Abstract
Cellular and physiological responses to changes in dioxygen levels in metazoans are mediated via the posttranslational oxidation of hypoxia-inducible transcription factor (HIF). Hydroxylation of conserved prolyl residues in the HIF-α subunit, catalyzed by HIF prolyl-hydroxylases (PHDs), signals for its proteasomal degradation. The requirement of the PHDs for dioxygen links changes in dioxygen levels with the transcriptional regulation of the gene array that enables the cellular response to chronic hypoxia; the PHDs thus act as an oxygen-sensing component of the HIF system, and their inhibition mimics the hypoxic response. We describe crystal structures of the catalytic domain of human PHD2, an important prolyl-4-hydroxylase in the human hypoxic response in normal cells, in complex with Fe(II) and an inhibitor to 1.7 Å resolution. PHD2 crystallizes as a homotrimer and contains a double-stranded β-helix core fold common to the Fe(II) and 2-oxoglutarate-dependant dioxygenase family, the residues of which are well conserved in the three human PHD enzymes (PHD 1–3). The structure provides insights into the hypoxic response, helps to rationalize a clinically observed mutation leading to familial erythrocytosis, and will aid in the design of PHD selective inhibitors for the treatment of anemia and ischemic disease.
Keywords: erythropoietin, dioxygenase, hypoxic response, 2-oxoglutarate
In metazoans the α/β heterodimeric hypoxia-inducible transcription factor (HIF) (1) regulates the transcription of an array of genes including those coding for glycolytic enzymes, erythropoietin, and VEGF. The levels and transcriptional activity of the HIF-α, but not the HIF-β, subunit are regulated by oxygen. Hydroxylation of either Pro-402 or Pro-564 in human HIF-1α (2, 3) within the C-terminal oxygen-dependent degradation domain (CODDD) enables its binding to the von Hippel-Lindau protein (pVHL), a targeting element of the E3-ubiquitin ligase; subsequent ubiquitylation leads to proteasomal degradation of HIF-α (for reviews, see refs. 4–7). In humans, this mechanism is augmented by hydroxylation of an asparagine residue in the C-terminal transcriptional activation domain (8); this modification blocks interaction of HIF-1α with the CBP/p300 coactivator, thereby disabling HIF-mediated transcription.
Hydroxylation of HIF-1α is catalyzed by four 2-oxoglutarate (2OG) dioxygenases: three prolyl hydroxlyases (PHD 1, 2, and 3) (also known as HPH 3, 2, and 1 and EGLN 2, 1, and 3; refs. 9–11) and an asparaginyl hydroxylase [factor inhibiting HIF (FIH); refs. 12 and 13]. The available evidence implicates PHD2 as the most important HIF hydroxylase in down-regulating the hypoxic response during normoxia (5, 7, 14, 15).
The HIF hydroxylases are Fe(II) and 2OG-dependent dioxygenases (16, 17); their requirement for dioxygen has led to their characterization as cellular oxygen sensors (refs. 9–11, 18, and 19; Fig. 1a). The first 2OG dioxygenase to be identified was procollagen prolyl-hydroxylase, which like the PHDs catalyzes trans-4-hydroxylation reactions. Procollagen prolyl hydroxylation stabilizes the collagen triple helix structure. Because of its medical importance in collagen-related diseases, including scurvy, procollagen prolyl-hydroxylase has been the subject of therapeutic intervention by small molecules (20); however, no structural information for a prolyl-hydroxylase is yet available. 2OG dioxygenases also have been shown to play other roles in human cells, including lipid metabolism (21, 22), DNA repair (23, 24), and histone modification (25).
Fig. 1.
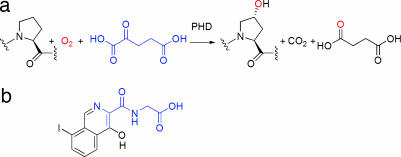
The PHD reaction and an inhibitor. (a) PHD catalyzed prolyl-4-hydroxylation of HIF-α; one of the oxygens (red) from the dioxygen cosubstrate is incorporated into proline to form trans-4-hydroxyproline, and the other is incorporated into succinate. (b) The structure of compound A.
A molecular understanding of the hypoxic response is important for developing a molecular understanding of genetic disorders, including Chuvash polycythemia and von Hippel-Lindau disease (4). The role of the PHDs as ubiquitous mediators of oxygen/hypoxia sensing has raised the question as to whether the HIF hydroxylases possess especially adapted structural and mechanistic features. We therefore initiated crystallographic studies on PHD2 (NCBI GI 13489073) and here describe structures, solved independently by two groups, of PHD2 in complex with Fe(II) and a 2OG-competitive isoquinoline inhibitor, {[(4-hydroxy-8-iodoisoquinolin-3-yl)carbonyl]amino}acetic acid (compound A), a derivative of known procollagen prolyl-hydroxylase and PHD inhibitors (Fig. 1b).
Results and Discussion
Crystallization of PHD2cat.
