Abstract
The yeast chromatin protein Sin1p/Spt2p has long been studied, but the understanding of its function has remained elusive. The protein has sequence similarity to HMG1, specifically binds crossing DNA structures, and serves as a negative transcriptional regulator of a small family of genes that are activated by the SWI/SNF chromatin-remodeling complex. Recently, it has been implicated in maintaining the integrity of chromatin during transcription elongation. Here we present experiments whose results indicate that Sin1p/Spt2 is required for, and is directly involved in, the efficient recruitment of the mRNA cleavage/polyadenylation complex. This conclusion is based on the following findings: Sin1p/Spt2 frequently binds specifically downstream of many ORFs but almost always upstream of the first polyadenylation site. It directly interacts with Fir1p, a component of the cleavage/polyadenylation complex. Disruption of Sin1p/Spt2p results in foreshortened poly(A) tracts on mRNA. It is synthetically lethal with Cdc73p, which is involved in the recruitment of the complex. This report shows that a chromatin component is involved in 3′ end processing of RNA.
Keywords: fir1, HMG1, poly(A), PAF complex
Sin1p is a yeast chromatin nonhistone protein that has been shown to function as a negative transcriptional regulator of several genes, including Suc2 (1), Ino1 (2), and SSA3 (3). Activity of the HO promoter, as measured from an HO promoter driving a lacZ gene (4), is also affected by Sin1. Sin1 (also known as spt2 in this context) mutants were identified as suppressors of Ty and insertions in the 5′ noncoding region of the HIS4 gene (5). Because the negative regulation of these genes is overcome by the SW1/SNF chromatin-remodeling complex (4, 6, 7) and the C-terminal domain of Sin1p interacts with Swi1p (8), it was suggested that a function of SIN1p is to somehow maintain chromatin compaction at specific loci in the chromatin. Peterson et al. (2) found a functional relationship between the C-terminal domain (CTD) of RNA polymerase II and Sin1p, but these data were not pursued further. Sequence analysis of Sin1p showed sequence similarity in two domains to HMG1 (6, 7), a known chromatin protein. Work from our laboratory showed that Sin1p can bind four-way junction and crossing DNA structures (9), supporting the idea that Sin1p binds DNA as it enters and exits the nucleosome.
In the context of a global mapping project, Tong et al. (10) reported that there is a synthetic lethal interaction between sin1 and cdc73, a member of the PAF complex. The PAF complex, which accompanies RNA polymerase II during elongation, was shown to have an important function in 3′ end formation and in polyadenylation (11–13). In addition, a functional interaction was demonstrated between Sin1p and Hpr1p, which is associated with the PAF complex (14).
Most recently, evidence has been presented indicating that Sin1p/Spt2p plays roles in transcription elongation, chromatin structure and genome stability (15). In that study, synthetic lethal interactions were reported between sin1 and paf1, and between sin1 and ctr9. In addition, growth defects were identified in sin1, rtf1 double mutants. Paf1p, Ctr9p and Rtf1p are all members of the PAF complex. In the same study, using ChIP analysis, Spt2p was found preferentially associated with DNA that is actively transcribed, binding to 3′ untranslated regions more frequently than expected. Indeed, careful examination of Sin1p-binding sites on the DNA identified previously (16) shows that Sin1p often binds DNA downstream of ORFs (see Results).
All these data indicated that there might be a role for Sin1p in 3′ end formation of mRNA in yeast. In this paper, we show that Sin1p is associated with Fir1p, a component and positive regulator of the RNA cleavage/polyadenylation complex (17). Furthermore, deletion of Sin1p, or mutations in its C terminus, result in foreshortened poly(A) stretches at the 3′ termini of mRNA. We present a model suggesting that the polyadenylation machinery is recruited to the nascent RNA transcript by interactions with the heavily phosphorylated CTD of RNA polymerase II at serine-2 (18, 19), the PAF complex that accompanies the polymerase (11–13), and Sin1p that is bound to the chromatin just downstream of the ORF of many yeast genes.
Results
Sin1p Binds Downstream of Many Genes, Just Upstream of a Major Polyadenylation Site.
