Abstract
A reverse transcriptase PCR (RT-PCR) assay using conserved primers deduced from the core-envelope 1 (C-E1) region of the hepatitis C virus (HCV) genome was developed for subtyping purposes. The sensitivity and specificity of this assay tested against two HCV reference panels containing genotype 1 through 5 subtypes were similar to those of an RT-PCR assay from the 5′-untranslated region (5′-UTR). The sensitivity of the RT-PCR typing assay in the more variable C-E1 region was, however, lower than that of the RT-PCR in the highly conserved 5′-UTR when testing multiple clinical samples. Thus, 71 (88%) of 81 consecutive samples from hospitalized Danish patients positive for HCV antibodies and RNA (5′-UTR) were positive also in the C-E1 RT-PCR assay. Phylogenetic analysis of the E1 sequences obtained by direct sequencing of HCV from two reference panels and 71 Danish patients allowed us to readily distinguish the subtypes. In contrast, phylogenetic analysis of their corresponding 5′-UTR sequences was able to predict only major genotypes. Three different genotypes and four subtypes were identified among Danish samples: 1a (43%), 1b (11%), 2b (6%), and 3a (39%). An isolate from a Somalian refugee was identified as a new HCV type related to Somalian isolates described as subtype 3h. The most common genotype in Denmark is genotype 1 (53%), which is the most difficult to treat. However, Denmark had the highest prevalence in Europe of subtype 3a, which responds more favorably to treatment. The described C-E1 RT-PCR with sequencing is suggested as an easy routine assay for definitive genotyping and subtyping of HCV.
Hepatitis C virus (HCV) is an important cause of chronic liver disease. HCV causes 20% of acute hepatitis cases, 70% of all chronic hepatitis cases, 40% of all cases of cirrhosis of the liver, 60% of hepatocellular carcinomas, and 30% of liver transplants in Europe (11). This virus has a positive-sense single-stranded RNA genome of about 9.6 kb containing one long open reading frame (ORF) with untranslated regions (UTR) at both ends (9). The polyprotein is processed into structural and nonstructural proteins. The core and the two envelope proteins (E1 and E2) are part of the virion. Different HCV isolates show considerable sequence diversity in the two envelope proteins, while the 5′-UTR and, to a lesser extent, the core region are highly conserved (4, 6, 14). HCV isolates from around the world can be divided into distinct major groups or genotypes with about 66 to 69% nucleotide similarity, which can in turn be divided further into subtypes with about 77 to 80% nucleotide similarity (3). Phylogenetic analysis may aid in the separation of sequences into distinct types (30). So far six major genotypes (HCV-1 to HCV-6) have been described, each containing multiple subtypes (e.g., 1a, 1b, etc.). The isolates formerly published as genotypes 7 to 11 are now considered subtypes within genotypes 3 (subtype 10) and 6 (subtypes 7, 8, 9, and 11) (38). In general, sequence characteristics of a particular subtype are found throughout the HCV genome. Thus, the HCV genotype has been determined primarily based on analysis of partial genome sequences. The most extensive database exists for the 5′-UTR, core, E1, and NS5B (3, 5, 13, 17, 26, 28). Whereas the 5′-UTR is highly conserved and therefore preferred for diagnosis, the core, envelope, and NS5B regions are less conserved and therefore highly discriminative and may be preferred for subtyping. Although the 5′-UTR contains characteristic sequence motifs of some genotypes, analysis of this region may not accurately predict all genotypes or subtypes.
Many different typing methods have been used, all of which depend on a few relatively specific nucleotide changes: RT-PCR using type-specific amplification (18, 42), hybridization of RT-PCR products with type-specific probes (34), restriction fragment length polymorphism of RT-PCR amplicons (27), or heteroduplex mobility analysis of RT-PCR amplicons (41). However, these methods were often found not to be definitive, as more sequence data became available. Thus, there is a need for developing sequence-based genotyping assays that can be used to definitively determine the genotypes and subtypes of existing and emerging HCV isolates. The responsiveness to standard therapy with interferon and ribavirin, the pathogenesis of infection, the demographic profile of patients, and the performance of diagnostic assays may all vary according to HCV genotype or subtype (16, 43). Thus, typing of HCV is important as an epidemiological marker, as an aid in deciding upon therapy regimes, and as a predictor of responses to therapy.
