Abstract
Terpene synthases are a mechanistically intriguing family of enzymes that catalyze complex, multistep reactions that are capable of generating hundreds of structurally diverse hydrocarbon and oxygenated scaffolds of biological and commercial importance. Interestingly, distantly related terpene synthases from fungi to plants all contain an invariant three-dimensional fold, and molecular comparisons of their active sites indicate that they are enriched with relatively inert amino acid residues that do not react directly with the reaction intermediates. Therefore, catalytic specificity appears to rely on the contour and dynamics of the active site created by the positioning of amino acid backbones and side chains on this catalytic surface and by supporting layers of residues surrounding the synthase active site cavity. Despite the high degree of structural relatedness among terpene synthases, previous studies suggest that no clear relationship between phylogenic organization and catalytic specificities is easily deciphered. We now report on the reciprocal interconversion of catalytic specificities between two distinct yet evolutionarily related terpene synthases based on the systematic identification and mutational replacement of variable residues within and surrounding the active site. Furthermore, we uncover previously undocumented biosynthetic activity during the interconversion, activity that could have been present in a common ancestor of these two highly related synthases. These results provide a simplified means for mapping structural features that are responsible for functional attributes and a strategy for identifying residues that differentiate divergent biosynthetic properties in phylogenetically related terpene synthases.
Keywords: phylogenetic relationships, rational design, sesquiterpene, structure–function
Terpenes comprise the most diverse collection of natural products known. Terpene hydrocarbon scaffolds are generated by the action of mono-, sesqui-, and diterpene synthases that catalyze multistep reactions with diphosphorylated substrates of 10 (geranyl diphosphate), 15 [farnesyl diphosphate (FPP)] or 20 (geranylgeranyl diphosphate) carbons (1). The reactions catalyzed by terpene synthases are unparalleled relative to other classes of enzymes because they often consist of a series of stereochemically complex steps. These reactions include ionization of the diphosphate substituent creating an acyclic and reactive carbocation intermediate. In some cases, the isomerization of the all-trans substrate configuration to a cis-trans isomer also occurs before subsequent reactions. Additional steps add increasing complexity and include regio- and stereospecific formation of single or multiple rings, proton eliminations to form double bonds, water quenching of carbocations to create terpene alcohols, and stereospecific hydride, methyl, and methylene migrations. Equally intriguing, the three-dimensional structure of terpene synthases from fungi to plants is highly conserved, including a “terpene fold” composed largely of inert amino acids lining the active site (2–4).
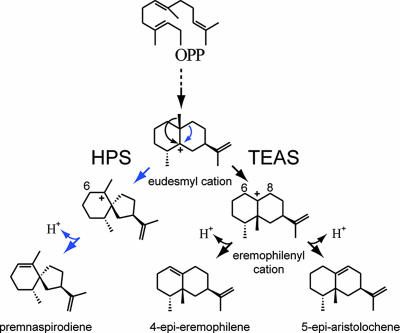
Molecular comparisons of Nicotiana tabacum 5-epiaristolochene synthase (TEAS) and Hyoscyamus muticus premnaspirodiene synthase (HPS), two sesquiterpene synthases from solanaceous plants that share 72% amino acid identity, have proven particularly useful in identifying structural elements underlying the evolution of product specificity in terpene synthases (5). TEAS catalyzes the initial ionization of FPP, which then undergoes two rounds of cyclization reactions, terminating with the migration of the C14 methyl group across the ring plane and neutralization through abstraction of a proton from C8 to yield 5-epi-aristolochene, a eudesmane-type sesquiterpene (Scheme 1). HPS catalyzes a series of reactions that is identical to those catalyzed by TEAS, up to but not including migration of the C14 methyl moiety. Instead, HPS catalyzes a migration of a methylene group followed by deprotonation at C6, yielding the vetispirane-type sesquiterpene premnaspirodiene. Domain-swapping experiments based on primary amino acid comparisons of TEAS and HPS previously demonstrated that specificity for the final reaction steps resides between amino acids 386 and 449 of HPS and amino acids 261 and 342 of TEAS, corresponding to exons 4 and 6, respectively, and that chimeric enzymes harboring both of these domains catalyze the biosynthesis of both the eudesmane and spirane-type reaction products (5).
