers. We sought to characterize Her2 signal transduction pathways further by using MS-based quantitative proteomics. Stably transfected cell lines overexpressing Her2 or empty vector were generated, and the effect of an EGFR and Her2 selective tyrosine kinase inhibitor, PD168393, on these cells was characterized. Quantitative measurements were obtained on 462 proteins by using the SILAC (stable isotope labeling with amino acids in cell culture) method to monitor three conditions simultaneously. Of these proteins, 198 showed a significant increase in tyrosine phosphorylation in Her2-overexpressing cells, and 81 showed a significant decrease in phosphorylation. Treatment of Her2-overexpressing cells with PD168393 showed rapid reversibility of the majority of the Her2-triggered phosphorylation events. Phosphoproteins that were identified included many known Her2 signaling molecules as well as known EGFR signaling proteins that had not been previously linked to Her2, such as Stat1, Dok1, and δ-catenin. Importantly, several previously uncharacterized Her2 signaling proteins were identified, including Axl tyrosine kinase, the adaptor protein Fyb, and the calcium-binding protein Pdcd-6/Alg-2. We also identified a phosphorylation site in Her2, Y877, which is located in the activation loop of the kinase domain, is distinct from the known C-terminal tail autophosphorylation sites, and may have important implications for regulation of Her2 signaling. Network modeling, which combined phosphoproteomic results with literature-curated protein–protein interaction data, was used to suggest roles for some of the previously unidentified Her2 signaling proteins.
Keywords: breast cancer, cellular networks, proteomics, signal transduction, tyrosine kinase inhibitors
Her2/neu (Her2) is a receptor tyrosine kinase belonging to the EGF receptor (EGFR)/ErbB family (1). Her2 is distinguished from other members of this family, EGFR, Her3, and Her4, by its lack of a known ligand. Under normal physiologic conditions, Her2 heterodimerizes with other ErbB family members after that family member binds its ligand. Overexpression of Her2 results in constitutive, ligand-independent activation of tyrosine kinase signaling and is found in 20–30% of human breast cancers, where it is associated with a more aggressive course and poorer prognosis (2). Targeting Her2 with a monoclonal antibody, trastuzumab, is an effective therapy for Her2-positive breast cancer, but relapse or resistance to trastuzumab occurs, and clinical trials of tyrosine kinase inhibitors (TKIs) and other antibodies are under way to determine the best treatment for such patients (3). Like EGFR, Her2 recruits and activates multiple signaling proteins, including phospholipase C γ1 (PLCγ1), phosphatidylinositol 3-kinase (PI3K), Shc-Grb2-SOS, RasGAP, and Stat (signal transducer and activator of transcription) 5 (1). This recruitment occurs by means of autophosphorylation sites in the C-terminal tail of the molecule. Detailed biochemical, proteomic, and modeling studies of the EGFR signaling pathways have demonstrated a highly complex signaling network, and Her2 is likely also to use a similarly complex network to mediate cell proliferation and transformation (4–6). Further knowledge of the Her2 signaling network could potentially identify drug targets or approaches to treat trastuzumab resistance.
MS-based quantitative proteomics is a valuable tool for characterizing signaling pathways and generally utilizes stable isotopes to label cellular proteins. Stable isotopes are incorporated either by metabolic labeling, as in the SILAC (stable isotope labeling with amino acids in cell culture) method, or by chemical derivatization (7). By these methods, 113 tyrosine-phosphorylated proteins involved in EGF and platelet-derived growth factor signaling in mesenchymal stem cells and 78 EGF-induced phosphorylation events in a breast cell line have been quantified (4, 5). Here, we describe a quantitative proteomic analysis of Her2 signaling by using SILAC. We identified multiple previously unrecognized Her2 signaling proteins and observed activation loop phosphorylation in the kinase domain of Her2. Proteomic measurements were combined with literature-curated protein–protein interaction (PPI) data to suggest roles for some of the previously unidentified Her2 signaling proteins.
Results
Her2 Transfectants and PD168393 Dose Response.