Sin1p is a chromatin component whose presence affects the transcription of a number of genes as is described above. Harbison et al. (16) mapped DNA-binding transcriptional regulators, including Sin1p (Spt2p), to the genome by using a chromatin immunoprecipitation assay. Careful examination of their data indicates that Sin1p often binds downstream of ORFs. In many cases, the binding site is downstream of two ORFs that are on opposing strands, precluding the possibility that Sin1p is binding promoter sequences at those positions. We compared the locations of the Sin1p-binding sites on 12 different genes having high affinity to Sin1p to predicted (20) and actual polyadenylation sites. The actual polyadenylation sites were determined experimentally by using 3′ RACE (see Materials and Methods). It is apparent from the results shown in Fig. 1 that in almost all cases, Sin1p binds ≈50 to 100 bp upstream of the first polyadenylation site after the ORF. In some cases, the binding is a bit further downstream, but usually, in the context of chromatin, not more than one nucleosome away from the 3′ end of the coding region. These data suggested that Sin1p might play a role in 3′ processing events of the nascent mRNA transcript.
Fig. 1.
Schematic representation of Sin1p binding site and polyadenylation sites on selected genes. The last 500 nucleotides of the coding region plus 500 nucleotides of the 3′ noncoding region are displayed. The numbers on the abscissa are relative to the 3′ end of the ORF. All genes selected scored with a high confidence level for Sin1p binding (16). The thick green line denotes the ORF. The black vertical arrow denotes the Sin1p binding site. The black and blue graphs denote predicted polyadenylation sites using the maximum-likelihood (ML) and the discrete state-space (DSM) models, respectively (20). The red arrows denote actual polyadenylation sites determined by 3′ RACE. Major polyadenylation sites are marked with a thick arrow, while more minor sites are marked with a thin arrow. A black horizontal arrow denotes the position of the left primer used in the 3′ RACE. The gel at the right of each graph shows a sample 3′ RACE lane for each gene. Marker sizes are 1000, 900, 800, 700, 600, 500, 400, 300, 250, 200, 150, 100, and 50 bp.
Sin1p Interacts Directly with Fir1p.
We previously reported results from a two-hybrid screen (21, 22) by using Sin1p as bait. In addition to recovering the previously reported colonies containing Cdc23 and Sap1, we also recovered colonies encoding amino acids 178–876 of Fir1p in the same screen. A direct interaction between the two proteins was confirmed by showing that 35S-labeled, in vitro translated Fir1p (amino acids 173–876) bound GST-Sin1p fusion proteins derived from the N terminus of the protein, whereas GST alone did not bind (Fig. 2). A foreshortened peptide containing amino acids 173–810 bound the GST-Sin1p equally well (data not shown). This data supports the possible involvement of Sin1p in the 3′ processing of nascent mRNA transcripts, because Fir1p was identified recently as a positive regulator of polyadenylation (17).
Fig. 2.
Association between Sin1p and Fir1p peptides in vitro. GST or GST/Sin1p bacterially produced fusion proteins were immobilized on 200-μl glutathione agarose beads. Fir1p peptides labeled with [35S]methionine were produced in the Promega TNT-coupled transcription-translation system. Ten microliters of the reaction was incubated with the glutathione agarose beads. After washing, bound proteins were eluted with glutathione, and three-quarters of the eluted volume was subjected to SDS/PAGE and autoradiography. The multiple bands represent partial Fir1p molecules that result from premature transcriptional and translational termination, internal transcriptional initiation, and RNA and protein degradation. The arrow shows the full-length protein.