Scheme 1.
Proposed reaction mechanisms for eremophilane (TEAS)-type and vetispiradiene (HPS)-type sesquiterpene synthases. TEAS and HPS share reaction steps leading up to the eudesmyl carbocation but then bifurcate. HPS catalyzes a methylene migration followed by deprotonation at C6 to generate premnaspirodiene, whereas TEAS catalyzes a methyl migration followed by deprotonation at C8 yielding 5-epi-aristolochene. Mutants of TEAS described herein catalyze the biosynthesis of 4-epi-eremophilene, a reaction product that arises from deprotonation at C6 rather than C8 of the eremophilenyl cation intermediate.
The association of separate structural elements of TEAS and HPS with distinct catalytic pathways for the final steps of FPP transformation was unexpected. Moreover, molecular modeling of HPS by using the structural coordinates for TEAS indicated that those residues in immediate contact with the initial substrate were nearly identical. Together, these results argued that catalytic specificity in these phylogenetically related terpene synthases must be modulated at a distance by residues surrounding the active site. This phenomenon has been observed in other enzyme systems where energetic pathways linking active site residues to distal sites of structure (described in refs. 6 and 7). The precedence for contributions of residues within and in supporting layers of the active site surface of terpene synthases to catalysis has been reported as well (8–11). For example, Köllner et al. (8) observed that a single amino acid substitution within the active site of a pair of maize sesquiterpene synthases was sufficient to change the stereochemical configuration of a methyl substituent for a dominant reaction product. Most recently, Yoshikuni et al. (9) furthered this observation by demonstrating that active-site saturation mutagenesis of γ-humulene synthase, a promiscuous sesquiterpene synthase generating >50 reaction products, yielded mutant synthases with narrower product specificity often possessing enhanced production of specific reaction products and much lower activities for others. Mutations of outer-tier residues of oxidosqualene cyclase, a mechanistically related triterpene synthase, suggested that residues residing outside the active site might influence catalysis by altering the position and/or dynamic properties of active site residues relative to reaction intermediates, either through alterations of hydrogen-bonding networks or direct physical interactions (10). Similarly, Hyatt and Croteau (11) reported that combinations of amino acid substitutions within (−)-pinene synthase, a monoterpene cyclase, located both in and around the active site altered the reaction product profile in subtle and distinct ways. Molecular modeling suggested that these mutations might induce changes in the active site shape and volume that are sufficient to reposition reactive intermediates for alternative reaction pathways.
Despite reports of how individual or groups of amino acid residues contribute to reaction cascades of terpene synthases, several of these studies have actually reported on the premature termination of multistep reactions and the release of neutral reaction intermediates (11–13). In contrast, domain-swapping experiments have illustrated the potential for engineering alternative catalytic pathways into the terpene synthases by using phylogenetic information, but without the resolution of defining the minimal structural changes required for altering or controlling discrete catalytic steps (5). Hence, a general model that uses comparative methods based on natural variation of terpene synthases to ascribe functional roles for particular amino acid positions has not yet been achieved. If such a model were possible, it should serve as a means to identify amino acids within and, importantly, surrounding the active site that contribute directly and indirectly to catalysis. It also should provide sufficient rationale to suggest a role(s) for each residue’s involvement in a catalytic mechanism, and it should predict how alternative or new catalytic activities and specificities can be rationally engineered into terpene synthases. Finally, although Yoshikuni et al. (9) argue that functional remodeling of terpene synthases based on phylogenetic comparisons is difficult at best, a unifying method of analysis and enzyme engineering should fundamentally recapitulate the sometimes subtle and difficult to discern lineages that are selected for during the evolution of biosynthetic diversity in terpene synthases.