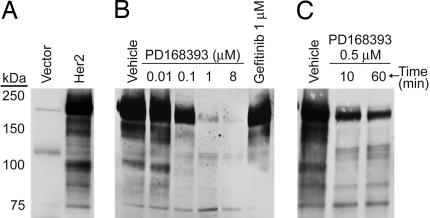
NIH 3T3 cells were stably transfected with Her2 cDNA or empty vector construct. 3T3 cells were used because they have low basal levels of ErbB family members and are a classical model for studying Her2 signaling (8). The resulting 3T3-Her2 cells had high levels of Her2 expression and displayed a transformed morphology (Fig. 7, which is published as supporting information on the PNAS web site), consistent with prior reports (8). As expected, intense tyrosine phosphorylation of multiple proteins was seen in 3T3-Her2 cells, indicating constitutive activation of Her2 signaling (Fig. 1A). PD168393 is a Her2 and EGFR selective TKI, with a reported IC50 of 5 nM in heregulin-treated MDA-MB-453 breast cancer cells (9). Treatment of 3T3-Her2 cells with PD168393 showed rapid and potent inhibition of Her2-induced tyrosine phosphorylation with half-maximal inhibition at ≈100 nM and substantial inhibition at 10 min after drug addition (Fig. 1 B and C). In contrast, 1 μM gefitinib, an EGFR selective TKI, showed minimal effect on tyrosine phosphorylation in 3T3-Her2 cells (Fig. 1B). For subsequent experiments, 100 nM PD168393 for 60 min was chosen, because this dose was expected to reduce nonspecific effects by the inhibitor and still allow detection of peaks for MS quantitation.
Fig. 1.
Effect of Her2 and PD168393 on protein tyrosine phosphorylation. (A) Western blot with anti-phosphotyrosine antibody on lysates from 3T3-Her2 or empty vector cells. (B) 3T3-Her2 cells treated for 1 h with PD168393, gefitinib, or DMSO vehicle were blotted as in A. (C) 3T3-Her2 cells treated with 0.5 μM PD168393 for the indicated times were blotted as in A.
MS with SILAC.
The SILAC protocol was performed (Fig. 2A) with normal arginine (light Arg), 13C6-Arg (medium Arg, +6-Da shift), and 13C615N4-Arg (heavy Arg, +10-Da shift) as the stable isotope label (6, 10, 11). Cell lysates were combined, and phosphotyrosine-containing proteins were purified by using anti-phosphotyrosine antibodies. Phosphoproteins were separated by SDS/PAGE, visualized by silver staining, digested with trypsin, and subjected to liquid chromatography–tandem MS. Peptide sequence was obtained from the MS/MS spectra, and phosphorylation was quantified from the relative intensities of the light, medium, and heavy Arg-labeled tryptic peptides in the MS spectra (10). For example, the Her2 peptide, ITDFGLAR, showed a 24-fold difference in the intensity of the heavy Arg and light Arg peaks and a 4-fold difference in the intensity of the heavy Arg and medium Arg peaks (Fig. 2B Upper). This finding implies that there is a 24-fold increase in phosphorylated Her2 in 3T3-Her2 cells and 4-fold inhibition of Her2 phosphorylation by 100 nM PD168393. Because tryptic digestion is performed after pull-down with anti-phosphotyrosine antibodies, the ratios of any Arg-labeled peptide can provide information on the tyrosine phosphorylation level of the protein (6, 11, 12).
Fig. 2.
Schematic for SILAC and representative MS spectra. (A) Schematic for SILAC. (B) The MS and MS/MS spectra of a tryptic peptide derived from Her2. Identified y- and b-ions are indicated. R** represents heavy Arg. (C) MS spectra showing the four major patterns of phosphorylation observed. Peptide sequences are VGQAQDILR (Dok1), VAGQSSPSGIQSR (Fyb), FFEILSPVYR (FAK), and HDGAFLIR (Grb2).
By using SILAC, 462 proteins were identified and quantified. In total, 4,000 peptide mass spectra were used to identify proteins, and, of these, 33% contained Arg and provided data for quantification. On average, proteins were identified by 8.6 peptides and quantified by 2.9 peptides. Four major patterns of change in protein phosphorylation were seen with Her2 transfection and PD168393 treatment (Fig. 2C). Like Her2, the adaptor protein Dok1 showed increased phosphorylation in 3T3-Her2 cells and inhibition of phosphorylation by PD168393. Another adaptor protein, Fyn-binding protein (Fyb), showed increased phosphorylation in 3T3-Her2 cells but no significant change in phosphorylation with PD168393. Focal adhesion kinase (FAK) showed a decrease in the 3T3-Her2 cells, and Grb2 showed no significant changes in phosphorylation. This decrease in FAK was unexpected but is consistent with observations that FAK and p130-Cas/BCAR1 are dephosphorylated in EGF-treated A431 cells (13). Grb2 is a well known Her2 interacting protein, and although some studies suggest that it can be phosphorylated, our results are consistent with the original report on Grb2, which showed that Grb2 binds to, but is not phosphorylated by, EGFR (14).