In the present study, we have developed and tested an HCV genotyping assay based on a region spanning both the core and the envelope 1 regions (C-E1). The results obtained with this sequence-based assay were compared with those obtained by analysis of corresponding 5′-UTR sequences from two HCV reference panels and from 71 consecutive HCV-positive patient samples from Denmark submitted for typing. The HCV genotype distribution in the Danish population is reported.
MATERIALS AND METHODS
HCV-infected patients.
Sera investigated in this study were obtained from 81 consecutive Danish patients. After collection, these samples were sent to the Statens Serum Institut in Copenhagen for diagnosis of HCV infection and subtyping and found positive for anti-HCV antibodies in an enzyme-linked immunosorbent assay (Ortho HCV 3.0 ELISA; Chiron) and HCV RNA in a commercial 5′-UTR-based RT-PCR test (Amplicore HCV test; Roche Diagnostic Systems Inc., Branchburg, N.J.). Most patients were hospitalized at departments of infectious diseases, internal medicine, hepatology, and pediatrics. Fifty-seven percent of the samples were from males, and 43% were from females. The mean age of the patients was 40 years, ranging from 6 to 71 years of age.
HCV genotype reference panels.
One HCV genotype reference panel was obtained from the World Health Organization Hepatitis Center, Robert Koch Haus, Universitätsklinikum, Essen, Germany, and contained nine human serum samples representing five genotypes and nine subtypes (subtypes 1a, 1b, 2a, 2b, 2c, 2i, 3a, 4d, and 5a). Their genotypes were established by sequencing a 216-nucleotide (nt) fragment of the core region (40). These samples were all diluted with HCV-negative pooled plasma to a final titer of approximately 105 genome equivalents/ml or 4 × 105 IU/ml. Another HCV genotype panel was obtained from the National Institutes of Health (NIH) and contained plasma samples from acutely infected chimpanzees (J. Bukh, unpublished data). These samples titrated with chimpanzees represented five genotypes and six subtypes (genotypes 1b, 2a, 2b, 3a, 4a, and 5a). Their genotypes were established by sequence analysis of complete ORF or C-E1 sequences. Their genome titers ranged from approximately 104 to 106 IU/ml.
Specific HCV RNA extraction.
Viral RNA was extracted from 100 μl of serum or plasma by using a lysis buffer from an RNA isolation kit from Stratagene (La Jolla, Calif.) according to the manufacturer's protocol. The specific HCV RNA was captured from the lysis suspension by using a 5′-UTR-specific DNA oligonucleotide (capture probe) (Table 1) immobilized to magnetic beads (Dynabeads; Dynal Inc., Oslo, Norway). The capture probe is identical to the outer reverse primer in the 5′-UTR RT-nested PCR (4). One milliliter of Dynabeads stock (M-280 Streptavidin Dynabeads) was washed three times in phosphate-buffered saline (pH 7.4)-0.1% bovine serum albumin for 1 min followed by removal of the supernatant. The Dynabeads were subsequently resuspended in 1 ml of coupling buffer (10 mM Tris-HCl, 1 mM EDTA, 1 M NaCl [pH 7.5]), and 20 μg of the biotin-coupled DNA oligonucleotide capture probe was added. After 10 min of incubation, the solution was washed once in coupling buffer and twice in a blocking buffer (20 mM Tris-HCl, 1 M LiCl, 2 mM EDTA [pH 7.4]). One hundred twenty microliters of the extracted RNA was added to 100 μl of the oligonucleotide-coupled Dynabeads, followed by incubation at 20°C for 30 min. The Dynabeads were washed twice in the blocking buffer and once in a 0.1 M Tris buffer (pH 7.4), and the RNA was subsequently dissolved in 10 μl of sterile RNase-free water. The immobilized capture probe had the same sequence as the external antisense primer used in the 5′-UTR PCR and was therefore used also as a primer for the RT in the in-house 5′-UTR RT-PCR assay. For comparison, RNA was also extracted from 100 μl of serum by guanidine isothiocyanate lysis with silica gel membrane purification by using the Qiagen (Valencia, Calif.) RNeasy kit.
TABLE 1.