In moving toward such a model for terpene synthase catalysis, the current study provides a simplified strategy for identifying amino acids that contribute to discrete reaction steps within terpene synthase reaction coordinates and then demonstrates the utility of this method for the reciprocal interconversion of two distinct classes of terpene synthase activities, those of eremophilene and vetispirane synthases. Most significantly, the successful interconversion of TEAS and HPS activities establishes a minimal set of residues responsible for divergent biosynthetic properties in eremophilene and vetisprirane synthases. In the process of systematically interconverting activities and identifying a previously unidentified activity, a framework facilitating the elucidation of the evolutionary relationships between sequence diversity of terpene synthases and the chemical complexity found in nature has also been established.
Results
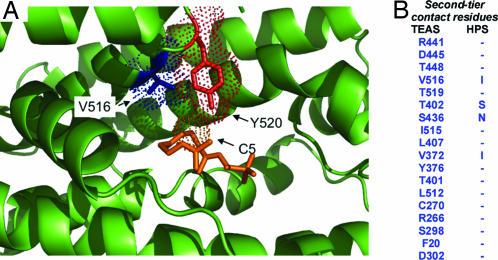
A contact mapping strategy was developed to formulate a more systematic means of identifying amino acids that are within or more removed from the active site surface but are poised to contribute to catalytic outcomes (Fig. 1). The map was generated by using the three-dimensional coordinates of TEAS complexed with farnesyl hydroxy-phosphonate (FHP), an unreactive FPP analog (2). The process commenced by identifying concentric “tiers” of amino acid contacts radiating outward from each carbon atom of FHP. Ten residues within van der Waals radii (≈3.5 Å) of FHP defined the first tier, whereas the second tier encompassed the next 18-residue shell of contacts (Fig. 1). The HPS amino acid sequence was then threaded onto the TEAS template structure and energy-minimized, and the identities of residues at each position were compared. According to the contact map that was generated, the first-tier residues of HPS and TEAS are identical, whereas only 4 of the corresponding 18 second-tier residues, 2 of which line the active site cavity, differed (Fig. 1).
Fig. 1.
Illustration of the contact mapping strategy to identify amino acid residues that differentiate the reaction specificity between terpene synthases. (A) Using the TEAS crystal structure complexed with the substrate analog farnesyl hydroxy-phosphonate (2) (orange), those amino acid residues within van der Waals radii (dotted spheres) for each carbon of the substrate analog were mapped and designated as first-tier residues (shown in red). Second-tier residues (shown in blue) surrounding and potentially influencing active site geometry were mapped as residues within van der Waals radii of the first-tier residues. (B) Using an energy-minimized model of HPS rendered onto the molecular coordinates of TEAS, second-tier residue differences between TEAS and HPS were identified. No differences in first-tier residues and only four second-tier differences were observed between these two synthases. However, it is especially important to note that second-tier residues do not simply equate to residues surrounding the active site. For instance, although second-tier residues T402 and V516 are beyond van der Waals radii of one possible position of the substrate analog’s farnesyl chain as observed in the TEAS structure (2), these residues are positioned to contribute directly to the active site surface and architecture. Depending on the dynamics of the farnesyl chain and its position after cleavage from its diphosphate tether, it may directly contact side chains at positions 402 and/or 516.