In 3T3-Her2 cells, 198 proteins showed significant (>1.5-fold) increases in phosphorylation, and 81 proteins showed a significant (<0.66-fold) decrease (Fig. 3A). As expected, the largest increase in phosphorylation was seen in Her2 itself (Fig. 3A and Table 1). The effect of PD168393 on all proteins was also quantified (Fig. 3B). Of those showing increased phosphorylation, 83 proteins were inhibited >1.5-fold by 100 nM PD168393, and 27 proteins showed a smaller degree of inhibition (1.3- to 1.5-fold), suggesting that 110 of these 198 proteins are affected by this TKI. Under these conditions, 79 proteins were not affected by PD168393, including Fyn and three subunits of PI3K (Table 1). This observation raises the question of whether different arms of the Her2 signaling pathway have differential inhibitor sensitivity. Of the 183 proteins with no significant change with Her2 transfection, only 13 showed a decrease in phosphorylation by 100 nM PD168393, suggesting that, under these conditions, few nonspecific effects of the inhibitor are seen.
Fig. 3.
Quantification of phosphorylation by SILAC. (A) Summary of fold change with Her2 for all 462 proteins, with several individual proteins highlighted. Proteins with a ratio >1.5 (upper red line) are considered as increased in their tyrosine phosphorylation level, and those with ratios <0.66 (lower red line) are considered as decreased in their tyrosine phosphorylation level. (B) Effect of PD168393 on proteins that show increased phosphorylation, no change, or decreased phosphorylation in 3T3-Her2 cells. The number of proteins in each category is shown. Areas without numbers have fewer than five proteins.
Table 1.
Effect of Her2 overexpression and PD168393 treatment on protein phosphorylation: A list of selected proteins
| Protein name | Fold change with Her2 | Fold inhibition with 100 nM PD168393 |
|---|---|---|
| Increased phosphorylation | ||
| Known Her2 signaling proteins | ||
| Her2/neu | 24.5 | 3.1 |
| HSP90-α | 4.1 | 2.2 |
| HSP90-β | 3.0 | 1.7 |
| PLCγ1 | 2.6 | 2.0 |
| PI3K p85-β | 2.2 | 0.8 |
| PI3K p85-α | 1.9 | 1.1 |
| PI3K p110-β | 1.7 | 1.0 |
| Fyn | 1.7 | 0.8 |
| Known EGFR signaling proteins | ||
| Dok1 | 4.2 | 2.0 |
| RIN1 | 3.8 | 2.0 |
| STAT1 | 3.5 | 1.6 |
| δ-Catenin | 3.5 | 2.1 |
| RAP1a | 2.8 | 0.8 |
| RhoGDI-α | 2.2 | 0.7 |
| Cortactin | 2.1 | 1.1 |
| p120-rasGAP | 1.7 | 1.5 |
| Previously unidentified Her2 signaling or effector proteins | ||
| Emerin | 9.9 | 5.8 |
| SKAP55 homologue | 8.0 | 2.1 |
| Fyn-binding protein (Fyb) | 5.3 | 1.1 |
| Semaphorin 7A | 5.8 | 0.7 |
| Transmembrane protein 33 | 4.0 | 1.6 |
| 17-β hydroxysteroid dehydrogenase 12 | 3.8 | 1.9 |
| RAB18 | 3.2 | 1.3 |
| AxI receptor tyrosine kinase | 2.8 | 1.4 |
| RIKEN cDNA 2310079N02 | 2.6 | 2.2 |
| Moesin | 2.5 | 1.2 |
| PDCD6/ALG-2 | 2.4 | 1.4 |
| Calcineurin B homologous protein 1 | 1.8 | 1.4 |
| Poly(A)-binding protein cytoplasmic 1 (PABPC1) | 1.6 | 0.9 |
| No significant change | ||
| GRB2 | 0.8 | 1.0 |
| Decreased phosphorylation | ||
| Flotillin 1 | 0.6 | 0.8 |
| p130-Cas (BCAR1) | 0.5 | 0.8 |
| Caveolin 1 | 0.5 | 0.9 |
| FAK (PTK2) | 0.5 | 1.1 |
| Growth arrest specific 1 | 0.4 | 1.0 |
Her2 Signaling Proteins.