Several hypothetical functional domains in Sin1p have been defined based on genetic, structural, and biochemical considerations (6, 9, 21, 23–25). These domains include two HMG1-like domains, (amino acids 26–88 and 98–159), an acidic domain (amino acids 224–304), and a basic C-terminal domain (amino acids 303–333). To address which of these domains interacts with Fir1p, we synthesized portions of Sin1p fused to GST based on the functional domains that have been suggested, bound them to glutathione-agarose beads, and asked whether the radiolabeled Fir1p peptide 173–810 would bind to Sin1p fragments. As can be seen in Fig. 2, peptides containing the first HMG1-like domain (amino acids 41–99) bound the Fir1 peptide, whereas molecules containing the other domains did not bind Fir1p. A fusion peptide containing only a part of this HMG1-like domain (amino acids 1–64) was able to bind Fir1p, but less well than those peptides containing the entire domain. In previous work, we showed that a Sin1p peptide containing this HMG1-like domain could bind four-way junction DNA (9). Taken together, these results demonstrate that the first HMG1-like domain of the protein can interact with both Fir1p and with DNA. Because it is unlikely that this peptide interacts with both DNA and Fir1p simultaneously, we suggest that interaction between Fir1p and Sin1p probably results in Sin1p displacement from the DNA.
Lack of a Functional Sin1p Results in Foreshortened poly(A).
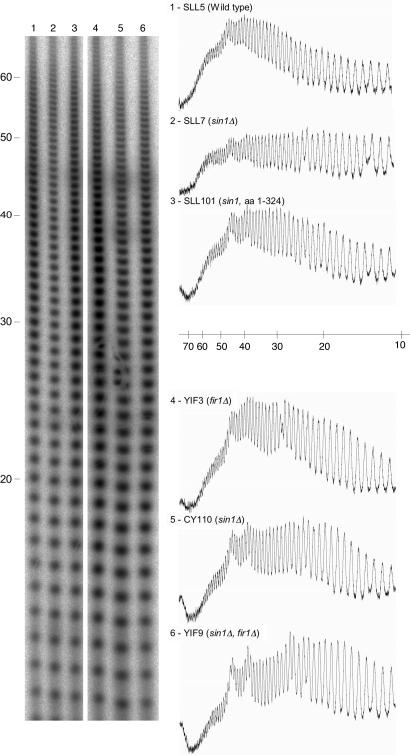
To test whether the interaction between the chromatin component (Sin1p) and the polyadenylation regulator (Fir1p) has functional significance, we compared average poly(A) length in yeast strains carrying deletions in one or both of these proteins. In our assay, we isolated whole-cell RNA from each strain and labeled the 3′ termini with [32P]pCp by using RNA ligase. We then digested all of the RNA in each sample except for the poly(A) by using an RNase T1/RNase A mix. Reaction products were electrophoresed on sequencing gels and autoradiographed. The results can be seen in Fig. 3. Examination of lanes 1 and 2 clearly demonstrate that Sin1 deletion results in a skewed distribution of poly(A) tails that are significantly shorter in the sin1 deletion than in the wild type. These results indicate that Sin1p, a chromatin protein, is required for normal polyadenylation.
Fig. 3.
Poly(A) length determination of total cell RNA. End-labeled RNA was digested with RNase and loaded on a 15% polyacrylamide sequencing gel. Ten percent of each reaction described in detail in Materials and Methods was loaded per lane. After autoradiography, the phosphorimage was quantitated by using imagej (see, which are published as supporting information on the PNAS web site).
Previous genetic and biochemical evidence (6, 9) showed that the C terminus of Sin1p has functional significance. We therefore tested a strain lacking the nine terminal amino acids of Sin1p (Fig. 3, lane 3) in our polyadenylation assay. As can be seen, this strain gave intermediate poly(A) lengths.
Fir1 deletions are not lethal but result in foreshortened poly(A) tracts by 5–7 nt when compared with wild type in an in vitro polyadenylation reaction by using cell extracts from those strains (17). Our in vivo results agree with those in vitro results, showing that fir1 deletions result in poly(A) tracts that are somewhat foreshortened (Fig. 3, compare lanes 1 and 4). Strikingly, a strain carrying the sin1 deletion together with a fir1 deletion had significantly shorter poly(A) tracts than the fir1 deletion alone, suggesting that the Sin1p might contact additional polyadenylation complex components in addition to Fir1p.
Neither Sin1p nor Fir1p Affect poly(A) Site Selection.