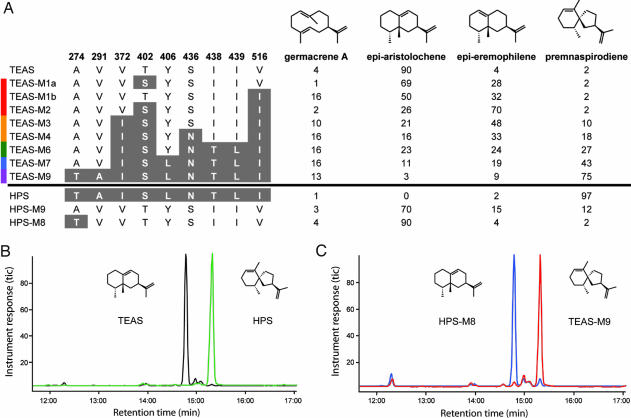
Mutational replacement of TEAS second-tier residues to the corresponding residues in HPS was performed, expressed in bacteria, and the biochemical properties of the mutant enzymes were examined by using in vitro assays, including GC-MS determinations of the reaction product profiles. Stepwise mutagenesis of the TEAS second-tier residues to those corresponding to HPS demonstrated an almost additive effect on reaction product specificity and significantly up-regulated a reaction product previously overlooked (Fig. 2). Single mutants, T402S and V516I, likely located near the farnesyl chain of FPP or a subsequently formed reaction intermediate and contributing directly to the active site cavity surface, resulted in enzymes yielding 4-epi-eremophilene as an abundant (>25%) reaction product, whereas the double mutant (TEAS T402S/V516I) resulted in a more complete transmutation of TEAS into a “synthetic” epi-eremophilene synthase. Epi-eremophilene was recently produced as a side product in the synthetic preparation of 5-epi-arisotolchene from capsidiol (14). Notably, to date, no biological source of epi-eremophilene has been discovered. Both 4-epi-eremophilene and 5-epi-aristolochene are predicted to arise from a common eremophilenyl intermediate with a carbocation centered at C7 and the final proton abstracted from C6 or C8, respectively (Scheme 1). The deprotonation and carbocation quenching from C6 mirrors the reaction step observed in HPS. Therefore, the 402/516 double mutant identifies a discreet and potentially modular structural element within the active site that controls the regiospecificity of the final deprotonation step of the reaction cascade but does not affect the preceding alkyl migration. Interestingly, this result indicates that the alkyl migration is actually controlled by residues lying outside of the active site surface. Mutation of the two remaining second-tier differences, V372I and S436N, resulted in the appearance of greater amounts of premnaspirodiene (10–18% of total reaction products) but not a full transmutation of activity.
Fig. 2.
Comparison of reaction product profiles for wild-type TEAS and HPS with profiles of mutants of contact map residues. (A) In vitro-generated reaction products were examined directly by GC-MS. The percentage of each reaction product was determined from total ion chromatographs as illustrated by the GC traces in B and C. Minor reaction products of <1% were not considered in these analyses.
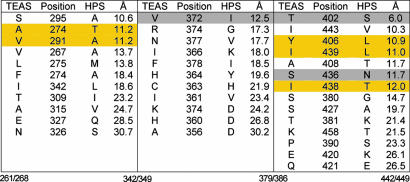
Because mutations of the residues that were identified as differing between TEAS and HPS in the first contact map iteration provided for a significant yet incomplete transmutation to the premnaspirodiene synthase activity, extension of the contact mapping strategy was necessary. To prioritize additional residue positions for subsequent mutagenesis, the outermost amino acid residue in the initial contact map that influenced reaction specificity, Val-372, was chosen as the distance boundary for considering additional residue differences between TEAS and HPS (Fig. 3). Val-372, with its Cα positioned 12.5 Å from the geometric center of the active site, when mutated to Ile, significantly increased the amount of the premnaspirodiene reaction product formed during in vitro assays (Fig. 2). Eleven residues differing between TEAS and HPS within 12.5 Å of the active site center were thus targeted for further evaluation (Fig. 3). Visual inspection of the position and orientation of these residues within the TEAS crystal structure excluded Ser-295, Ile-443, and Ala-408 because they are either localized on the surface of the enzyme or oriented in a manner unlikely to impact the network of residues making up the active site cavity. Three of the remaining eight residues were identified in the initial contact map, leaving five residues for further consideration.
Fig. 3.
Extending the contact mapping strategy to a 12.5-Å sphere. Three peptide regions previously correlated with catalytic specificity of terpene synthases (5) contain 36 residue differences between TEAS and HPS ranging from 6 to 31 Å from the active site center. The amino acid positions bracketing each region of TEAS and the corresponding regions in HPS are noted at the bottom. Several of the residues identified in the initial contact map (Fig. 1) are found within these contiguous linear domains (highlighted in gray) and serve to define a sphere of residues (12.5 Å from the active site center) for experimental assessment. Residues within 12.5 Å and pointing toward the active site or poised to influence active site architecture were visually identified and targeted for mutagenesis (highlighted in yellow).