Secondary structure prediction indicates that PHD2 (426 residues, calculated molecular mass 46 kDa) contains two structural domains. The N-terminal domain (≈21–58) has homology to MYND zinc finger domains and the catalytic C-terminal domain (≈181–426) has homology to the 2OG dioxygenases (9, 10). Because crystallization of the full-length PHD2 was unsuccessful, a series of truncated constructs were made. N-terminally truncated forms of PHD2, such as (PHD2181–426) and (PHD2181–417), were catalytically viable (26) and only produced one crystal form in complex with bicyclic aromatic inhibitors such as compound A (Fig. 1b).
Structure Solution.
The structure of PHD2181–426 in complex with Fe(II) and compound A was solved by using single-wavelength anomalous diffraction (SAD) based on the anomalous signal from an Fe, I, and several protein S atoms by using a rotating anode generator. The crystal structure of PHD2181–417 in complex with Fe(II) and compound A was solved by combining phases from the single isomorphous replacement with anomalous scattering and multiple wavelength anomalous diffraction methods. The final refinement of PHD2181–417 with a native data set collected to 1.7 Å yielded Rcryst = 0.216/Rfree = 0.253. The crystal structures of PHD2181–426 and PHD2181–417 are near identical (rms deviation = 0.43 Å) with residues from 188 to 403 visible in the maps and included in the refined models (1.7 Å). These structures are jointly referred to as PHD2cat.
Overall Architecture of PHD2cat.
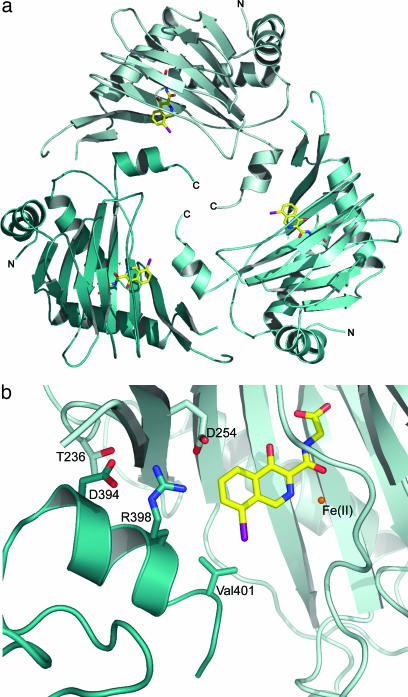
PHD2cat crystallizes as a homotrimer (Fig. 2a) with intermolecular contacts between the C-terminal α-helix (α4) and the surrounding active site residues of a neighboring subunit (Fig. 2b). This head-to-tail arrangement causes the C-terminal helix of one monomer to cap the active site of a neighbor. The homotrimer is stabilized by electrostatic and H bond interactions between Asp-254 Oδ2 and Arg-398 NH1 (2.8 Å), Thr-236 Oγ1 and Asp-394 Oδ2 (2.7 Å), and the hydrophobic interaction between the Val-401 side chain of the C-terminal helix from another subunit and compound A. The loop present after helix α3 interacts with a “finger-like” loop of a neighbor that links β2 and β3. Although the buried surface area (2,432 Å2) between monomers is potentially adequate for biologically significant oligomerization, the complementarities of residues buried at the interface is not (27); analytical ultracentrifugation of PHD2181–426 implies the enzyme exists predominantly as monomer in solution (Emma Longman, personal communication).
Fig. 2.
Crystallographic oligomerization of PHD2. (a) Ribbons representation demonstrating the way PHD2cat crystallizes as a trimer (each monomer is colored a different shade of blue). (b) Close up view of C-terminal interactions in the active site (light cyan) with the inhibitor (yellow) and neighboring molecule (dark teal).
The position of the C-terminal helix relative to the active site identifies PHD2 as a member of a distinct subfamily of 2OG oxygenases that includes enzymes involved in the biosynthesis of the cephalosporin family of β-lactam antibiotics (28). The PHDs thus differ from the FIH subfamily, where, at least for FIH, the C-terminal helices enable dimerization (Fig. 6, which is published as supporting information on the PNAS web site; refs. 29–31). The head-to-tail mediated crystal packing of PHD2cat has also been observed in other 2OG oxygenases (28, 32); however, the trimeric state observed with PHD2cat is unique.
The double-stranded β-helix (DSBH) fold of 2OG oxygenases contain eight β-strands (I to VIII) (33). DSBH β-strand II of PHD2cat, which immediately precedes two of the Fe(II) coordinating residues, has φ/ψ angles within the β region of the Ramachandran plot but does not maintain the antiparallel H bond pairing with its neighboring β-strand VII, as observed in other 2OG oxygenase structures; β-strands I (β4), III (β5), IV (β6), V (β7), VI (β8), VII (β9), and VIII (β10) complete the DSBH (Fig. 3a). A conserved N-terminal helix-strand-helix-strand motif, common to DAOCS, is present in PHD2; strands β1, β2, and β3 of this motif extend the DSBH major β-sheet (16, 28), comprised of β-strands I (β4), VIII (β10), III (β5), and VI (β8). The minor β-sheet is comprised of strands II, VII (β9), IV (β6), and V (β7).
