Abstract
Cyanobacteria, blue-green algae, are the most abundant autotrophs in aquatic environments and form the base of all aquatic food chains by fixing carbon and nitrogen into cellular biomass. The single most important nutrient for photosynthesis and growth is nitrate, which is severely limiting in many aquatic environments particularly the open ocean. It is therefore not surprising that NrtA, the solute-binding component of the high-affinity nitrate ABC transporter, is the single-most abundant protein in the plasma membrane of these bacteria. Here, we describe the structure of a nitrate-specific receptor, NrtA from Synechocystis sp. PCC 6803, complexed with nitrate and determined to a resolution of 1.5 Å. NrtA is significantly larger than other oxyanion-binding proteins, representing a previously uncharacterized class of transport proteins. From sequence alignments, the only other solute-binding protein in this class is CmpA, a bicarbonate-binding protein. Therefore, these organisms created a solute-binding protein for two of the most important nutrients: inorganic nitrogen and carbon. The electrostatic charge distribution of NrtA appears to force the protein off the membrane while the flexible tether facilitates the delivery of nitrate to the membrane pore. The structure not only details the determinants for nitrate selectivity in NrtA but also the bicarbonate specificity in CmpA. Nitrate and bicarbonate transport are regulated by the cytoplasmic proteins NrtC and CmpC, respectively. Interestingly, the residues lining the ligand binding pockets suggest that they both bind nitrate. This implies that the nitrogen and carbon uptake pathways are synchronized by intracellular nitrate and nitrite.
Keywords: ABC transporter, carbon, nitrogen assimilation, regulation
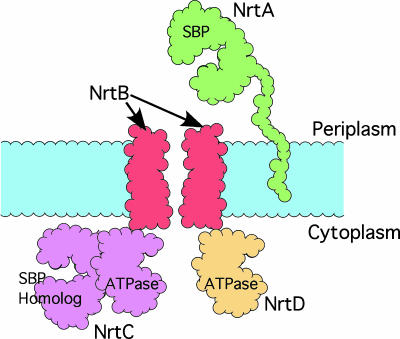
The single most important nutrient for photosynthesis and growth is nitrate, which is severely limiting in many aquatic environments such as the open ocean (1, 2). Therefore, the cyanobacteria have developed a high-affinity ABC transport system that is composed of four polypeptides (Fig. 1): a high-affinity periplasmic solute-binding lipoprotein (NrtA), an integral membrane permease (NrtB), a cytoplasmic ATPase (NrtD), and a unique ATPase/solute-binding fusion protein (NrtC) that regulates transport (3). NrtA binds both nitrate and nitrite (Kd = 0.3 mM) and is necessary for cell survival when nitrate is the primary nitrogen source (4). The role of NrtA is to scavenge nitrate/nitrite from the periplasm for delivery to the membrane permease, NrtB. The passage of solute through the transmembrane pore is linked to ATP hydrolysis by NrtC and NrtD. NrtD consists of a single ATPase domain. In contrast, NrtC contains both an ATPase domain and a C-terminal solute-binding domain that shares 50% amino acid sequence similarity with NrtA, and is required for the ammonium-mediated inhibition of nitrate transport (5, 6). Aside from the homologous transporter for bicarbonate, CmpABCD, there are no other known examples of ABC transporters that have an ATPase/solute-binding fusion component.
Fig. 1.
Cartoon representation of the assembled NrtABCD nitrate transporter. NrtA is tethered to the periplasmic membrane by a flexible linker and captures nitrate/nitrite in the periplasm for delivery to the transmembrane complex created by NrtB. In many ABC transporters, the transmembrane pore is created by a dimer of two transmembrane spanning polypeptides. NrtC and NrtD are ATPases that couple ATP hydrolysis to nitrate/nitrite transport through the pore. NrtC is unique in that it contains a C-terminal solute-binding domain homologous to NrtA.