Sequential mutagenesis at these five positions culminated in a nine amino acid mutant TEAS (TEAS-M9), with four amino acids defined by the initial contact map plus five additional residues within a 12.5-Å sphere of the active site. TEAS-M9 catalyzes the formation of 75% premnaspirodiene (Fig. 2A) and possesses a reaction profile resembling that of the wild-type HPS enzyme (Fig. 2 B and C). As observed with the initial contact map mutants, the newly constructed mutants gained premnaspirodiene synthase specificity in a stepwise manner at the expense of 5-epi-aristolochene and 4-epi-eremophilene accumulation without a significant alteration in the accumulation of the reaction intermediate, germacrene A. Importantly, the TEAS-M9 mutant possessed steady-state kinetic constants (Km = 6.24 μM and kcat = 0.130) comparable to the wild-type TEAS and HPS enzymes (Km = 3–7 μM and kcat = 0.02–0.103) (12, 15). The catalytic efficiencies for the intermediate TEAS mutants were also directly comparable to the wild-type TEAS (kcat/Km = 0.012). For instance, kcat/Km = 0.006 for TEAS-M1a, kcat/Km = 0.008 for TEAS-M1b, and kcat/Km = 0.004 for the double mutant TEAS-M2.
Further corroboration of the contact mapping strategy and its ability to systematically identify evolutionarily reciprocal changes in terpene synthases should encompass transmutations of other sesquiterpene synthases, most notably the reciprocal engineering of HPS, to assess how these spatially equivalent positions might influence catalytic activity. Successful conversion of HPS into a 5-epi-aristolochene synthase was achieved through introduction of the reciprocal nine mutations (Fig. 2A). An intermediate mutant containing eight of the nine amino acid substitutions within the HPS background gene was an even more efficient 5-epi-aristolochene synthase, with ≈90% specificity for the 5-epi-aristolochene reaction product and an overall product profile almost identical to TEAS (Fig. 2 B and C).
Discussion
The current results have identified residues within, surrounding, and distal to the active site that contribute directly to the reaction specificity of terpene synthases. The stepwise alteration in the reaction product profiles presented here is not consistent with one amino acid residue playing a greater role in catalytic outcome than another; rather, it supports a simple model wherein the identified residues play a shared role in catalytic outcome. A logical inference would be that the amino acids identified in the contact mapping strategy help shape the active site geometry, possibly modulate the active site dynamics, and thus influence the catalytic trajectory down which intermediates navigate in route to a final reaction product (Fig. 4).
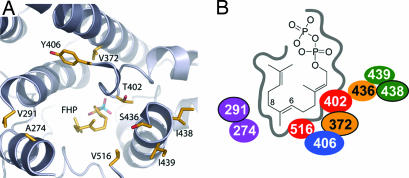
Fig. 4.
Identity and spatial relationships of residues that distinguish TEAS and HPS activities. (A) Structure of the TEAS active site cavity as viewed from the back of the active site looking out the entrance, with several loops and helices cut away for ease of viewing. (B) Schematic depiction of the active site and positioning of the nine residues that control reaction product specificity (see Scheme 1). For example, residues V516 and T402 (red) reside on opposite sides of the reaction pocket and, when mutated to V516I and T402S, are predicted to impinge on the eremophilenyl intermediate in a manner that hinders deprotonation at C8 either because of physical hindrance or by altering the dynamics of deprotonation at C6. Outer-tier residues that are unable to directly contact the reaction intermediates [positions 274 and 291 (purple), 372 and 436 (orange), 406 (blue), and 438 and 439 (green)] are positioned to potentially favor particular conformations of the eudesmyl intermediate that are conducive to a specific reaction trajectory through modulation of the dynamics and steric shape of the active site cavity. The models were created with the TEAS atomic coordinates reported by Starks et al. (2) by using pymol software.