Fig. 3.
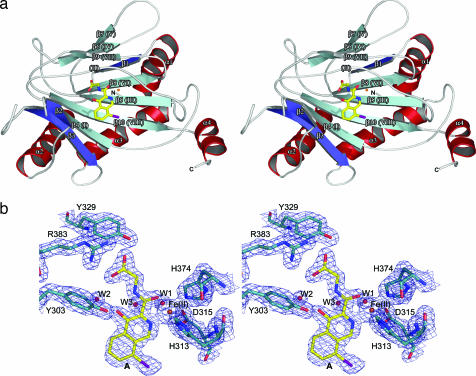
Overall view of the crystal structure of a PHD2 monomer and representative electron density. (a) Stereoview ribbons representation of PHD2cat with compound A and Fe(II) (orange sphere). Secondary structure is numbered sequentially and is color coordinated with that of Fig. 10. Roman numerals in parenthesis indicate the eight strands of the DSBH according to Stirk et al. (33). (b) Stereoview of iron and inhibitor binding to PHD2cat (cyan stick) with 2Fo − Fc electron density contoured to 2σ (blue).
Three α-helices (α1, α2, and α3) pack along the major β-sheet and stabilize the DSBH. The C-terminal α-helix (α4) extends from strand VIII (β10) into the active site of a neighboring molecule (Fig. 2b), which blocks access to the active site in the crystals. Hydrophobic patches, primarily present along the exposed minor β-sheet (Tyr-197, Ile-207, Val-209, Ile-251, Val-241, Val-311, Val-338, Ile-342, Phe-346, Phe-353, Ala-354, Ile-356, Pro-378, and Tyr-380), may enable interaction of PHD2 with HIF-1α or other known PHD2-interacting proteins (e.g., inhibitor of growth family member 4; ref. 34) and/or, possibly, where the full-length PHD2 MYND domain associates with the catalytic domain.
The Active Site.
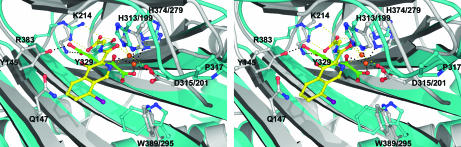
The active site, located between the major and minor β-sheets, comprises a relatively deep pocket compared to other 2OG oxygenases. There is a single metal ion coordinated in an octahedral manner by His-313, Asp-315, His-374, compound A, and a water molecule (Fig. 3b). Atomic absorption spectroscopy and particle-induced x-ray emission analyses of PHD2181–426 samples used for crystallization (notably purified in the absence of nickel) demonstrated that PHD2cat copurified with Fe(II), although zinc also was present in protein samples (35). Compound A binds to the Fe(II) via bidentate coordination through N1 of its isoquinoline ring (N1-Fe(II); 2.2 Å) and O11 of the amide carbonyl (O11-Fe(II); 2.2 Å) forming a ca. planar 5 membered chelate ring. The amide carbonyl O11 of compound A coordinates the iron ca. trans to the Asp-315 side chain and its isoquinoline nitrogen is trans to His-374 Nε2. The identity of the three Fe(II) coordinating residues as His-313, Asp-315, and His-374 confirms predictions from mutational and sequence comparison studies (9). A chain of five H-bonded waters, bridged by Thr-387 Oγ, extends from the iron through the proposed 2OG-binding site alongside compound A (in order from Fe– Wat1– Wat3– Thr-387– Wat2– Wat17– Wat13).
With the exception of the Fe(II) and 2OG-binding residues, the active site is predominantly lined by hydrophobic residues, as suggested in ref. 36. These residues are derived from the β-strands (β3) [Ile-256], I (β4) [Met-299, Ala-301, and Tyr-303], II (Tyr-310), III (β5) [Thr-325, Ile-327, and Tyr-329], IV (β6) [Leu-343], VI (β8) [Phe-366], VII (β9) [Val-376], and VIII (β10) [Ala-385, Thr-387, and Trp-389] (Fig. 7, which is published as supporting information on the PNAS web site). The bicyclic aromatic rings and I atom of compound A project through the active site opening and are sandwiched between the side chains of Tyr-310, Met-299, and Trp-389 (Fig. 4a). A hydrophobic shelf formed by the side chains of Ile-256 and Trp-258 leads to the active site opening. The hydrophobic nature of residues at the active site may reflect a requirement of the enzyme for protection from potential oxidative damage because they are less susceptible to oxidation via Fenton type chemistry mediated by reactive species that leak from the iron in unproductive reactions. Indeed, compared with some other 2OG dioxygenases/related dioxygenases (37, 38), PHD2 is resistant to damage mediated by Fe(II), ascorbate, and oxygen (data not shown).