The 198 proteins showing increased phosphorylation included several known Her2 signaling proteins and modulators of signaling, such as PLCγ1, regulatory and catalytic subunits of PI3K (p85β, p85α, and p110β), Src family member Fyn, RasGAP, and heat shock protein 90 (Table 1). Identification of known Her2 signaling proteins provides proof of concept for this approach. Several known EGFR signaling proteins, which had not been previously implicated in Her2 signaling, were identified, including Stat1, Dok1, and δ-catenin. Importantly, many previously unrecognized potential Her2 signaling proteins were identified, including the receptor tyrosine kinase Axl, the adaptor proteins SKAP55 homologue and Fyb, the small GTPases Rap1A and Rab 18, the calcium-binding protein apoptosis-linked gene 2/programmed cell death 6 (Pdcd-6/Alg-2), and the axonal guidance protein semaphorin 7A (Table 1) (15, 16). The 81 phosphoproteins that decreased in 3T3-Her2 cells include FAK, p130-Cas/BCAR1, and caveolin 1. All quantified proteins and all Arg-containing peptides that were identified are listed in Tables 2 and 3, which are published as supporting information on the PNAS web site.
Confirmation of phosphorylation status for a subset of proteins was performed by immunoprecipitation and Western blotting. PLCγ1, a known direct target of Her2 (1), was used as a positive control and showed increased phosphorylation in the 3T3-Her2 cells as compared with empty vector cells (Fig. 4A). PLCγ1 phosphorylation was inhibited by PD168393, and total PLCγ1 protein levels were unaffected. Stat1 is an SH2 (Src homology 2) domain-containing transcription factor that is phosphorylated in response to cytokines and growth factors. EGFR has been shown to activate STAT 1, 3, 5a, and 5b, whereas Her2 has thus far only been shown to activate STAT 3 and 5b (17, 18). Analogous to PLCγ1, Stat1 is phosphorylated in 3T3-Her2 cells, and phosphorylation is inhibited by PD168393 (Fig. 4A). The adaptor protein Dok1 contains a phosphotyrosine-binding domain and binds EGFR at residues pY1086 and pY1148 (19). δ-Catenin is known to be phosphorylated by EGFR and Src and forms a complex with cadherins and α-, β-, and γ-catenins, regulating cell adhesion (20). Phosphorylation of both Dok1 and δ-catenin in 3T3-Her2 cells and inhibition by PD168393 was seen (Fig. 4A).
Fig. 4.
Confirmation of protein phosphorylation by immunoprecipitation and Western blot. 3T3 transfectants (A) or BT-474 cells (B) were treated with PD168393 for 1 h and then lysed as described in ref. 11.
The phosphorylation status of two potential previously unidentified Her2 signaling molecules was also examined by Western blotting. The receptor tyrosine kinase Axl is expressed in multiple cell types, including breast epithelial cells, and can transform NIH 3T3 cells by activating PI3K and Src (21, 22). As predicted by MS, Axl phosphorylation was seen in 3T3-Her2 cells and inhibited by PD168393. In contrast, based on the MS data, the adaptor protein Fyb showed a different pattern, exhibiting increased phosphorylation in 3T3-Her2 cells, which was unaffected by PD168393 (Fig. 2C). Western blotting confirmed that Fyb is hyperphosphorylated in 3T3-Her2 cells, but we also observed that the protein level of Fyb was increased in 3T3-Her2 cells (Fig. 4A), possibly reflecting gene expression changes associated with constitutive Her2 signaling. Of the six proteins verified by immunoprecipitation and Western blotting, five showed increased phosphorylation without changes in total protein level. In comparing stably transfected cell lines, changes in protein expression can sometimes occur and may potentially affect the ratios measured by SILAC. As judged by these validation experiments, such circumstances are a minority, and the effect of short-term inhibitor treatment may provide clues as to when such expression level changes occur.
To assess the relevance of these proteins to Her2 signaling in other cell lines, we used BT-474 breast cancer cells, which overexpress endogenous Her2. Phosphorylation of PLCγ1 and Stat1 and inhibition of phosphorylation by PD168393 was seen in BT-474 cells (Fig. 4B). Thus, induction of Stat1 phosphorylation in response to Her2 can be demonstrated in a second cell line, suggesting that phosphoproteins identified by performing SILAC on 3T3-Her2 cells may be applicable to other Her2-overexpressing cell lines.
Identification of Her2 Activation Loop Phosphorylation.