The extended contact mapping strategy is sufficient to identify the amino acid differences between TEAS and HPS that mediate the final reaction product specificities. However, as seen with the M8/M9 HPS mutants, it does not identify the minimum number of residues or the specific combinations of mutations that are necessary to provide for the interconversion between these two enzyme activities. Determining the minimum and specific combinations of residues mediating final product specificity will be important for a complete mechanistic understanding of these two enzymes. Such a study would entail the generation of a large set of site-directed mutant enzymes (512 mutants; nine sites with two different amino acids in all possible combinations) and will benefit from recent advances for robust mutagenic methods (16) and high-throughput analytical assays (17).
Yoshikuni et al. (9) recently reported a mutagenic approach for assessing the plasticity of residues comprising the active-site contour of γ-humulene synthase. Their approach disregarded any phylogenetic or evolutionary relationship among cyclases, relying simply on the saturation mutagenesis of the entire active site surface followed by the development of an algorithm to refine subsequent rounds of mutagenesis. Ultimately, mutagenesis of 19 active site residues in various combinations resulted in catalytically competent enzymes that enhanced and suppressed the yield of specific reaction products observed in the parent enzyme or released neutral reaction intermediates. Unlike the concentric tiers of residues up to 12.5 Å outside the active site that were shown to direct unique (transmutation between methyl and methylene migrations) and previously uncharacterized (generation of 4-epi-eremophilene) catalytic specificity in the current study, Yoshikuni et al. (9) were able to tease out contributions of the corresponding first-tier residues to 7 of 52 different reaction products specific to the γ-humulene parent enzyme. However, whereas Yoshikuni et al. (9) argued that phylogenetic relationships are difficult at best to discern in closely related synthases and use as guiding principles in a design process, we have shown that subtle phylogenetic variation in residues located beyond the immediate active site surface play fundamental roles in directing the reaction trajectory of functionally distinct yet closely related terpene synthases.
Additional testing of the contact mapping strategy based on apparent phylogenetic relationships as described here is necessary before its full applicability and generality can be determined in both closely and more distantly related terpene synthases. TEAS and HPS, for instance, are members of a synthase family of enzymes that direct the all-trans form of FPP down a complex reaction cascade through many structurally diverse reaction intermediates and yield reaction products retaining the all-trans stereochemistry. The current results validate the contact mapping strategy for identifying amino acids and positions within these enzymes that dictate the penultimate reaction steps; however, extending the contact mapping strategy to synthases that catalyze reactions diverging at earlier steps represents an important future target. Long-term, identifying the structural features that govern reaction product specificity for various members of the terpene synthase families and classes should provide a unique perspective into the evolution of this family of enzymes (18, 19) and the potential for using this information in the rational design and engineering of different catalytic activities into existing terpene synthase enzymes (20, 21).
Materials and Methods
Molecular Modeling.
Mutant enzyme structures were built with the program modeler 4.0 (University of California, San Francisco) by using TEAS coordinates (Protein Data Bank entry 5EAT) as a template. The five lowest-energy structures were subsequently used for docking studies. Presumptive reaction intermediates were constructed by using chemdraw ultra 7.0.1 and were mopac energy-minimized by using chem3d ultra 7.0.0 (CambridgeSoft, Cambridge, MA). Docking was performed by using Genetic Optimization for Ligand Docking (gold; CCDC Software, Cambridge, U.K.). Images were made by using chimera (University of California, San Francisco) and pymol (DeLano Scientific, South San Francisco, CA).
Mutagenesis and Isolation of Synthase Protein.