The specificity of the nitrate transporter is conferred by NrtA (3). NrtA is ≈49% identical (60% similar) in amino acid sequence to the bicarbonate receptor CmpA. In its entirety, it does not have significant homology to any other known protein. To elucidate the molecular determinants of nitrate specificity, we determined the crystal structure of the Synechocystis 6803 NrtA to 1.5 Å. Although the general shape of NrtA is akin to that of other solute-binding proteins, NrtA clearly represents a unique structural variant of these “C-clamp” proteins. From this structure and sequence alignments of other bicarbonate and nitrate transporters, the molecular basis for solute selectivity is clear and suggests that regulatory domains of both bicarbonate and nitrate transport systems bind nitrate. Based on these findings, a model is presented that demonstrates how such synergistic regulation of bicarbonate and nitrate transport is important in conserving energy during the process of carbon fixation and nitrogen assimilation.
Results
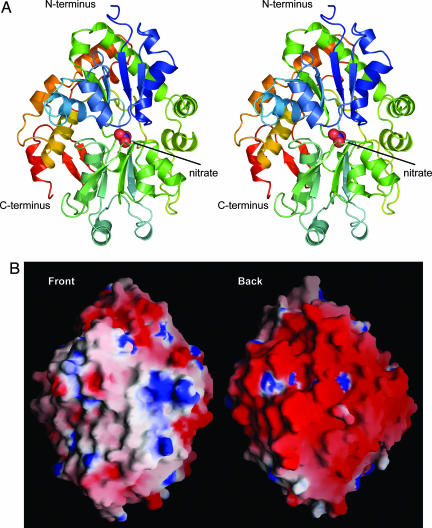
NrtA is an α/β protein composed of two domains (I and II) organized with a C-clamp shape (Fig. 2A). Nitrate occupies the cleft between the two domains. Both domains consist of a five-stranded mixed β-sheet surrounded by α-helices. The order of the β-strands (21354) within domain I suggests NrtA belongs to class II of the periplasmic-binding protein (PBP) superfamily (7) that also includes the oxyanion-binding proteins for phosphate (8, 9), sulfate (10), and molybdate (11, 12). Like NrtA, these structures consist of two globular domains of mixed β-sheet flanked by α-helices, with the solute-binding cleft located between the two domains. However, NrtA has an extra 100 aa extending from the C terminus. These residues comprise several α-helices and a two-stranded antiparallel β-sheet that cradle the back of the C-clamp. This architectural feature may provide structural support for the nitrate-binding pocket or play a role in the conformational changes associated with solute transport.
Fig. 2.
The structure of NrtA. (A) Stereo ribbon representation of the NrtA crystal structure colored blue to red as the chain extends from the N terminus to the C terminus. NrtA consists of two α/β domains arranged with a C-clamp shape, with nitrate, depicted as spheres, bound in the cleft between the two domains. The view is of the front of the C-clamp, which opens to the nitrate-binding cleft. (B) Electrostatic surface potential of the front and back of the NrtA structure, with positive and negative charge shown in blue and red, respectively.
Like most bacterial periplasmic-binding proteins, NrtA is tethered to the membrane surface by a lipidated cysteine (C30) and a very long flexible linker (residues 31–56) rich in glycine and serine (3). Therefore NrtA is akin to a “balloon on a string” with its solute-binding domain capturing nitrate/nitrite in the periplasm and transporting it to the NrtB transmembrane permease. This process appears to be partially driven by an unusual and asymmetric distribution of electrostatic potential (Fig. 2B). The entire back of NrtA is enveloped by negative charge that may facilitate NrtB association with the front of NrtA by preventing unproductive association with the phospholipid bilayer. By forcing NrtA off the surface of the membrane, the likelihood of solute capture and appropriate contact with NrtB is increased.