Fig. 4.
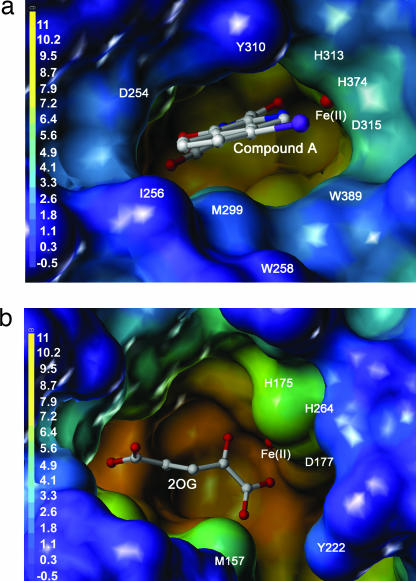
Surface representations of PHD2cat (a) and phytanoyl CoA hydroxylase (b) (21) comparing the entrances to the active site cavities. The surfaces are colored by depth with a gradient from blue (outermost) to orange (innermost).
Compound A’s carboxylate side chain is bound in a predominantly hydrophobic pocket with the exception of the Arg-383 and Tyr-329 side chains, with which it forms electrostatic and H bonding interactions (Arg383NH1-O3, 2.9 Å; Arg383NH2-O1, 2.7 Å; and Tyr329OH-O1, 2.6 Å). Despite soaking and cocrystallization efforts, it was not possible to replace the inhibitor with 2OG or its unreactive analogue, N-oxalylglycine, an observation possibly attributable to the “trapping” of the inhibitor by the C-terminal helix of a neighboring molecule in the crystal of which the Val-401 side chain makes contact with compound A (2.6 Å) (Fig. 2b). Evidence that compound A binds in the 2OG binding pocket came from kinetic and soft ionization electrospray mass spectrometric analyses demonstrating competitive binding with 2OG (Fig. 8, which is published as supporting information on the PNAS web site) and from mutagenesis of Arg-383 to Ala leading to almost complete inactivation of the protein (<5% by 2OG turnover assays with CODDD556–574 peptide substrate) (unpublished data). The assignment of Arg-383 as a 2OG-binding residue confirms a prior study in which mutation of the equivalent Arg in PHD1 (Arg-367) to Ala was shown to ablate activity (19).
Inhibitor Binding.
Although many 2OG oxygenase inhibitors are aromatic heterocycles (39), the only previously reported structures for 2OG oxygenases in complex with inhibitors have used N-oxalylamino acids (40), which bind iron in a bidentate manner (29, 41). A series of 2-hydroxybenzoate inhibitors related to compound A have been proposed to bind to the PHDs via the carboxylate and phenolic oxygens (42), and compound A has the potential to bind in a similar bidentate manner via its amide carbonyl and phenolic oxygens. The structure reveals that although the amide carbonyl oxygen of compound A coordinates the Fe(II), the second Fe(II)-coordinating atom is the aromatic isoquinoyl nitrogen rather than the phenolic oxygen. This mode of binding enables a hydrogen bond between the phenolic oxygen of compound A and the side-chain hydroxyl of the conserved Tyr-303 residue (Fig. 5; see also Fig. 9, which is published as supporting information on the PNAS web site). However, the Tyr303Phe mutant retains enzymatic activity and is inhibited by compound A (unpublished data), demonstrating that the hydroxyl of Tyr-303 is not catalytically essential. One role of the phenolic oxygen of compound A may be to increase the electron density at the isoquinoyl nitrogen, thereby stabilizing the observed binding mode. However, during crystallographic refinement, the ring atoms of compound A were restrained to be coplanar. If the tautomeric state in which the ketone form is partially present in bound compound A, the sp2 hybridization of the phenolic oxygen may enable an intramolecular hydrogen bond with the amide NH of its glycine side chain.
Fig. 5.
Stereoview of the superimposed active sites of PHD2cat (blue) and FIH (gray) (PDB ID code 1H2N; ref. 29). Compound A (yellow) bound to PHD2cat and 2OG (green) bound to FIH, Fe(II) is represented by an orange sphere, yellow dashes indicate selected ligand interactions in the PHD2cat structure, and black dashes in FIH. Note the different coordination modes for compound A in PHD2cat versus 2OG for FIH. The weaker inhibition of FIH by compound A over that of PHD2 may be explained by a steric clash with FIH’s Gln-147 side chain; the corresponding residue in PHD2 is Ala-301. The site of a clinically observed mutation in PHD2, P317R (43) is close to the iron coordination site.
Comparison of PHD2cat with FIH.
The opening to the active site of PHD2cat is narrower than the human 2OG oxygenases FIH and PAHX (Fig. 4) and especially so compared with the crystal structure of bacterial proline-3-hydroxylase (44). The narrow active site opening may have significance for the role of PHD isozymes as sensors because it may rationalize the tight binding constants for Fe(II) and 2OG (<1 μM and <2 μM respectively) and its copurification with Fe(II) and 2OG (35).