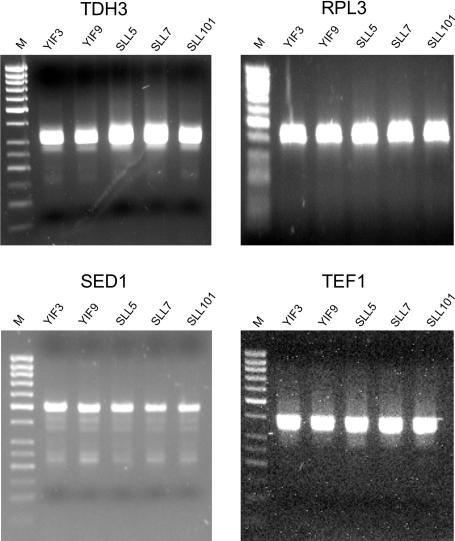
Because our results intimated that Sin1p is involved in polyadenylation, we asked whether it or Fir1p are involved in selection of the site at which the mRNA is cleaved and polyadenylated. This question was addressed by performing 3′ RACE on RNA samples that had been isolated from wild-type, sin1 and fir1 strains, and a sin1, fir1 double mutant. Of the 21 reactions performed, four sample reactions are displayed in Fig. 4. No significant differences were seen between the samples, indicating that neither Sin1p nor Fir1p participate in determination of the cleavage and polyadenylation site.
Fig. 4.
Sin1p does not affect poly(A) site selection. 3′ RACE was performed on total RNA samples isolated from wild-type and mutant yeast. Relevant genotypes are YIF3, fir1Δ; YIF9, sin1Δ, fir1Δ; SLL5, wild type; SLL7, sin1Δ; SLL101, sin1-324 (missing the C-terminal amino acids). Primers are listed in Table 1, which is published as supporting information on the PNAS web site.
Discussion
Almost all mRNA transcripts in eukaryotes are polyadenylated at their 3′ ends by a complex of proteins that are largely conserved throughout evolution. Polyadenylation has been shown to be important for mRNA stability, its protection from ribonucleases, transcription termination, export from the nucleus, and translation (26). Polyadenylation occurs in conjunction with transcriptional termination in several steps. Before or concurrent with termination, the transcript is cleaved at specific sites, a tract of adenylate residues is added, and it is then trimmed in a transcript-specific manner. These reactions are highly coordinated and are carried out by a host of factors. In yeast, these factors include poly(A)-binding protein (Pab1p), cleavage and polyadenylation factors CFIA, CFIB, CPF, and poly(A) nuclease PAN (17). Each factor with the exception of CFIB is encoded by multiple polypeptides. Additional regulatory factors are associated with these complexes. These factors include Fir1p (876 aa) that positively regulates mRNA polyadenylation and Ref2p that regulates it negatively.
Recent progress has been made that addresses how the cleavage/polyadenylation complex is recruited to the nascent RNA. The CTD of RNA polymerase II, which becomes progressively more phosphorylated at serine-2 as it progresses during elongation, interacts with Pcf11 (18, 19) and serves as a termination factor (27).
Sin1 is synthetically lethal with cdc73, paf1, and ctr9 (10, 15), all components of the PAF elongation complex. Recent data indicate that the PAF complex facilitates the linkage of transcriptional and posttranscriptional events, particularly polyadenylation (12). Although the PAF complex associates with RNA polymerase II during initiation and elongation, it is required for proper polyadenylation of at least a subset of transcripts (13). In the absence of Cdc73p, the remaining PAF components are not associated with the transcribing polymerase, and polyadenylation is foreshortened. It has been suggested that the PAF complex functions in 3′ end formation by recruitment of the 3′ end-processing factors (11). The synthetic lethality of Sin1 with components of this complex, and the fact that cdc73 and sin1 mutants individually result in foreshortened poly(A) tracts, imply that Sin1p and components of the PAF complex have required overlapping functions.
Nourani et al. (15) have shown that Sin1p is frequently associated with the coding region of actively transcribed genes and that its presence is important for the maintenance of histone H3 at transcribed regions. In addition, sin1Δ mutants display increased intrachromosomal recombination in actively transcribed regions (15). These data suggested that the nonsequence specific association of Sin1p with the transcribed DNA helps maintain the integrity of the chromatin during transcription.