Sequences of the primers used for the amplification and sequencing of HCV isolates in the C-E1 region and the capture probe used for RNA extraction, deduced from 5′-UTR
| Name | Position (nt)a | Sequenceb |
|---|---|---|
| Capture probe (5′UTR a2)b | (−13)-(−42) | 5′CGAGACCTCCCGGGGCACTCGCAAGCACCC3′ |
| Outer C-E1 forward (493 S-H77) | 493-518 | 5′GCAACAGGGAACCTTCCTGGTTGCTC3′ |
| Outer C-E1 reverse (987 R-H77) | 987-964 | 5′CGTAGGGGACCAGTTCATCATCAT3′ |
| Inner C-E1 forward (502 S-H77) | 502-527 | 5′AACCTTCCTGGTTGCTCTTTCTCTAT3′ |
| Inner C-E1 reverse (975 R-H77) | 975-952 | 5′GTTCATCATCATATCCCATGCCAT3′ |
| Chimer forwardc | 5′AGCGGATAACAATTTCACACAGGAAACCTTCCTGGTTGCTCTTTCTCT3′ | |
| Chimer reversec | 5′CGCCAGGGTTTTCCCAGTCACGACGTTCATCATCATATCCCATGCCAT3′ | |
| 1223 forward sequencing | 5′AGCGGATAACAATTTCACACAGGA3′ | |
| 1224 reverse sequencing | 5′CGCCAGGGTTTTCCCAGTCACGAC3′ |
Numbered according to the first nucleotide in the ORF of the HCV reference strain H77 (GenBank accession no. AF009606) (3).
The capture probe is identical to the outer reverse primer in the 5′UTR RT-nested PCR.
Underlined sequences of Chimer forward and Chimer reverse correspond to those of the commercial sequencing primers 1223 and 1224 (Promega), respectively.
RT-nested PCR of the C-E1 region of HCV.
For the specific extraction of HCV the immobilized conserved 5′-UTR capture probe (Table 1) was used, but the reverse transcription and first round of PCR were performed with the external C-E1 primers 493S H77 and 987R H77, and the second PCR was performed with the inner C-E1 primers 502S H77 and 975R H77 (Table 1). The primer names refer to ORF nucleotide locations on the HCV reference strain H77, and the primers were deduced from relatively conserved regions in the core and E1, respectively (Fig. 1). The final reaction mixture (volume, 50 μl) contained 7 mM MgCl2, 1× PCR buffer II (Applied Biosystems), 0.8 mM deoxynucleoside triphosphate, 40 U of RNase inhibitor, 100 U of RT (Perkin-Elmer), 1.25 U of Taq DNA polymerase, and a 0.1 μM concentration (each) of external sense and antisense primers. Reverse transcription was performed under the following conditions: 42°C for 1 h, and then 94°C for 4 min followed by 35 cycles of 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min. For the nested second round of PCR, 10 μl of the first-round PCR product was added to a new reaction mixture to a total volume of 100 μl of reaction mixture containing 2 mM MgCl2, 1× PCR buffer II, 0.2 mM deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase, and a 0.2 μM concentration (each) of the internal sense and antisense primers. Thirty-five additional PCR cycles were then performed under the same conditions as above.
FIG. 1.
Positions of the C-E1 RT-PCR primers in the HCV reference strain H77 and alignment with published full-length HCV subtypes (http://s2as02.genes.nig.ac.jp). The sequences shown correspond to the positive strand. Positions of the distal nucleotides within each primer refer to the ORF. For each sequence the GenBank accession number, strain name, and subtype are provided. Additional strains published only as partial sequences were included in order to cover all subtypes. For genotype 4, the GenBank accession numbers are U10235 and L16677 for Z1 (a), U10238 and L16678 for Z6 (b), and U10192 and L16656 for DK13 (c).
Reverse transcription and PCR of the 5′-UTR of HCV.
The HCV 5′-UTR primers used in the RT-nested PCR have been described previously (4). Reverse transcription and the first round of PCR were performed together by using external sense and antisense primers, a1 and a2 (4). The Dynabead-captured specific lysis extract (10 μl) or 10 μl of the Qiagen column extract was added to a 40-μl reaction mixture. The reaction mixtures and cycle conditions were the same as for the C-E1 assay, except that the annealing temperature was 55°C in the first PCR and 45°C in the second PCR.