Comparison of PHD2cat and FIH reveals that the conformations of the two coordinating histidines are near identical but that of the aspartic acid differs (Fig. 5). In the FIH.Fe(II).2OG.substrate structure (PDB ID code 1H2L) (29), one of the Asp carboxylate oxygens coordinates the iron (Asp-201 Oδ2-Fe(II); 2.1 Å), whilst the other carboxylate oxygen hydrogen bonds to the backbone amide of Asn-803 of the substrate (FIH Asp-201 Oδ1–HIF Asn-803 NH; 3.1 Å). In PHD2cat, one of the Asp-315 oxygens coordinates the iron (Asp-315 Oδ1-Fe(II); 2.2 Å) and the other carboxylate oxygen hydrogen bonds to the well defined water molecule (Asp-315 Oδ2-Water1; 2.6 Å) that completes the octahedral coordination of Fe(II) (Fig. 3b). Superposition of PHD2cat and FIH reveals that the side chain amide carbonyl of compound A occupies the same coordination position to the iron in PHD2 as the 2-oxo oxygen of 2OG. However, the heterocyclic nitrogen has a different coordination position compared with the 2-carboxylate of 2OG (Fig. 5). This difference is interesting because of uncertainty regarding the relative coordination position of the 2OG 1-carboxylate in the mechanism and it may be that it is inverted for different enzymes (in some cases, both modes may operate for the same enzyme; ref. 45). Although generally similar, the 2OG-binding site of PHD2 differs significantly in detail to that of FIH in employing basic and Tyr residues to bind the 2OG 5-carboxylate. For the basic residue, PHD2cat employs Arg-383 from DSBH β-strand VIII (β10) rather than a lysine in FIH (Lys-214) from β-strand IV (β6). The tyrosine, Tyr-329 in PHD2, is from β-strand III (β5), whereas Tyr-145 of FIH is not from one of the core DSBH strands, but from the N-terminal strand (β6). Although their iron-binding sites use similar ligands, the residue differences in the 2OG-binding site and the overall structural differences described above define the PHDs and FIH as belonging to structurally distinct human 2OG oxygenase subfamilies, suggesting different evolutionary paths for the two families.
The structure rationalizes why compound A inhibits PHD2 with severalfold greater potency than FIH (Ki > 1 mM for FIH) by using a 2OG turnover assay by structural comparison. In FIH, Gln-147 is equivalent to Ala-301 of PHD2, the longer side chain of which would clash with the aromatic ring of compound A (Fig. 5). The shape of the PHD2cat 2OG-binding pocket also explains why the l-enantiomer of N-oxalylalanine is a better PHD inhibitor than the corresponding d-enantiomer: because the latter would clash with the Val-376 side chain (36). N-Oxalyl-d-phenylalanine, which is selective for FIH over the PHDs (41), likely does not inhibit PHD2 for the same reason. Thus, the development of potent small-molecule inhibitors that bind at the active site and are selective for PHDs over FIH should be possible. However, homology modeling indicates that obtaining inhibitors based on 2OG analogues that are specific for the individual PHD isoforms may be more difficult to obtain because of their high degree of similarity (Figs. 10 and 11, which are published as supporting information on the PNAS web site).
Comparison of PHDs.
Sequence comparisons and modeling studies indicate that the PHD2cat active site is highly conserved among the three human PHDs, suggesting that specificity differences are not solely due to the regions proximate to the 2OG and Fe(II) binding sites (Figs. 10 and 11). It is possible that the substrate specificity of the PHDs, in part, is determined by regions relatively remote from the iron center and may involve the variable N- and C-terminal regions. The sequence of the “β2-β3 finger” motif described earlier is not well conserved among the three PHD isozymes, which suggests a role in distinguishing their functions. In the structure of PAHX, a similar finger exists and is found antiparallel to the same finger of a symmetry-related molecule across a crystallographic 2-fold axis (21). These regions, in particular the C-terminal region, are likely to be involved in determining substrate specificity (46). Other regions that vary between the PHDs are the 2 N-terminal helices α1 and α2, which are relatively far from the active site. Thus, the different physiological roles for the PHDs are likely to be reflected in different modes of regulation rather than solely on their biochemical properties. The sequence alignment of PHD homologues from different organisms indicates that the iron and cosubstrate-binding sites are very similar (Fig. 9); the strictly conserved residues within the active site of these PHD homologues include Asp-254, Trp-258, Tyr-303, Tyr-310, His-313, Asp-315, Leu-343, Phe-366, His-374, Val-376, Arg-383, and Trp-389.
Biological Significance and Clinical Relevance.