Mutations were made with a pET-28b vector (Novagen) construct harboring the coding sequence for TEAS by using the standard QuikChange mutagenesis method (Stratagene). Specific mutagenic primers (with mutagenic bases shown in boldface) and corresponding templates used to generate the specific mutants were as follows. T402S (TEAS-M1a): primer, CCT AAG CAA TGC ACT AGC AAC TAG CAC ATA TTA CTA CCT CGC GAC; template, pET-28b-TEAS. V516I (TEAS-M1b): primer, CCT ATT CTC AAT CTT GCT CGT ATT ATT GAG GTT ACA TAT ATA CAC; template, pET-28b-TEAS. T402S/V516I (TEAS-M2): primer, CCT AAG CAA TGC ACT AGC AAC TAG CAC ATA TTA CTA CCT CGC GAC; template, pET-28b-TEAS-M1b. V372I/T402S/V516I (TEAS-M3): primer, GCA ATA GAA AGA ATG AAA GAA ATA GTA AGA AAT TAT AAT GTC GAG TCA ACA TGG; template, pET-28b-TEAS-M2. S436N/T402S/V516I/V372I (TEAS-M4): primer, CCA AAA ATT CTT GAA GCT AAT GTA ATT ATA TGT CGA GTT ATC; template, pET-28b-TEAS-M3. S436N/I438T/I439L/T402S/V516I/V372I (TEAS-M6): primer, CCA AAA ATT CTT GAA GCT AAT GTA ACT CTA TGT CGA GTT ATC GAT GAC ACA GC; template, pET-28b-TEAS-M3. Y406L/S436N/I438T/I439L/T402S/V516I/V372I (TEAS-M7): primer, CTA GCA ACT AGC ACA TAT TAC TTG CTC GCG ACA ACA TCG TAT TTG GGC ATG; template, pET-28b-TEAS-M6. V291A/S295A/Y406L/S436N/I438T/I439L/T402S/V516I/V372I (TEAS-M9): primer, GCT CTC TCA AGC TCG CGT CAT GCT CGC TAA GAC CAT AGC AAT GAT TTC GAT TG; template, pET-28b-TEAS-M7.
The HPS-M8 and -M9 constructs were made by using the SCOPE methodology (16) with pH9-HPS, an in-house Gateway expression vector, as the template and the following mutagenic primers (mutant bases are shown in boldface). Forward, GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAATATCAATAGAATCC; T281A, CACCCCCATCGCCCAAAAGTAGCAC; A298V, CATTGCTATAGTCTTAACAAGCATGACACG; I379V, CAATAAAATAGTTTCCCACAACCTCCTTCATTCTTTC; S409T/L413Y, GCACTAGCTACTACCACATATTACTATCTAACTACGAC; N443S/T445I/L446I, CTGGAAGCTAGTGCTATAATATGCCGAGTTGTTG; I523V, CCTGGCTCGAATTGTAGATGTCACTTACAAGC; reverse, GGGGACAAGTTTGTACAAAAAAGCAGGCTTACTGGTTCCGCGTGGATCCATGGCCCCAGCTATAGTG.
All mutations were verified by automated nucleotide sequencing. Plasmid DNA constructs were transformed into BL21 (DE3) cells, and expression of the synthase genes was induced by IPTG addition to the cultures as described in refs. 12 and 17. The constructs provided N-terminal histidyl tags, which afforded >90% pure protein upon Ni+ affinity chromatography in all cases (15). Protein amounts were quantified by using the Bradford method (Bio-Rad) and IgG as the protein standard.
Synthase Assays.
Fifty-microliter reactions containing 200 mM Tris·HCl (pH 7.5), 40 mM MgCl2, 160 nM enzyme, and variable amounts of [1-3H]FPP (radioactive FPP was from NEN; nonradioactive FPP was from Sigma) were used for rate determinations. Briefly, 10 μl of FPP (sufficient for a final concentration of 0.7–23 μM) was rapidly mixed with 40 μl of enzyme solution at room temperature (23°C) and allowed to incubate for 1 min. The reaction was terminated by addition of 150 μl of a 0.2 M KOH/0.1 M EDTA stop solution. Reactions were extracted with 500 μl of hexane, and an aliquot was taken for determination of radiolabeled hydrophobic product by means of liquid scintillation counting. Reaction products were not purified with silica before counting, because background dpms were <1% of the reaction products and because the synthase mutants could possibly generate reaction products containing hydroxyl substituents. Kinetic constants were determined from direct fits of the Michaelis–Menton equation to the data by using graphpad prism 2.01.
Reaction Product Analysis.