Sequencing of C-E1 and 5′-UTR amplicons of HCV.
For the purification of PCR products for cycle sequencing, we used the S400 column from Pharmacia Biotech. These columns were used on the 272-bp PCR products from the 5′-UTR. For purification of inner PCR products from the C-E1 region, either S400 or silica-based Qia Quick purification was used. Both DNA strands were sequenced by using the rhodamine dye terminator cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) and an automatic DNA sequencer (ABI-377; Applied Biosystems). The internal PCR primers were used for cycle sequencing. Before sequencing of the C-E1 region, in most cases we performed an additional PCR of 10 cycles with chimeric primers containing the internal C-E1 primer sequences at the 3′ end and a new sequence at the 5′ end to introduce the primer sequences of the commercial sequencing primers 1224 and 1223 (Promega, Madison, Wis.) (Table 1). This was done to also allow dye primer sequencing by the commercially available dye primers 1233 and 1224. The hybrid primers were called Chimer forward and Chimer reverse (Table 1).
Data analysis.
Sequences were edited by using the computer program Edit SEQ from the DNA-star Lasergene program. Primer sequences were deleted, and the terminal ends were trimmed to include only sequences from both DNA strands. The sequences obtained were aligned with reference sequences by using Clustal X, version 1.8 (15). The alignments were edited and trimmed by using Genedoc, version 2.6.002. Gaps were manually rearranged to obtain codon-based nucleotide alignments of the C-E1 sequences. Phylogenetic trees were generated by use of the PHYLIP package, version 3.57c (12). Genetic distance matrices were calculated with the DNADIST program, using the maximum-likelihood model with a transition-to-translation ratio of 4.25 (33). For amino acid alignments the distance matrices were calculated with the PROTDIST program by using the Dayhoff PAM model. The Neighbor program was used to obtain neighbor-joining trees (12). Bootstrap 1000 values indicating similarities of multiple alignments assessed the robustness of the neighbor-joining phylogenetic tree. The consensus tree was constructed by using Consense. The graphical presentation of the tree was done with the Treeview program, version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). A total of 83 HCV sequences, which included both 5′-UTR and the E1 sequences of known genotypes, were selected from the literature (5, 6, 8, 19, 20, 21, 22, 23, 29, 31, 35, 36, 37, 39, 40) and used with the results obtained from the Essen and NIH typing panels (15 samples) for the phylogenetic analysis in order to evaluate the performance of the typing assays.
Nucleotide sequence accession numbers.
The 81 5′-UTR and the 71 E1 HCV sequences from the Danish patients have been submitted to GenBank; the 5′-UTR sequences were given accession numbers AY188091 through AY188171, and the E1 sequences were given accession numbers AY177816 through AY177886.
RESULTS
Specificity of the C-E1-based method for genotyping HCV.
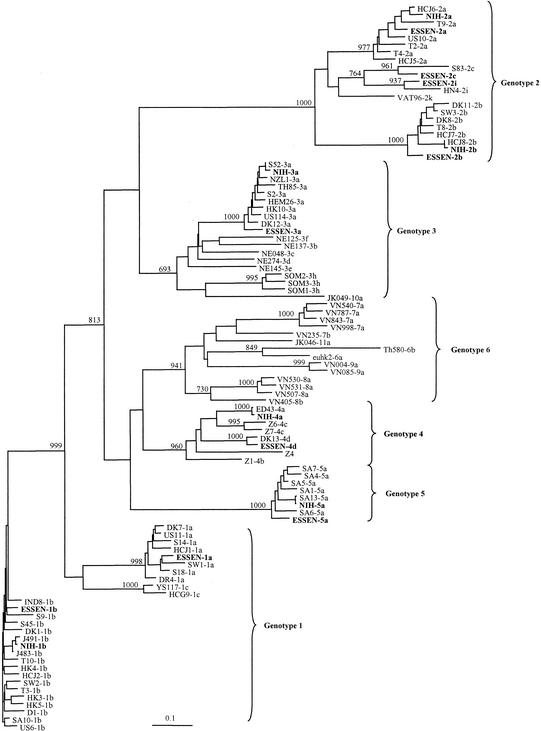
For RT-nested PCR of the 5′-UTR of HCV, we used previously published highly conserved primers (4). For the RT-nested PCR of C-E1, we designed relatively conserved primers based on alignments of all published full-length HCV sequences representing the six major genotypes of HCV and some partial sequences in order to include all subtypes (6, 7) (Fig. 1). Although sequence variation was expected within the primers, the 3′ ends from which the elongation takes place were fully conserved. When tested on 10-fold serially diluted RNA extracted from HCV isolates