Structural information on the components of the HIF system helps to rationalize polymorphisms, causing human genetic diseases involving erythropoeisis. The functional effects of pVHL mutations leading to von Hippel-Lindau disease have been rationalized by the structures of pVHL in complex with the HIF(hyp564) peptide (47, 48). However, not all cases of familial erythrocytosis involve pVHL mutations, implying other lesions to the HIF system. An inherited mutation in PHD2 linked with a familial erythrocytosis occurs at Pro-317 of PHD2 (43). Pro-317 is located two residues from the Fe(II)-binding aspartate at the i + 3 position of a type VIII β-turn and close to the active site entrance implying that mutation to Arg at this position is likely to alter both Fe(II) and substrate binding (Fig. 5). To date, only this clinically relevant PHD2 mutation has been identified (43), and no mutants of procollagen prolyl hydroxylase have yet been reported. However, in the case of PAHX, which plays a role in enzymatic degradation of the phytol side chain of chlorophyll, multiple mutations leading to Refsum disease have been identified and shown to cluster in the region of its iron and 2OG-binding site (21, 22).
The recognition of the role of posttranslational hydroxylation as a central mechanism for the regulation of the HIF-signaling pathway raises questions regarding a general role for posttranslational hydroxylation in other signaling pathways and the involvement of 2OG oxygenases, which possess special features with respect to oxygen/hypoxia sensing. Biological data and sequence analyses suggest that the HIF system is ubiquitous in metazoans. Direct experimental data are not as readily available on putative 2OG oxygenases that might catalyze protein hydroxylation, but the number of close sequence homologues suggests that PHDs are highly conserved through eukaryotes and prokaryotes, more so than FIH.
Sequence searches and comparisons in light of the PHD2 structure reveal that the catalytic cores are well conserved in a range of organisms that also possess HIF, and some that do not, including Dictyostelium and several prokaryotic pathogens, such as Pseudomonas aeruginosa and Vibrio cholerae (Fig. 9). The PHD homologue in Dictyostelium catalyzes the hydroxylation of SKP1, a homologue of the FBOX component of the E3 ubiquitin ligase (49). Furthermore, most of the active site features, other than those involved in iron and 2OG binding, that appear to differentiate PHD2 from other characterized 2OG oxygenases are well conserved. Although the evolutionary origins of the bacterial PHD-like enzymes are unclear and could be due to horizontal gene transfer from metazoans, they raise the possibility that the role of 2OG oxygenases in regulating cellular responses to oxygen are ancient.
Materials and Methods
PHD2181–426 was produced in Escherichia coli and purified by cation exchange chromatography (29). A PHD2181–417 construct was expressed in E. coli and purified on a Ni-NTA column followed by gel filtration.
Structure Solution by SAD.
PHD2181–426 crystals were grown anaerobically by using the hanging-drop method with 4-μl drops (2 μl of 20 mg·ml-1 protein/2 mM compound A/50 mM Tris·HCl, pH 7.5/2 μl of well solution) over well solution containing 1.6–2.0 M (NH4)2SO4, 2–8% dioxane, 100 mM MES (pH 6.5), and 1 mM Fe(II)SO4. Crystals were transferred directly to 50% NaMalonate pH 7.5 and frozen in liquid N2. A highly accurate and redundant data set was collected by using a single crystal (0.5 × 0.2 × 0.2 mm) mounted in an Oxford Cryosystems Cobra cryostream at 100 K by using a X8 Proteum (Bruker-AXS) equipped with a Microstar microfocus rotating anode x-ray generator producing CuKα radiation, four circle KAPPA goniometer, and SMART 6000 CCD detector. Data collection strategy (optimized for SAD) and indexing were performed by using proteumplus aiming for a target redundancy of 40 in fine-slice mode (0.25°). Data were integrated using saint and scaled by using sadabs followed by analysis and manipulation in xprep (Bruker-AXS, 2005). The structure was solved by using solve/resolve 2.08 (50). An Fe, I, and six sulfur sites were sufficient to obtain initial phases refined to a figure of merit of 0.31 (Fig. 12, which is published as supporting information on the PNAS web site). Density modification in resolve increased the figure of merit to 0.58 and resulted in a high quality electron density map to 2.2 Å. Model building was done by using the graphical software coot 0.26 (51) and crystallographic refinement was performed by using cns 1.1 (52).
Structure Solution by Single Isomorphous Replacement with Anomalous Scattering/Multiple Wavelength Anomalous Diffraction.