To identify the particular reaction products, synthase assays were scaled up to 2.5 ml with 2 μM enzyme and 80 μM unlabeled FPP. The reactions were incubated for 1 h and extracted twice with 2 ml of pentane. Pooled extracts were dried to 50 μl under a stream of nitrogen gas and analyzed by GC-MS analysis as described in ref. 12. 4-Epi-eremophilene was verified against a synthetic standard prepared by Zhao et al. (14). Alternatively, the vial assay as described in ref. 16 was used to analyze wild-type and mutant cyclase product spectra by GC-MS.
Acknowledgments
We thank Yuxin Zhao, David Schenk, and Bob Coates for helpful discussions, Scott Kinison, and Arthur Malone and MaryAnn Bowman for excellent technical support. This work was supported by National Institutes of Health Grant GM054029 (to J.P.N. and J.C.). J.P.N. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- TEAS
Nicotiana tabacum 5-epi-aristolochene synthase
- HPS
Hyoscyamus muticus premnaspirodiene synthase
- FPP
farnesyl diphosphate.
Footnotes
Conflict of interest statement: J.P.N. and J.C. are scientific cofounders of and have a financial interest in Allylix. B.T.G. is a current employee of Allylix, a biotechnology company that produces specialty chemicals.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Davis E. M., Croteau R. Vol. 209. Berlin: Springer; 2000. Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids–Topics in Current Chemistry; pp. 53–95. [Google Scholar]
- 2.Starks C. M., Back K. W., Chappell J., Noel J. P. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 3.Lesburg C. A., Guangzhi Z., Cane D. E., Christianson D. W. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 4.Rynkiewicz M. J., Cane D. E., Christianson D. W. Proc. Natl. Acad. Sci. USA. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Back K. W., Chappell J. Proc. Natl. Acad. Sci. USA. 1996;93:6841–6845. doi: 10.1073/pnas.93.13.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin M. B., Bowman M. E., Ferrer J. L., Schroder J., Noel J. P. Chem. Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 7.Suel G. M., Lockless S. W., Wall M. A., Ranganathan R. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 8.Köllner T. G., Schnee C., Gershenzon J., Degenhardt J. Plant Cell. 2004;16:1115–1131. doi: 10.1105/tpc.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshikuni Y., Ferrin T. E., Keasling J. D. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 10.Lodeiro S., Segura M. J., Stahl M., Schulz-Gasch T., Matsuda S. P. T. ChemBioChem. 2004;5:1581–1585. doi: 10.1002/cbic.200400086. [DOI] [PubMed] [Google Scholar]
- 11.Hyatt D. C., Croteau R. Arch. Biochem. Biophys. 2005;439:222–233. doi: 10.1016/j.abb.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Rising K. A., Starks C. M., Noel J. P., Chappell J. J. Am. Chem. Soc. 2000;122:1861–1866. [Google Scholar]
- 13.Yoshikuni Y., Martin V. J. J., Ferrin T. E., Keasling J. D. Chem. Biol. 2006;13:91–98. doi: 10.1016/j.chembiol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y. X., Schenk D. J., Takahashi S., Chappell J., Coates R. M. J. Org. Chem. 2004;69:7428–7435. doi: 10.1021/jo049058c. [DOI] [PubMed] [Google Scholar]
- 15.Mathis J. R., Back K., Starks C., Noel J., Poulter C. D., Chappell J. Biochemistry. 1997;36:8340–8348. doi: 10.1021/bi963019g. [DOI] [PubMed] [Google Scholar]
- 16.O’Maille P. E., Tsai M. D., Greenhagen B. T., Chappell J., Noel J. P. Methods Enzymol. 2004;388:75–91. doi: 10.1016/S0076-6879(04)88008-X. [DOI] [PubMed] [Google Scholar]
- 17.O’Maille P. E., Chappell J., Noel J. P. Anal. Biochem. 2004;335:210–217. doi: 10.1016/j.ab.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Trapp S. C., Croteau R. B. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aubourg S., Lecharny A., Bohlmann J. Mol. Genet. Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 20.Dwyer M. A., Looger L. L., Hellinga H. W. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 21.Morley K. L., Kazlauskas R. J. Trends Biotechnol. 2005;23:231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]