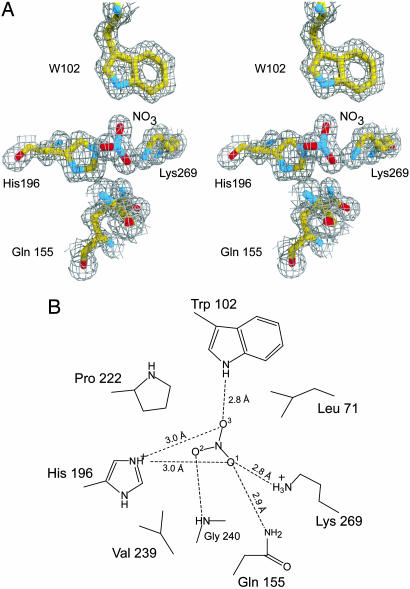
As suggested by previous biochemical analysis (4), one molecule of nitrate is bound per NrtA protein (Fig. 3). The nitrate is completely occluded from solvent by several hydrophobic side chains at the entrance to the binding site: L71, P222, and V239. In an aqueous environment, the resonance state of the nitrate anion evenly distributes the negative charge among the three oxygen atoms. However, the environment surrounding the nitrate oxygens in the NrtA binding pocket is clearly asymmetric (Fig. 3 A and B). O1 of the nitrate anion is proximal to the positive charge from the Nζ of K269 and Nε2 of H196 (2.8 and 3.0 Å, respectively), and is 2.9 Å from the Nε2 of Q155. In contrast, O2 is hydrogen-bonded to the amide nitrogen of G240 but is otherwise surrounded by the hydrophobic side chains of P222 and V239. O3 is hydrogen-bonded to the Nε of W102 (2.8 Å) and is within 3.0 Å of the Nε2 of H196. The result is a polarization of the charge distribution of nitrate, with O1 having a greater partial negative charge than both O2 and O3. Thus, NrtA selects for one of the nitrate resonance states through this asymmetric environment. From the number and types of interactions, it seems likely that the O1 and O3 interactions will remain the same but that the O2 atom and its interactions with the protein will be absent in the case of nitrite. O2 in the bound nitrate molecule has relatively few interactions and, hence, may be why NrtA binds nitrate and nitrite with approximately the same affinity.
Fig. 3.
The nitrate-binding cavity. (A) Representative electron density of nitrate and coordinating residues. The map shown was calculated with the coefficients (2Fo − Fc), where Fo is the experimentally observed structure factor amplitude and Fc is the calculated structure factor amplitude from the model. For the sake of clarity, G240 was omitted. (B) Schematic representation of the nitrate-binding site. Shown here are the protein–ligand interactions between NrtA and nitrate with all potential hydrogen-bonding and electrostatic interactions depicted as dashed lines.
The closest homologue of NrtA is CmpA, a bicarbonate-binding protein found in several freshwater species of cyanobacteria (e.g., Synechocystis 6803) (Table 1). The high degree of sequence homology between these proteins suggests that they share a similar fold and common ancestry but have diverged to create highly selective binding pockets for their respective ligands. To better understand the structural determinants for ligand specificity, the sequences of NrtA and CmpA were aligned and the putative binding pockets were compared. The most notable difference between NrtA and CmpA is the substitution of K269 of NrtA with a glutamate in CmpA. In NrtA, K269 complements the negative charge from O1 of the nitrate and aids in polarizing the nitrate anion (Fig. 3B). The substitution to glutamate in CmpA would abrogate nitrate binding due to charge repulsion between the O1 of nitrate and the carboxylic acid side chain. Conversely, glutamate in CmpA provides a hydrogen-bonding acceptor for the hydroxyl group of bicarbonate. This finding suggests that the anion selectivity for these two proteins is largely based on a single amino acid substitution. It is not clear whether the threonine at position 239 in CmpA, as opposed to valine in NrtA, would interact with bicarbonate and affect ligand specificity. However, it should be noted that this residue is either valine or isoleucine in CmpC. The final notable difference in the putative binding cleft is the conservative change from H196 in NrtA to glutamine in CmpA. The likely defining factor for nitrate versus bicarbonate binding is the positive versus negative charge at residue 269. This sophisticated level of substrate discrimination, based on the deletion or insertion of a hydrogen-bonding acceptor, has been observed in the strict solute preference of the Escherichia coli phosphate-binding protein (8).
Table 1.