Crystals of the PHD2181–417-(His)6 in complex with compound A were grown by the hanging drop vapor diffusion method from 28% polyethylene glycol 8000/200 mM (NH4)2SO4/100 mM Mes, pH 6.4 at a protein concentration of 15 mg/ml. The crystal structure of PHD2181–417 was determined by combining phases from single isomorphous replacement with anomalous scattering and multiple wavelength anomalous diffraction. solve located five Hg sites in the HgAc2 derivative, the Fe atom site in the native data set with anomalous differences, and four Se sites in the SeMet-derivatized PHD2181–417-(His)6 crystal from multiple wavelength anomalous diffraction data collected at three different wavelengths around the Se absorption edge. Phase refinement resulted in an initial figure of merit of 0.43. Density modification by resolve increased the figure of merit to 0.67 and a produced a very good-quality map by using phases to 2.5 Å. Model building was done by using quanta2000.2 (Accelrys, Inc., San Diego) and refinement with cnx2000.12 (Accelrys, Inc.) and native data collected to 1.7 Å. For detailed methods and a table of crystallographic data and refinement statistics, see Supporting Text and Table 1, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank P. J. Ratcliffe and C. W. Pugh for discussion and encouragement; K. Harlos and T. Walter for assistance with crystallization robotics; E. F. Garman and E. D. Lowe for assistance SAD data collection; V. L. Rath of Reciprocal Space Consulting for data collection at Advanced Light Source; C. Tegley and S. Mercede of Amgen for compound A; S. Elliott, R. Hungate, P. Tagari, and J. Zhang of Amgen for discussion; and D. Schnieder, A. Saxena, and A. Soares at National Synchrotron Light Source beamline X12B for contributions to preliminary experiments with PHD2181–426. Wellcome Trust and the Biotechnology and Biological Sciences Research Council provided partial funding for the research.
Abbreviations
- DSBH
double stranded β-helix
- FIH
factor inhibiting HIF
- HIF
hypoxia-inducible factor
- 2OG
2-oxoglutarate
- PHD
prolyl hydroxylase
- SAD
single-wavelength anomalous diffraction.
Footnotes
Conflict of interest statement: C.J.S. is a cofounder and K.S.H. and L.A.M. are employees of ReOx, Ltd., Oxford, and R.S.S., V.L., J.Z., J.L., R.J.M.K., E.Y., and S.J. are employees of Amgen, Inc. Both ReOX and Amgen are developing therapeutic inhibitors of the HIF hydroxylases.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2G19 and 2G1M).
References
- 1.Semenza G. L., Nejfelt M. K., Chi S. M., Antonarakis S. E. Proc. Natl. Acad. Sci. USA. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivan M., Kondo K., Yang H. F., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin J., William G. Biochem. Biophys. Res. Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 5.Dann C. E., III, Bruick R. K. Biochem. Biophys. Res. Commun. 2005;338:639–647. doi: 10.1016/j.bbrc.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 6.Hirota K., Semenza G. L. Biochem. Biophys. Res. Commun. 2005;338:610–616. doi: 10.1016/j.bbrc.2005.08.193. [DOI] [PubMed] [Google Scholar]
- 7.Schofield C. J., Ratcliffe P. J. Biochem. Biophys. Res. Commun. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 8.Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 9.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O’Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., et al. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Bruick R. K., McKnight S. L. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Gunzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., Kaelin W. G., Jr. Proc. Natl. Acad. Sci. USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y.-M., Bullock A. N., Welford R. W. D., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., et al. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 13.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berra E., Benizri E., Ginouves A., Volmat V., Roux D., Pouyssegur J. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 16.Clifton I. J., McDonough M. A., Ehrismann D., Kershaw N. J., Granatino N., Schofield C. J. J. Inorg. Biochem. 2006;100:644–649. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Hausinger R. P. Crit. Rev. Biochem. Mol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 18.Hewitson K. S., McNeill L. A., Elkins J. M., Schofield C. J. Biochem. Soc. Trans. 2003;31:510–515. doi: 10.1042/bst0310510. [DOI] [PubMed] [Google Scholar]
- 19.McNeill L. A., Hewitson K. S., Gleadle J., Horsfall L. E., Oldham N. J., Maxwell P., Pugh C. W., Ratcliffe P. J., Schofield C. J. Bioorg. Med. Chem. Lett. 2002;12:1547–1550. doi: 10.1016/s0960-894x(02)00219-6. [DOI] [PubMed] [Google Scholar]