We observed tyrosine phosphorylation in Her2 at position Y877, which is in the kinase domain and is distinct from the five known C-terminal tail autophosphorylation sites (Fig. 5A). Her2 Y877 is homologous to Src Y416, insulin receptor Y1162, and EGFR Y869, also termed Y845 in an alternate numbering system (Fig. 5B). These conserved tyrosine residues reside in the activation loop, a region that is autophosphorylated in many protein kinases and is a regulator of kinase activity (23, 24). To verify this phosphorylation site by an independent method, a phosphospecific antibody against this site was used, and strong phosphorylation of Y877 was detected in 3T3-Her2 and BT-474 cells (Fig. 5C). Y877 phosphorylation was inhibited by PD168393, implying that this phosphorylation event depends on Her2 kinase activity.
Fig. 5.
Identification of the Y877 phosphorylation site in the Her2 kinase domain. (A) MS/MS spectra of the peptide LLDIDETE(pY)HADGGKVPIK, which bears the phosphotyrosine residue at Y877 of Her2. The charge state of the parent ion is +3, thus yielding both +1 (upper line) and +2 (lower line) charged daughter ions on fragmentation. The mass difference corresponding to the phosphotyrosine residue, y11 minus y10 ion, is seen in both +1 and +2 charged fragment series. (B) clustalw 1.82 alignment of the kinase domains of human Her2, EGFR, Src, and insulin receptor. The phosphopeptide identified by MS in A is underlined. Black and gray arrows mark the conserved tyrosine residue and EGFR L858, respectively. The activation loop is indicated by horizontal arrows. (C) 3T3 transfectants or BT-474 cells were treated as in Fig. 4, and Western blotting was performed with phospho-Y877-specific Her2 antibody or anti-Her2 antibody.
Network Modeling by Expert Literature Curation.
To understand the role of proteins identified here, in Her2 signaling, we combined the above MS data with PPI information available in the Human Protein Reference Database (HPRD). HPRD currently contains 33,710 experimentally reported PPIs extracted from the scientific literature (25). Proteins identified by MS were overlaid on a literature-curated EGFR/ErbB pathway obtained from HPRD, and PPIs linking the previously unidentified and known signaling proteins were sought. In this manner, two previously unidentified Her2 signaling proteins were directly connected to the EGFR/ErbB signaling pathway (Fig. 6A). Pdcd-6/Alg-2 forms a complex with CIN85/SETA [Casitas B-lineage (Cbl)-interacting protein of 85 kDa/SH3-domain encoding, expressed in tumorigenic astrocytes] and AIP1/Alix, and they, in turn, bind Cbl and EGFR (26, 27). AIP1/Alix inhibited EGFR internalization and degradation; therefore, we hypothesize that Pdcd-6/Alg-2 protects Her2 from Cbl-mediated degradation. Similarly, poly(A)-binding protein cytoplasmic 1 (PABPC1) binds and colocalizes with paxillin, and disruption of this interaction inhibited cell migration (28). In 3T3-Her2 cells, Pabpc1 showed increased phosphorylation (Table 1), and paxillin showed borderline decreased phosphorylation (fold change of 0.68; Table 2), suggesting that Her2-induced phosphorylation of Pabpc1 may affect focal adhesions.
Fig. 6.
Network modeling. (A) A focused view of a module of the EGFR/ErbB signaling pathway linking two proteins identified in this study, Pdcd6 and Pabpc1. Gene symbols used are from HPRD. Yellow and purple boxes represent known and previously unrecognized Her2 signaling proteins identified in this study, respectively. White boxes represent connecting proteins not identified by MS but known to be involved in EGFR signaling. (B) Bayesian network model showing a directed graph of influence on protein phosphorylation. Blue hexagons, tan ovals, and white ovals represent ErbB receptor tyrosine kinases, other protein kinases, and other proteins, respectively. Edges in green, yellow, and red are conserved in >400, >300, and >200 of the 500 highest scoring networks, respectively. Based on the underlying assumptions of Bayesian networks, loops are not permitted, and red edges represent alternate routes taken by the simulation on different runs.
Network Modeling by Machine Learning.