Sequence alignment of the putative ligand-binding residues within the nitrate- and bicarbonate-binding proteins
| Protein* | Ligand-binding residues† |
% identity/% similarity‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 71 | 102 | 155 | 196 | 222 | 239 | 240 | 269 | ||
| NrtA | |||||||||
| S. 6803 | L | W | Q | H | P | V | G | K | 100/100 |
| S. 7942 | L | W | Q | H | P | V | G | K | 56/72 |
| P. laminosum | P | W | Q | H | P | V | G | K | 67/79 |
| T. elongatus | L | W | Q | H | P | V | G | K | 66/77 |
| CmpA | |||||||||
| S. 6803 | I | W | N | Q | P | T | G | E | 48/61 |
| S. 7942 | I | W | N | Q | S | T | G | E | 45/61 |
| T. elongatus | I | W | N | Q | P | T | G | E | 48/63 |
| NrtC§ | |||||||||
| S. 6803 | L | W | N | Q | P | V | G | K | 33/51 |
| S. 7942 | G | W | G | P | P | A | G | K | 29/46 |
| P. laminosum | L | W | N | H | P | V | G | K | 34/54 |
| T. elongatus | L | W | N | H | P | V | G | K | 33/52 |
| CmpC§ | |||||||||
| S. 6803 | L | W | N | H | P | V | S | K | 29/49 |
| S. 7942 | L | W | N | H | P | I | G | K | 29/51 |
| T. elongatus | L | W | N | H | P | V | G | K | 30/52 |
*Protein sequences were from the following cyanobacteria: Synechocystis 6803, Synechococcus 7942, Phormidium laminosum, and Thermosynechococcus elongatus BP-1.
†Based on overall amino acid sequence alignments by clustalw.
‡Percent amino acid identity and similarity is given in relation to the Synechocystis 6803 NrtA sequence.
§Only the last 380 residues were used.
The flux of nitrate through the NrtABCD transporter is controlled by the unique ATPase/solute-binding protein NrtC (5, 6). The C-terminal domain of NrtC (380 residues) is >30% identical and 50% similar to NrtA and inhibits nitrate uptake when ammonium is added to the culture media. Mutants lacking the C-terminal domain of NrtC do not have this feedback inhibition. However, the mechanism of this inhibition and the specificity of the NrtC binding pocket is unknown. To ascertain the preferred ligand of NrtC, the C-terminal domain of NrtC was aligned to NrtA (Table 1). Interestingly, these two binding sites are nearly identical, with only conservative differences at residues 155 and 196 (NrtA numbering). With the exception of NrtC from Synechococcus sp. PCC 7942, the overall pattern of hydrogen-bonding donors and acceptors is the same. Based on both the overall sequence similarity and the conservation of hydrogen-bonding donors in the putative binding pocket, it seems more likely that NrtC binds nitrate/nitrite than ammonium.
Surprisingly, the solute-binding pocket of CmpC appears to also favor nitrate binding. Like NrtC, CmpC is thought to play a role in regulating bicarbonate uptake, although the mechanism of inhibition is unknown. The solute-binding domain of CmpC is ≈50% similar to NrtA, and the alignment of the solute-binding domains shows that the proposed binding pocket of CmpC is almost identical to that of NrtA. In particular, the presence of a lysine in CmpC at the equivalent position as K269 of NrtA suggests that nitrate, and not bicarbonate, is the likely solute.