We propose that as the transcription complex approaches a potential cleavage/polyadenylation site, the cleavage/polyadenylation complex is recruited by three components, all associated with the RNA polymerase II (Fig. 5): (i) The CTD of RNA polymerase II, which becomes increasingly phosphorylated at serine-2 (18, 28), interacts with Pcf11 in the CFIA component of the polyadenylation complex (19). (ii) Members of the PAF complex that are associated with the RNA polymerase II interact with as-yet-undetermined members of the polyadenylation complex (suggested by ref. 13). (iii) Sin1p, which may accompany the elongation complex (15), specifically binds a DNA sequence in the 3′ noncoding region, near the cleavage/polyadenylation site, and interacts with Fir1p in the cleavage/polyadenylation complex. If any of these three interactions is ablated (by gene disruption), polyadenylation is less efficient, and average cellular poly(A) is shorter than in the wild type. If any two of the “recruiters” are missing, polyadenylation is curtailed, and the result is lethality. This idea is supported by reported synthetic lethalities: (i) between ctk1, whose gene product phosphorylates the CTD at serine-2, and any of the PAF complex components (11); and (ii) between cdc73, paf1, or ctr9, whose gene products are part of the PAF complex, and Sin1 (10).
Fig. 5.
Model for polyadenylation complex recruitment/activation. The polyadenylation complex is recruited or activated in three parallel interactions denoted by arrows: (i) Hyperphosphorylated CTD on serine-2 contacts Pcf11p that is part of CFIA. (ii) The PAF complex contacts unknown members of the polyadenylation complex. (iii) Sin1p, specifically associated with the DNA just upstream of a polyadenylation site, contacts Fir1p that positively regulates polyadenylation. To allow the transcription apparatus to proceed, Sin1p must be displaced from the DNA by Hpr1p, the SWI/SNF complex, and the ADA/GCN5 complex. The recruited/activated polyadenylation complex cleaves the mRNA downstream of where the Sin1p was bound and polyadenylates the transcript. If a component of any of the pathways is missing, polyadenylation is less efficient, and average poly(A) lengths are shorter. Blue denotes core factors required for pre-mRNA 3′ processing. Yellow denotes polyadenylation factors. Pink denotes poly(A)-trimming factors. Stippled shapes denote factors that displace Sin1p from the DNA. The striped bar denotes the ORF. The line after the ORF denotes the 3′ noncoding region.
Although our data indicate that Sin1p participates in the recruitment of the mRNA processing/polyadenylation machinery and ChIP analysis (16) shows that it preferentially binds DNA just upstream of the polyadenylation site, Sin1p is not required to identify the polyadenylation site.
Hpr1p, which appears to be involved in chromatin remodeling, has been shown to be associated with the PAF complex (14). At 37°C, a strain lacking Hpr1p and expressing a semidominant sin1-2 allele (29) is unable to grow, whereas strains with a sin1 deletion grow almost normally. It was suggested in that paper that in the wild type, Hpr1p might function to facilitate the removal of Sin1p from the chromatin and that in its absence, in the sin1-2 strain, sin1-2p may be bound to the chromatin more tightly, causing toxicity. That interpretation is consistent with another observation that overexpression of the C-terminal of Sin1p (where the sin1-2 mutation is found) is toxic (8), but that this toxicity is suppressed in spt4, spt5, and spt6 strains. Each of these proteins, like Hpr1p, has been implicated in modification of chromatin structure. The Spt4/Spt5 complex interacts genetically and physically with Spt6, presumably to mediate transcription through chromatin (30, 31). Interestingly, recent data have shown a functional connection between Spt2p and Spt6p (15), and spt6 mutations can alter mRNA 3′ end formation at least in one documented case (32).
Sin1p has been isolated in a protein complex that includes Pub1p, a poly(A)+ RNA-binding protein, Npl3, a poly(A)+ RNA-transport protein, TIF4631 and TIF4632, proteins that associate with the poly(A)-binding protein Pab1p, and Sgn1p, another RNA-binding protein (33). These data also support Sin1p involvement in RNA processing.