For broader understanding of Her2 signaling pathways, we used a form of machine learning called Bayesian networks, which are probabilistic models depicting the influence of one variable on another with a graph of nodes and directed edges (29). In this case, each node represents the phosphorylation level of a protein, and each edge represents the causal influence of an upstream protein on the phosphorylation level of a downstream protein. Arrows indicate the direction of the edges, and they should not be interpreted as direct kinase–substrate relationships. The Bayesian network (Fig. 6B) was constructed by using the phosphorylation levels measured here by SILAC combined with three published proteomic studies of EGFR signaling (4–6). Integration of multiple studies was needed, because Bayesian network modeling requires measurements of each node under multiple conditions. The 90 proteins measured in at least two data sets (Fig. 8A, which is published as supporting information on the PNAS web site) were used to initiate network modeling. Bayesian network edges were restricted to PPIs in HPRD connecting pairs of the 90 proteins, yielding 157 edges connecting 55 proteins; the 35 proteins not connected to others in the set were dropped from the model. The search space was sampled by scoring 8 billion possible networks, and the common directed edges present in the 500 highest scoring networks were compiled (Fig. 6B). The inferred network bears similarity to the known biochemical signaling pathway, and interesting predictions arising from it include the following: (i) Axl influences the phosphorylation of the PI3K regulatory subunit (PIK3R2), which is supported by the literature (22) and (ii) phosphorylation of ephrin receptor A2 (EphA2) in response to EGFR or Her2 involves Grb2 or Shc1. EphA2 phosphorylation may also involve Src, as suggested by a recent proteomic study on Src (12). The advantage of the Bayesian network modeling performed here is that it combines PPI data with quantitative proteomic data to infer de novo a network that both recapitulates known portions of the signaling pathway and suggests new relationships between proteins.
Discussion
Use of quantitative proteomics to study signal transduction permits a comprehensive strategy to characterize protein networks and pathways. In this study, we obtained quantitative measurements on 462 proteins in Her2-transfected cells and, by simultaneously comparing three conditions, measured the effect of a Her2-targeted TKI. PD168393 is a preclinical compound used in the design of CI-1033, a TKI that is currently in clinical trials (30); therefore, this approach can be applied to drugs that are in clinical use or development to understand their effects on cellular networks. The identified phosphoproteins included many known Her2 and EGFR signaling proteins, as well as multiple previously unidentified Her2 signaling proteins, which should significantly advance the understanding of Her2. Evidence of Her2 activation loop phosphorylation at Y877 was obtained by MS and confirmed by phosphospecific antibody. Finally, two network modeling approaches were used to infer possible relationships between proteins identified by MS.
The role of the activation loop in regulating kinase activity has been studied by many groups. Autophosphorylation of the activation loop in protein kinase A, insulin receptor tyrosine kinase, and Src yields a 5- to 500-fold increase in kinase activity (23, 24). Mutations of other residues in the EGFR activation loop, such as the L858R mutation seen in human lung cancer and the mouse gain-of-function mutation L861Q, have dramatic effects on kinase activity, downstream signaling, and small-molecule inhibitor sensitivity (31–33). Although a role for activation loop phosphorylation in EGFR and Her2 has been controversial (34–37), our demonstration of Her2 Y877 phosphorylation warrants renewed interest in this site.
Although MS studies can identify previously uncharacterized proteins involved in a signaling pathway, significant issues of determining the proteins' function and role remain. Bioinformatics and computational approaches can streamline this process. We present two complementary network modeling methods that offer different insights into the same data set: one relying on expert literature curation and the other relying on machine learning through Bayesian networks. The expert literature curation method suggested roles for previously unidentified proteins within Her2 signaling pathways. In contrast, the Bayesian network approach generated de novo a probabilistic network representing core aspects of Her2 and EGFR signaling. The Bayesian approach can integrate multiple proteomic data sets and should become more powerful, given the anticipated growth of data resources. Both network modeling approaches are intended to generate hypotheses, and experimental validation of their inferences will be needed.
In conclusion, this study extends our knowledge of Her2 signaling by identifying previously uncharacterized downstream signaling proteins, demonstrating activation loop phosphorylation in Her2, and using network modeling to generate hypotheses about the role of several previously unidentified proteins. Given the importance of Her2 in breast cancer and other diseases, this study provides valuable leads for designing future therapies.
Materials and Methods
Cell Lines and Transfection.
Her2 cDNA (a gift from Dan Leahy, Johns Hopkins University School of Medicine) was cloned into pIRES-neo3 (BD Biosciences Clontech). NIH 3T3 cells (American Type Culture Collection) were transfected with Lipofectamine 2000 (Invitrogen), and G418-resistant clones were selected. PD168393 (Calbiochem) or gefitinib (Qventas, Branford, CT) was dissolved in DMSO, and cells were treated as indicated. 3T3 and BT-474 cells (American Type Culture Collection) were incubated in serum-free media for 4 h and overnight, respectively, before all experiments.
MS.