Discussion
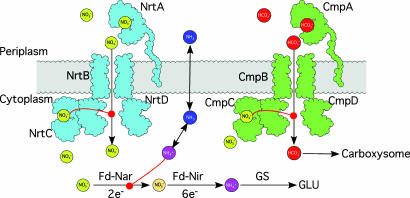
In cyanobacteria, carbon sequestration via photosynthesis and nitrogen fixation are interdependent (13). The comparison of the NrtA and putative NrtC and CmpC binding sites suggests that all three proteins bind nitrate. It is possible that NrtC and CmpC bind nitrate as a way to coordinate nitrogen and carbon uptake as summarized in Fig. 4. Under normal conditions, intracellular nitrate is immediately reduced to nitrite and then ammonium, requiring a total of eight electrons from reduced ferredoxin, the first electron acceptor of photosystem I (14). Ammonium, when available, is the preferred nitrogen source over nitrate because it can be directly incorporated into cellular biomass without expending valuable reducing equivalents (13). In wild-type cells, ammonium not only inhibits nitrate uptake but also halts nitrate utilization by inhibiting nitrate reductase (6). Mutants lacking the solute-binding domain of NrtC continue nitrate uptake in the presence of ammonium but can no longer reduce nitrate because of the inhibition of nitrate reductase (6). The inhibition of nitrate reduction leads to an increase in intracellular nitrate. As intracellular nitrate levels rise, CmpC and NrtC may bind nitrate to allosterically control nitrate and bicarbonate uptake. In the aquatic environment of most cyanobacteria, inorganic nitrogen is more limiting than carbon. Therefore, efficient control of the intracellular nitrogen and carbon balance will rely on the concentration of the more limiting nutrient. Without such regulation, a great deal of energy would be wasted in reducing nitrate to ammonium that, if unused, will dissipate across the cellular membrane. It should be noted that there are several bicarbonate transport systems in the cyanobacteria and that nitrate regulation of the Cmp and Nrt import systems may not be completely responsible for controlling the carbon and nitrogen balance. However, mutagenesis studies in Synechococcus have shown that the Cmp system has the highest affinity for bicarbonate (15) and therefore this model may be relevant at low bicarbonate and nitrate concentrations that are often found in the natural environments where cyanobacteria grow.
Fig. 4.
Model for synergistic regulation of nitrate and bicarbonate uptake. As shown in this model, both the Nrt (cyan) and Cmp (green) complexes are inhibited by nitrate (yellow balls). If energy or nutrient supplies were to become limiting, both nitrogen and carbon fixation would be blocked and the levels of ammonium (mauve balls) would rise. This, in turn, increases the concentration of nitrate and shuts down both bicarbonate and nitrate uptake. It is logical to have nitrate act as the feedback regulator because its reduction to membrane permeable ammonia (dark blue balls) would waste a great deal of cellular reductive potential if not fully used by carbon and nitrogen fixation pathways.
The 1.5-Å structure of NrtA is the first structure of a nitrate receptor and reveals the molecular determinants of ligand specificity. The similarity of NrtA to CmpA, NrtC, and CmpC not only sheds light on the possible differences between nitrate and bicarbonate binding but also reveals a previously unknown link between nitrate and bicarbonate uptake in cyanobacteria. It remains unclear, however, as to why the bicarbonate and nitrate solute-binding domains are so highly conserved and yet different from other oxyanion-binding proteins.
Materials and Methods
Cloning and Protein Expression.
The solute-binding domain of nrtA (residues 30–447) from Synechocystis 6803 was cloned and expressed in E. coli. The NrtA gene was expressed with an N-terminal 6× His-tag in pET-28a (Novagen) that was modified such that the thrombin cleavage site was mutated to a recombinant tobacco etch virus (rTEV) cleavage site. E. coli Rosetta(DE3)pLysS (Novagen) cells were transformed with the pET28a-nrtA plasmid. Selenomethionine-substituted protein was expressed via the methionine inhibitory pathway (16). Native and selenomethionine-substituted NrtA were purified in an identical manner via Ni2+ affinity chromatography. Protein-containing fractions were pooled based on SDS/PAGE and dialyzed against 20 mM Hepes (pH 7.5) with 100 mM NaCl. The rTEV protease was added to dialyzed NrtA at a molar ratio of 1 rTEV:100 NrtA, and the mixture was incubated for 16 h at ≈23°C. The rTEV protease contained an N-terminal 6× His-tag, which allowed the separation of cleaved NrtA from both His-tagged NrtA and rTEV via Ni2+ affinity chromatography. Purified NrtA was exhaustively dialyzed against 20 mM Hepes (pH 7.5) with 100 mM NaCl and concentrated to ≈19 mg/ml.