We propose that, concomitant with its recognition by the polyadenylation complex, Sin1p is removed from the chromatin, allowing the transcription apparatus to pass with the recruited 3′ processing machinery in tow. The notion that Sin1p is removed from the chromatin is supported by the fact that the same short domain of Sin1p binds both DNA and Fir1p, and it is unlikely that both can interact with Sin1p simultaneously.
Sin1 initially was discovered as a suppressor of mutations in the SWI/SNF complex (2, 7). Later, Perez-Martin and Johnson (8) showed that the C terminus of Sin1p physically associates with components of the SWI/SNF complex and that this interaction is blocked in the full-length Sin1p protein by the N-terminal half of the protein. Recently, it has been reported that SWI/SNF can stimulate transcription even after promoter clearance (34) during elongation. Pollard and Peterson (1) reported that members of the ADA/GCN5 complex that can acetylate both histones and Sin1p are synthetically lethal with swi1, a component of the SWI/SNF complex. Mutations in Sin1 restore viability. Taking all these data together, we propose that the SWI/SNF complex, the ADA/GCN5 complex (via acetylation of Sin1p), and Hpr1p each facilitate the removal of Sin1p from the chromatin at a site downstream of the ORF of many genes. Continued research is necessary to elucidate the precise role of each of these factors in mRNA transcriptional termination and processing. The removal of Sin1p from the chromatin might be a prerequisite for the passage of the transcription apparatus, without which there can be no further processing of the mRNA.
We show that a chromatin component is required to complete mRNA processing, providing a link between chromatin structure and RNA processing. As details of the gene expression machinery are explored, it is becoming apparent that sometimes unexpected functional relationships exist between the different components (35–38). Further research will be necessary to determine the details of how the mRNA processing machinery is recruited and activated.
Materials and Methods
Yeast Strains.
SLL5 (wild type with respect to Sin1), SLL7 (sin1Δ), and SLL101 (sin1 1-324) were gifts of L. Lefebvre (University of British Columbia, Vancouver; ref. 6). YIF3 is fir1::HIS3, leu2Δ, trp1Δ, ura3-52, his3-Δ200, ade2-101, lys2-801. YIF9 is YIF3 with the addition of sin1::LEU2. Cy110 is sin1::TRP1, leu2Δ, trp1Δ, ura3-52, his3-Δ200, ade2-101, lys2-801.
Two-Hybrid Screen and Verification of Protein Interactions.
The two-hybrid screen by using Sin1p as a bait was carried out as in Shpungin et al. (22). A plasmid termed p82 was isolated from the libraries and was found to encode amino acids 173–876 of Fir1p. Plasmids p178-876 and p178-746 were constructed by PCR amplification of the partial Fir1 coding region by using p82 as template and inserting them between the BamHI and PstI sites of pBluescript (Stratagene). Coupled transcription-translation of [35S]methionine-labeled Fir1p peptides was accomplished by using the Promega TNT system. Plasmids encoding the GST/Sin1p fusions were described in ref. 9. The in vitro-binding assay between the Sin1p and Fir1p peptides was carried out as in Novoseler et al. (9).
poly(A) Length Determination.
The determination of poly(A) length for total cell RNA was done essentially as described in ref. 39. Briefly, [32P]pCp was ligated to the 3′ ends of 0.5 μg of RNA in a 30-μl reaction by using 12 units of T4 RNA ligase (Fermentas, Burlington, ON, Canada). After a 24-h incubation at 4°C, 40 μg of yeast tRNA was added, and all but the poly(A) was digested by a mix of 80 units of RNase T1 and 32 μg of RNase A for 2 h at 37°C in a final volume of 80 μl. Finally, the enzymes were neutralized by proteinase K and phenol/chloroform extraction and ethanol precipitated. The samples then were run on a 15% polyacrylamide sequencing gel and autoradiographed. Quantitative evaluation of the autoradiogram was done by analysis of phosphorimager images by using imagej.
3′ RACE.
3′ RACE was performed on total cell RNA by using the FirstChoice RLM-RACE kit (Ambion, Austin, TX) according to the instructions of the manufacturer. The PCR uses an RNA template and amplifies a DNA fragment between a primer specific to a gene (see list in Table 1) and the 5′ end of the poly(A) segment at the 3′ end of the RNA molecule.