SILAC, phosphotyrosine immunoaffinity purification, and tryptic digests were performed as described in ref. 11. Equal numbers of cells (3.3 × 108) were used for each labeling state. Tryptic peptides were separated by a reverse-phase nano-liquid chromatography system (1100 Series HPLC system; Agilent Technologies, Palo Alto, CA) connected to an electrospray ion source and a Q-STAR Pulsar mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). The resulting peak list files were analyzed by mascot (Matrix Science, Boston) by using the National Center for Biotechnology Information RefSeq database, and peptides were quantified by using msquant open source software (10).
Antibodies, Immunoprecipitation, and Western Blotting.
Antibodies were purchased as follows: Her2 and Dok1 (Santa Cruz Biotechnology); phosphotyrosine (4G10) and Fyb/ADAP (Upstate Biotechnology, Lake Placid, NY); δ-catenin (Abcam, Inc., Cambridge, MA); and phospho-PLCγ1 (Y783), PLCγ1, phospho-STAT1 (Y701), STAT1, Axl, and phospho-Her2 Y877 (Cell Signaling Technology, Beverly, MA). All data shown are representative of at least two experiments.
Network Modeling.
EGFR/ErbB pathway and PPI files were downloaded from the HPRD web site. Proteins showing increased phosphorylation in 3T3-Her2 cells were searched against the PPI file, using customized Perl scripts, to find PPIs linking previously unidentified Her2 signaling proteins to a component of the EGFR/ErbB pathway. Visualization of a focused portion of the network was done by using genmapp (38).
Bayesian network modeling was conducted as follows. The four proteomic studies were joined by converting their protein database accession numbers to human gene symbols, and their phosphorylation measurements were represented as discrete states (increased, no change, or decreased). A total of seven steady-state sets of observation were obtained from these four studies, and a Bayesian network structure learning algorithm, banjo 1.0.5 (39), was run. The network edge list was constrained to the HPRD data set of PPIs, and EGFR and ERBB2 (Her2) were defined as the causal start of the network. banjo was run 500 times, with each run searching 16 million networks and returning the highest scoring network. These high-scoring networks were compared, and the common edges were compiled. Further details on network modeling are provided in Supporting Methods and Tables 4–7, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Dan Leahy and Aruna Sathyamurthy (Johns Hopkins University School of Medicine) for Her2 cDNA and helpful discussions and Suresh Mathivanan for assistance with programming. This work was supported by the Susan G. Komen Breast Cancer Foundation (R.B.); a joint award from the American Society of Clinical Oncology and Breast Cancer Research Foundations (to R.B.); Department of Energy Grant DE-FG0204ER25626 (to A.S.P.); the Whitaker Foundation (J.S.B.); and National Institutes of Health Grants U54RR020839 (to J.S.B. and A.P.), CA106424 (to A.P.), and CA74305 (to P.A.C.).
Abbreviations
- SILAC
stable isotope labeling with amino acids in cell culture
- TKI
tyrosine kinase inhibitor
- PPI
protein–protein interaction
- HPRD
Human Protein Reference Database
- EGFR
EGF receptor
- PLCγ1
phospholipase C γ1
- PI3K
phosphatidylinositol 3-kinase
- FAK
focal adhesion kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Yarden Y., Sliwkowski M. X. Nat. Rev. Mol. Cell. Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., McGuire W. L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 3.Spector N. L., Xia W., Burris H., III, Hurwitz H., Dees E. C., Dowlati A., O’Neil B., Overmoyer B., Marcom P. K., Blackwell K., et al. J. Clin. Oncol. 2005;23:2502–2512. doi: 10.1200/JCO.2005.12.157. [DOI] [PubMed] [Google Scholar]
- 4.Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. Mol. Cell. Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Blagoev B., Ong S. E., Kratchmarova I., Mann M. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 7.Ong S. E., Mann M. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 8.Di Fiore P. P., Pierce J. H., Kraus M. H., Segatto O., King C. R., Aaronson S. A. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]