representing genotypes 1 to 5 (NIH panel), the sensitivity of RT-nested PCR (4) was equivalent to or at most 10-fold lower with the C-E1 primers than with the 5′-UTR primers (data not shown). We tested the possibility of using these new primers in RT-nested PCR assays followed by sequence analysis to routinely determine the genotypes and subtypes of HCV isolates. We first tested two HCV panels: the Essen panel, consisting of nine subtypes (1a, 1b, 2a, 2b, 2c, 2i, 3a, 4d, and 5a), and the NIH panel, consisting of six subtypes (1b, 2a, 2b, 3a, 4a, and 5a). We obtained PCR products from all of these samples in RT-nested PCR with both the 5′-UTR primers and the C-E1 primers. However, we failed in three reruns to get a useful sequence in one of the samples from the 5′-UTR product (NIH subtype 5a). Figure 2 shows the phylogenetic analysis of E1 sequences obtained from the two HCV reference panels together with the 83 reference sequences of known HCV types (5, 6, 8, 19, 20, 21, 22, 23, 29, 31, 35, 36, 37, 39, 40). The sequences obtained from the HCV panels cluster together with the sequences of the expected genotype and subtype (Fig. 2). This confirms that this sequence-based method was useful for distinguishing the most common subtypes of genotypes 1 through 6.
FIG. 2.
Phylogenetic analysis of E1 sequences generated from two HCV genotyping panels: the Essen panel (1a, 1b, 2a, 2b, 2c, 2i, 3a, 4, and 5a) and the NIH panel (1a, 2a, 2b, 3a, 4a, and 5a). The 83 reference sequences were collected from the literature as follows: D, DK, DR, HK, IND, S, SA, SW, T, US6, US10, US11, Z (6, 7); HC-G9, YS (20); HC-J1 (19); HC-J5, HC-J7, HC-J8 (21); HC-J6 (22); HEM, NZL, TH, US114 (22); NE (32); VN (34); VAT69 (31); JK046 and JK049 (36); Th580 (37); ED43 (8); HN4 (GenBank X76411) (29); and SOM (1). The phylogenetic tree was constructed by using the maximum-likelihood algorithm and the neighbor-joining method in the PHYLIP package (12). The numbers on the branching represent bootstrap 1000 values.
Genotyping of HCV from clinical isolates from Danish patients.
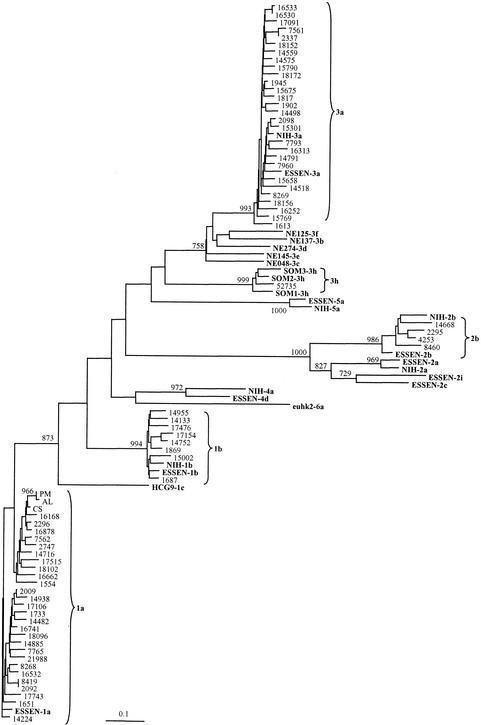
An inner C-E1 PCR product of the correct size (474 bp) was obtained from 71 (88%) of the 81 patients in RT-nested PCR with the C-E1 primers. Thus, 10 of the 5′-UTR RT-PCR-positive samples were negative when the C-E1 primers were used. Readable sequences are obtained at a variable distance from the sequencing primers. Therefore, the sequences obtained from both strings had to be trimmed in the alignment to 352 bp, corresponding to the E1 region (nt 577 through 928 in the ORF). This permitted the comparison of the exact same segments from all 71 patients and the 15 reference panel samples of known genotypes and subtypes and the construction of a phylogenetic tree (Fig. 3). All the patients' HCV sequences represented coding sequences of equal length without any gaps or stop codons. The nucleotide-based phylogenetic analysis performed by using the maximum-likelihood model gave sequence clustering similar to that obtained with the amino acid phylogenetic analysis performed with the Protdist program and Dayhoff PAM method (not shown). The E1 sequences from the Danish patients clustered with the genotypes 1, 2, and 3. Moreover, the analysis of the E1 region showed that the samples could be further classified into subtypes 1a and 1b, 2b, and 3a (Fig. 3). The distribution of HCV subtypes 1a, 1b, 2b, and 3a in Denmark is summarized in Fig. 4. One sample, 52735, was from a 35-year-old Somalian male refugee, and the sequence from this sample clustered with three published sequences from Somalia, SOM 1 to 3, that were tentatively classified as subtype 3h (1).