Supplementary Material
Acknowledgments
We thank L. Lefebvre for strains. This work was supported by internal Life Sciences Faculty funding at Bar Ilan University.
Abbreviation
- CTD
C-terminal domain.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pollard K. J., Peterson C. L. Mol. Cell. Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson C. L., Kruger W., Herskowitz I. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 3.Baxter B. K., Craig E. A. J. Bacteriol. 1998;180:6484–6492. doi: 10.1128/jb.180.24.6484-6492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 5.Winston F., Chaleff D. T., Valent B., Fink G. R. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebvre L., Smith M. Mol. Cell. Biol. 1993;13:5393–5407. doi: 10.1128/mcb.13.9.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruger W., Herskowitz I. Mol. Cell. Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Martin J., Johnson A. D. Mol. Cell. Biol. 1998;18:4157–4164. doi: 10.1128/mcb.18.7.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novoseler M., Hershkovits G., Katcoff D. J. J. Biol. Chem. 2005;280:5169–5177. doi: 10.1074/jbc.M406249200. [DOI] [PubMed] [Google Scholar]
- 10.Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., et al. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 11.Rosonina E., Manley J. L. Mol. Cell. 2005;20:167–168. doi: 10.1016/j.molcel.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 13.Penheiter K. L., Washburn T. M., Porter S. E., Hoffman M. G., Jaehning J. A. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Chang M., French-Cornay D., Fan H. Y., Klein H., Denis C. L., Jaehning J. A. Mol. Cell. Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nourani A., Robert F., Winston F. Mol. Cell. Biol. 2006;26:1496–1509. doi: 10.1128/MCB.26.4.1496-1509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J. B., Reynolds D. B., Yoo J., et al. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangus D. A., Smith M. M., McSweeney J. M., Jacobson A. Mol. Cell. Biol. 2004;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn S. H., Kim M., Buratowski S. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 19.Meinhart A., Cramer P. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 20.Graber J. H., McAllister G. D., Smith T. F. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberzon A., Shpungin S., Bangio H., Yona E., Katcoff D. J. FEBS Lett. 1996;388:5–10. doi: 10.1016/0014-5793(96)00500-5. [DOI] [PubMed] [Google Scholar]
- 22.Shpungin S., Liberzon A., Bangio H., Yona E., Katcoff D. J. Proc. Natl. Acad. Sci. USA. 1996;93:8274–8277. doi: 10.1073/pnas.93.16.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baxevanis A. D., Bryant S. H., Landsman D. Nucleic Acids Res. 1995;23:1019–1029. doi: 10.1093/nar/23.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yona E., Bangio H., Erlich P., Tepper S. H., Katcoff D. J. Mol. Gen. Genet. 1995;246:774–777. doi: 10.1007/BF00290726. [DOI] [PubMed] [Google Scholar]
- 25.Yona E., Bangio H., Friedman Y., Shpungin S., Katcoff D. J. FEBS Lett. 1996;382:97–100. doi: 10.1016/0014-5793(96)00159-7. [DOI] [PubMed] [Google Scholar]
- 26.Mangus D. A., Evans M. C., Jacobson A. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Gilmour D. S. Mol. Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Komarnitsky P., Cho E. J., Buratowski S. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y., Peterson C. L., Christman M. F. Mol. Cell. Biol. 1995;15:1698–1708. doi: 10.1128/mcb.15.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartzog G. A., Wada T., Handa H., Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartzog G. A., Speer J. L., Lindstrom D. L. Biochim. Biophys. Acta. 2002;1577:276–286. doi: 10.1016/s0167-4781(02)00458-x. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan C. D., Holland M. J., Winston F. J. Biol. Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 34.Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G. Mol. Cell. Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimaano C., Ullman K. S. Mol. Cell. Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed R. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 37.Burckin T., Nagel R., Mandel-Gutfreund Y., Shiue L., Clark T. A., Chong J. L., Chang T. H., Squazzo S., Hartzog G., Ares M., Jr. Nat. Struct. Mol. Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 38.Buratowski S. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Minvielle-Sebastia L., Winsor B., Bonneaud N., Lacroute F. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.