- 9.Fry D. W., Bridges A. J., Denny W. A., Doherty A., Greis K. D., Hicks J. L., Hook K. E., Keller P. R., Leopold W. R., Loo J., et al. Proc. Natl. Acad. Sci. USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong S. E., Kratchmarova I., Mann M. J. Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 11.Amanchy R., Kalume D. E., Pandey A. Sci. STKE. 2005 doi: 10.1126/stke.2672005pl2. pl2. [DOI] [PubMed] [Google Scholar]
- 12.Qiao Y., Molina H., Pandey A., Zhang J., Cole P. A. Science. 2006;311:1293–1297. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 13.Lu Z., Jiang G., Blume-Jensen P., Hunter T. Mol. Cell. Biol. 2001;21:4016–4031. doi: 10.1128/MCB.21.12.4016-4031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowenstein E. J., Daly R. J., Batzer A. G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E. Y., Bar-Sagi D., Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 15.Tarabykina S., Mollerup J., Winding P., Berchtold M. W. Front. Biosci. 2004;9:1817–1832. doi: 10.2741/1358. [DOI] [PubMed] [Google Scholar]
- 16.Pasterkamp R. J., Peschon J. J., Spriggs M. K., Kolodkin A. L. Nature. 2003;424:398–405. doi: 10.1038/nature01790. [DOI] [PubMed] [Google Scholar]
- 17.Olayioye M. A., Beuvink I., Horsch K., Daly J. M., Hynes N. E. J. Biol. Chem. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 18.Ren Z., Schaefer T. S. J. Biol. Chem. 2002;277:38486–38493. doi: 10.1074/jbc.M112438200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Yan Z., Farooq A., Liu X., Lu C., Zhou M. M., He C. J. Mol. Biol. 2004;343:1147–1155. doi: 10.1016/j.jmb.2004.08.072. [DOI] [PubMed] [Google Scholar]
- 20.Mariner D. J., Davis M. A., Reynolds A. B. J. Cell Sci. 2004;117:1339–1350. doi: 10.1242/jcs.01001. [DOI] [PubMed] [Google Scholar]
- 21.Berclaz G., Altermatt H. J., Rohrbach V., Kieffer I., Dreher E., Andres A. C. Ann. Oncol. 2001;12:819–824. doi: 10.1023/a:1011126330233. [DOI] [PubMed] [Google Scholar]
- 22.Goruppi S., Ruaro E., Varnum B., Schneider C. Mol. Cell. Biol. 1997;17:4442–4453. doi: 10.1128/mcb.17.8.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse M., Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 24.Adams J. A. Biochemistry. 2003;42:601–607. doi: 10.1021/bi020617o. [DOI] [PubMed] [Google Scholar]
- 25.Mishra G. R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T., et al. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B., Borinstein S. C., Gillis J., Sykes V. W., Bogler O. J. Biol. Chem. 2000;275:19275–19281. doi: 10.1074/jbc.M908994199. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M. H., Hoeller D., Yu J., Furnari F. B., Cavenee W. K., Dikic I., Bogler O. Mol. Cell. Biol. 2004;24:8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods A. J., Kantidakis T., Sabe H., Critchley D. R., Norman J. C. Mol. Cell. Biol. 2005;25:3763–3773. doi: 10.1128/MCB.25.9.3763-3773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman N. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 30.Smaill J. B., Rewcastle G. W., Loo J. A., Greis K. D., Chan O. H., Reyner E. L., Lipka E., Showalter H. D., Vincent P. W., Elliott W., et al. J. Med. Chem. 2000;43:1380–1397. doi: 10.1021/jm990482t. [DOI] [PubMed] [Google Scholar]
- 31.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paez J. G., Janne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T., et al. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 33.Fitch K. R., McGowan K. A., van Raamsdonk C. D., Fuchs H., Lee D., Puech A., Herault Y., Threadgill D. W., Hrabe de Angelis M., Barsh G. S. Genes Dev. 2003;17:214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segatto O., Lonardo F., Pierce J. H., Bottaro D. P., Di Fiore P. P. New Biol. 1990;2:187–195. [PubMed] [Google Scholar]
- 35.Gotoh N., Tojo A., Hino M., Yazaki Y., Shibuya M. Biochem. Biophys. Res. Commun. 1992;186:768–774. doi: 10.1016/0006-291x(92)90812-y. [DOI] [PubMed] [Google Scholar]
- 36.Biscardi J. S., Maa M. C., Tice D. A., Cox M. E., Leu T. H., Parsons S. J. J. Biol. Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H. T., O’Rourke D. M., Zhao H., Murali R., Mikami Y., Davis J. G., Greene M. I., Qian X. Oncogene. 1998;16:2835–2842. doi: 10.1038/sj.onc.1201820. [DOI] [PubMed] [Google Scholar]
- 38.Dahlquist K. D., Salomonis N., Vranizan K., Lawlor S. C., Conklin B. R. Nat. Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 39.Yu J., Smith V. A., Wang P. P., Hartemink A. J., Jarvis E. D. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.