FIG. 3.
Phylogenetic analysis of the E1 region of 71 HCV samples from Danish patients compared with the C-E1 sequences from the two HCV genotyping panels from Essen and NIH, respectively, and 10 reference sequences from Fig. 2 (shown in bold). The phylogenetic tree was constructed by using the maximum-likelihood algorithm and the neighbor-joining method in the PHYLIP package (12). The numbers on the branching represent bootstrap 1000 values.
FIG. 4.
HCV genotype distribution (in percentages) in Denmark based on phylogenetic analysis of the C-E1 region of the HCV genome of 71 patient samples. One sample (52735) from Somalia was classified as belonging to “other types.”
Specificity of the 5′-UTR-based method for genotyping HCV.
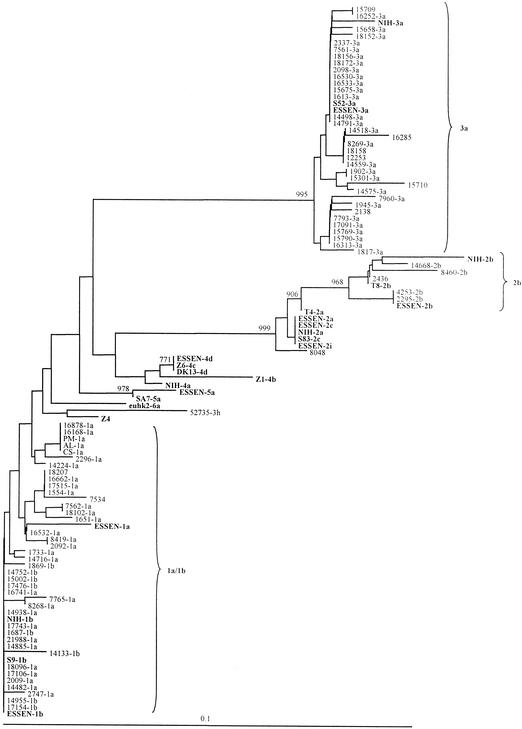
A PCR product of the correct size was obtained from each of the 81 patient sera in RT-nested PCR with the 5′-UTR primers. The 5′-UTR sequences from the Danish patients and reference HCV sequences of known genotypes were used in the construction of a phylogenetic tree (Fig. 5). In general, the panel sequences and the clinical samples clustered with the expected genotype isolates but not necessarily with the expected subtype sequences. For example, it was not possible to distinguish 1a and 1b, 2a, 2c and 2i, or 4a, 4b, 4c, and 4d, respectively (Fig. 5). Moreover, one isolate, Z4, listed as genotype 4 in the database as well as clustering with genotype 4 in our E1 analysis (Fig. 2), did not cluster with genotype 4 in the 5′-UTR but seemed related to the Somalian isolate 52735 (Fig. 5). Interestingly, the Somalian isolate 52735 that clustered close to genotype 3 in the E1 region did not cluster with genotype 3 in the 5′-UTR. As in the E1 analysis, the 5′-UTR sequences from all the Danish patients belonged to genotypes 1, 2, and 3. The 10 5′-UTR-positive but C-E1 RT-PCR-negative isolates shown in Fig. 5 most likely belonged to subtypes 2a or 2c (sample 8048), 2b (sample 2436), 1a and 1b (samples 18207 and 7534), or 3a (samples 15709, 12253, 18158, 16285, 2138, and 15710). This could suggest that the C-E1 assay is somewhat less sensitive for type 3 isolates.