Crystallization and Data Collection.
Native and selenomethionine-substituted NrtA crystals were grown via the hanging-drop method of vapor diffusion against 2.0–2.2 M ammonium sulfate, 20 mM NaNO3, and 100 mM succinate (pH 5.0). Crystals typically grew to dimensions of ≈0.5 mm × 0.5 mm × 0.3 mm and belonged to the space group P21 with unit cell dimensions of a = 55.9 Å, b = 53.8 Å, c = 65.9 Å, β = 107.7°. The solvent content of the crystals was ≈40%, with one monomer in the asymmetric unit. Crystals were harvested from the hanging-drop experiments and soaked for several hours to several days in synthetic mother liquor composed of 2.2 M ammonium sulfate, 300 mM NaCl, 25 mM NaNO3, and 50 mM succinate (pH 5.0). These crystals were serially transferred to a final cryoprotectant solution containing 20% ethylene glycol, 2.2 M ammonium sulfate, 300 mM NaCl, 25 mM NaNO3, and 50 mM succinate (pH 5.0). The crystals were flash-frozen with liquid nitrogen. X-ray data from both the native protein and the selenomethionine-substituted protein crystals were collected on a 3 × 3 tiled “SBC3” CCD detector at the Structural Biology Center 19-BM beamline (Advanced Photon Source, Argonne National Laboratory, Argonne, IL). The x-ray data were processed with hkl2000 and scaled with scalepack (17). The relevant x-ray data collection statistics are summarized in Table 2.
Table 2.
Data collection, phasing, and refinement statistics
| Parameter | Native | SeMet NrtA | ||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | ||
| Cell dimensions | ||||
| a, b, c, Å | 55.96, 53.8, 65.88 | 55.98, 53.76, 65.95 | ||
| α, β, γ, ° | 90, 107.7, 90 | 90, 107.7, 90 | ||
| Peak | Inflection | Remote | ||
| Wavelength | 0.97132 | 0.97899 | 0.97915 | 0.97132 |
| Resolution, Å | 50–1.51 | 50–2.01 | 50–2.01 | 50–1.99 |
| Rsym | 5.9 (15.0) | 8.4 (10.8) | 9.6 (12.4) | 6.4 (8.3) |
| I/σI | 20.3 (6.0) | 28.0 (18.3) | 28.5 (18.4) | 27.1 (18.7) |
| Completeness, % | 98.4 (99.4) | 97.4 (91.8) | 97.3 (91.0) | 97.2 (96.6) |
| Redundancy | 3.9 (3.7) | 4.0 (3.8) | 4.0 (3.8) | 3.9 (3.8) |
| Refinement | ||||
| Resolution, Å | 50–1.5 | |||
| No. of reflections | 57920 | |||
| Rwork/Rfree | 20.12/22.16 | |||
| No. of atoms | 3,236 | |||
| Protein | 3,017 | |||
| Ligand/ion | 4 | |||
| Water | 215 | |||
| B-factors | ||||
| Protein | 15.8 | |||
| Ligand/ion | 12.6 | |||
| Water | 24.3 | |||
| rms deviations | ||||
| Bond lengths, Å | 0.0139 | |||
| Bond angles, ° | 1.67 |
X-Ray Structural Analyses.
The structure of NrtA was determined via a traditional three-wavelength MAD experiment. The software package solve (18) was used to determine and refine the positions of the selenium atoms, and the program resolve (19) was then used to perform solvent flattening and initial protein model building. resolve successfully built 313 of 428 residues for NrtA, and the remaining residues were added manually by using the graphics package o (20). The experimental model was refined against the native data to 1.5 Å in cns (21). Further cycles of refinement in cns and manual model-building reduced the Rwork to 20.1% [Rfree (22) = 22%] for all measured x-ray data from 50 to 1.5 Å resolution. A Ramachandran plot of the coordinates in procheck shows that 90.9% of the residues are in the allowed regions, 7.9% are in the additionally allowed regions, 1.2% are in the generously allowed regions, and 0% are in the disallowed regions. Relevant maximum-likelihood refinement statistics are summarized in Tables 2 and 3. Representative electron density for the nitrate-binding site is shown in Fig. 3A.