- 20.Myllyharju J. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 21.McDonough M. A., Kavanagh K. L., Butler D., Searls T., Oppermann U., Schofield C. J. J. Biol. Chem. 2005;280:41101–41110. doi: 10.1074/jbc.M507528200. [DOI] [PubMed] [Google Scholar]
- 22.Jansen G. A., Waterman H. R., Wanders R. J. A. Hum. Mutat. 2004;23:209–218. doi: 10.1002/humu.10315. [DOI] [PubMed] [Google Scholar]
- 23.Trewick S. C., Henshaw T. F., Hausinger R. P., Lindahl T., Sedgwick B. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 24.Falnes P. O., Johansen R. F., Seeberg E. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 25.Tsukada Y.-i., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 26.McNeill L. A., Bethge L., Hewitson K. S., Schofield C. J. Anal. Biochem. 2005;336:125–131. doi: 10.1016/j.ab.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 27.Ponstingl H., Kabir T., Thornton J. M. J. Appl. Crystal. 2003;36:1116–1122. [Google Scholar]
- 28.Valegard K., van Scheltinga A. C. T., Lloyd M. D., Hara T., Ramaswamy S., Perrakis A., Thompson A., Lee H. J., Baldwin J. E., Schofield C. J., et al. Nature. 1998;394:805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 29.Elkins J. M., Hewitson K. S., McNeill L. A., Seibel J. F., Schlemminger I., Pugh C. W., Ratcliffe P. J., Schofield C. J. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 30.Dann C. E., III, Bruick R. K., Deisenhofer J. Proc. Natl. Acad. Sci. USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C., Kim S. J., Jeong D. G., Lee S. M., Ryu S. E. J. Biol. Chem. 2003;278:7558–7563. doi: 10.1074/jbc.M210385200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Ren J.-S., Clifton I. J., Schofield C. J. Chem. Biol. 2004;11:1383–1394. doi: 10.1016/j.chembiol.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Stirk H. J., Woolfson D. N., Hutchinson E. G., Thornton J. M. FEBS Lett. 1992;308:1–3. doi: 10.1016/0014-5793(92)81036-l. [DOI] [PubMed] [Google Scholar]
- 34.Ozer A., Wu L. C., Bruick R. K. Proc. Natl. Acad. Sci. USA. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeill L. A., Flashman E., Buck M. R. G., Hewitson K. S., Clifton I. J., Jeschke G., Claridge T. D. W., Ehrismann D., Oldham N. J., Schofield C. J. Mol. BioSys. 2005;4:312–324. doi: 10.1039/b511249b. [DOI] [PubMed] [Google Scholar]
- 36.Mole D. R., Schlemminger I., McNeill L. A., Hewitson K. S., Pugh C. W., Ratcliffe P. J., Schofield C. J. Bioorg. Med. Chem. Lett. 2003;13:2677–2680. doi: 10.1016/s0960-894x(03)00539-0. [DOI] [PubMed] [Google Scholar]
- 37.Ryle M. J., Liu A., Muthukumaran R. B., Ho R. Y., Koehntop K. D., McCracken J., Que L., Jr., Hausinger R. P. Biochemistry. 2003;42:1854–1862. doi: 10.1021/bi026832m. [DOI] [PubMed] [Google Scholar]
- 38.Barlow J. N., Zhang Z., John P., Baldwin J. E., Schofield C. J. Biochemistry. 1997;36:3563–3569. doi: 10.1021/bi962521y. [DOI] [PubMed] [Google Scholar]
- 39.Hewitson K. S., McNeill L. A., Schofield C. J. Curr. Pharm. Des. 2004;10:821–833. doi: 10.2174/1381612043452884. [DOI] [PubMed] [Google Scholar]
- 40.Cunliffe C. J., Franklin T. J., Hales N. J., Hill G. B. J. Med. Chem. 1992;35:2652–2658. doi: 10.1021/jm00092a016. [DOI] [PubMed] [Google Scholar]
- 41.McDonough M. A., McNeill L. A., Tilliet M., Papamicael C. A., Chen Q. Y., Banerji B., Hewitson K. S., Schofield C. J. J. Am. Chem. Soc. 2005;127:7680–7681. doi: 10.1021/ja050841b. [DOI] [PubMed] [Google Scholar]
- 42.Banerji B., Conejo-Garcia A., McNeill L. A., McDonough M. A., Buck M. R., Hewitson K. S., Oldham N. J., Schofield C. J. Chem. Commun. 2005:5438–5440. doi: 10.1039/b510707e. [DOI] [PubMed] [Google Scholar]
- 43.Percy M. J., Zhao Q., Flores A., Harrison C., Lappin T. R. J., Maxwell P. H., McMullin M. F., Lee F. S. Proc. Natl. Acad. Sci. USA. 2006;103:654–659. [Google Scholar]
- 44.Clifton I. J., Hsueh L. C., Baldwin J. E., Harlos K., Schofield C. J. Eur. J. Biochem. 2001;268:6625–6636. doi: 10.1046/j.0014-2956.2001.02617.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z., Ren J.-s., Harlos K., McKinnon C. H., Clifton I. J., Schofield C. J. FEBS Lett. 2002;517:7–12. doi: 10.1016/s0014-5793(02)02520-6. [DOI] [PubMed] [Google Scholar]
- 46.Lee H. J., Lloyd M. D., Harlos K., Clifton I. J., Baldwin J. E., Schofield C. J. J. Mol. Biol. 2001;308:937–948. doi: 10.1006/jmbi.2001.4649. [DOI] [PubMed] [Google Scholar]
- 47.Min J. H., Yang H. F., Ivan M., Gertler F., Kaelin W. G., Pavletich N. P. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 48.Hon W. C., Wilson M. I., Harlos K., Claridge T. D. W., Schofield C. J., Pugh C. W., Maxwell P. H., Ratcliffe P. J., Stuart D. I., Jones E. Y. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 49.van der Wel H., Ercan A., West C. M. J. Biol. Chem. 2005;280:14645–14655. doi: 10.1074/jbc.M500600200. [DOI] [PubMed] [Google Scholar]
- 50.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.