Table 3.
X-ray refinement statistics
| Resolution, Å | 50–1.5 |
| Rwork/Rfree, % | 20.12/22.16 |
| No. of atoms | 3,236 |
| Protein | 3,017 |
| Ligand/ion | 4 |
| Water | 215 |
| B-factors | |
| Protein | 15.8 |
| Ligand/ion | 12.6 |
| Water | 24.3 |
| rms deviations | |
| Bond lengths, Å | 0.0139 |
| Bond angles, ° | 1.67 |
Acknowledgments
We thank the staff of the Structural Biology Consortium (19-BM) at the Advanced Photon Source for assistance with data collection and data reduction, Dr. Maitrayee Bhattacharyya-Pakrasi for helpful advice throughout the course of these investigations, and Dr. James Thoden for providing the pET-28rTEV expression plasmid. This work is part of a Membrane Biology EMSL Scientific Grand Challenge project at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy Office of Biological and Environmental Research program located at Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated for the Department of Energy by Battelle.
Abbreviation
- rTEV
recombinant tobacco etch virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The x-ray coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2G29).
References
- 1.Guerrero M. G., Vega J. M., Losada M. Annu. Rev. Plant Physiol. 1981;32:169–204. [Google Scholar]
- 2.Redfield A. C., Ketchum B. H., Richards F. A. In: The Sea. Hill M. N., editor. Vol. 2. New York: Wiley; 1963. pp. 26–77. [Google Scholar]
- 3.Omata T. Plant Cell Physiol. 1995;36:207–213. doi: 10.1093/oxfordjournals.pcp.a078751. [DOI] [PubMed] [Google Scholar]
- 4.Maeda S., Omata T. J. Biol. Chem. 1997;272:3036–3041. doi: 10.1074/jbc.272.5.3036. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi M., Takatani N., Tanigawa M., Omata T. J. Bacteriol. 2005;187:498–506. doi: 10.1128/JB.187.2.498-506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi M., Rodriguez R., Lara C., Omata T. J. Biol. Chem. 1997;272:27197–27201. doi: 10.1074/jbc.272.43.27197. [DOI] [PubMed] [Google Scholar]
- 7.Dwyer M. A., Hellinga H. W. Curr. Opin. Struct. Biol. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Choudhary A., Ledvina P. S., Quiocho F. A. J. Biol. Chem. 1994;269:25091–25094. doi: 10.2210/pdb1pbp/pdb. [DOI] [PubMed] [Google Scholar]
- 9.Vyas N. K., Vyas M. N., Quiocho F. A. Structure (London) 2003;11:765–774. doi: 10.1016/s0969-2126(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 10.Pflugrath J. W., Quiocho F. A. J. Mol. Biol. 1988;200:163–180. doi: 10.1016/0022-2836(88)90341-5. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y., Rech S., Gunsalus R. P., Rees D. C. Nat. Struct. Biol. 1997;4:703–707. doi: 10.1038/nsb0997-703. [DOI] [PubMed] [Google Scholar]
- 12.Lawson D. M., Williams C. E., Mitchenall L. A., Pau R. N. Structure (London) 1998;6:1529–1539. doi: 10.1016/s0969-2126(98)00151-8. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero M. G., Lara C. In: The Cyanobacteria. Fay P., Van Baalen C., editors. Amsterdam: Elsevier Science; 1987. [Google Scholar]
- 14.Candau P., Manzano C., Losada M. Nature. 1976;262:715–717. [Google Scholar]
- 15.Omata T., Price G. D., Badger M. R., Okamura M., Gohta S., Ogawa T. Proc. Natl. Acad. Sci. USA. 1999;96:13571–13576. doi: 10.1073/pnas.96.23.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger T. C. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones T. A., Zou J.-Y., Cowan S. W. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A. T. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]