FIG. 5.
Phylogenetic analysis of the 5′-UTR region of 81 HCV samples from Danish patients compared with the sequences obtained from the two HCV genotyping panels from Essen and NIH, and 11 reference sequences from Fig. 2 (shown in bold). The genotype established by E1 analysis is indicated for 71 of the 81 clinical isolates. The phylogenetic tree was constructed by using the Kimura-2-parameter with the neighbor-joining method. The numbers on the branching represent bootstrap 1000 values.
DISCUSSION
We used extraction of the specific HCV RNA by a Dynabead-immobilized conserved probe from the 5′-UTR before RT-PCR. This resulted in no unspecific banding, but it did lead to the generation of “primer-dimers” of lower molecular weight, which were, however, removed by using the S400 column. However, only very low amounts of primer-dimers were seen with the C-E1 RT-PCR and here silica purification of PCR products worked satisfactorily. The C-E1 nested PCR primers were designed from relatively conserved regions in core and E1, respectively, as deduced by alignments of all available sequences. Although some variation within the primers was unavoidable, the 3′ ends were fully conserved. Moreover, the inner nested primers were designed to overlap the outer primers in order to generate optimal annealing for the inner primers. Together with the relatively low stringency obtained by the annealing temperature and magnesium concentration used in the first PCR, the assay should allow for amplification of all HCV subtypes. The use of an overlapping nested PCR design was employed to subsequently increase both the sensitivity and the specificity. Before sequencing of the C-E1 PCR product, a mutagenesis PCR was in most cases performed to introduce the sequences of the commonly used sequencing primers 1233 and 1224. By doing this, it is possible to use dye primer chemistry-based sequencing with the commercially available dye-labeled primers 1233 and 1224. The advantage lies in obtaining equally high chromatogram peaks, which can detect and roughly quantitate any heterogeneity stemming from mixed infections or quasispecies. Thus, we detected heterogeneity in the sera of several patients; however, since less than 4% of the 352 nucleotide positions in each individual sample showed microheterogeneity, this is most likely due to quasispecies rather than mixed infections.
Because the 5′-UTR is the most conserved region, it is preferred for diagnosis of HCV infection but it is less useful for genotyping. In our study, a phylogenetic analysis of the 5′-UTR sequences was able to identify genotype 1 but it did not distinguish subtype 1a from subtype 1b. In addition, analysis of all genotype 1 isolates for the putative subtype-specific mutation at position −99 (A/G) expected to distinguish 1a from 1b (18, 42) was in fact unable to distinguish 1a from 1b. Within genotype 1, nucleotide A at position −99 was always subtype 1a whereas nucleotide G on the same position was either subtype 1b (33%) or 1a (66%). Moreover, when the exact subtype designation was based on analysis of the E1 sequences and phylogenetic analysis was performed with the 5′-UTR, we could not distinguish subtypes 1a from 1b, or 2a from 2c or 2i, or 4a from 4b, 4c, or 4d. This is in accordance with the findings of others who were unable to distinguish these subtypes as well as genotype 1 and certain subtypes within genotype 6 (3). We also found that 5′-UTR analysis could not place the Somalian isolate 3h within genotype 3.
When performing phylogenetic analysis on the nonconserved E1 region of the HCV genome, a better resolution of subtypes is obtained. Thus, by assaying the genotype and subtype panel sera from Essen and NIH we demonstrated that we could correctly amplify and predict subtypes 1a, 1b, 2a, 2b, 2c, 2i, 3a, 4a, 4d, and 5a by using the C-E1-based method described here. The use of primers deduced from the more variable C-E1 region and/or the somewhat lower sensitivity might explain why only 71 (88%) of the 81 5′-UTR PCR-positive samples were positive in the C-E1 region. Thus, the C-E1 RT-PCR might be less useful for diagnosis of HCV infection but more accurate for typing. Assays of 71 C-E1 HCV-positive clinical sera from Denmark detected the presence of subtypes 1a, 1b, 2b, and 3a. These same HCV types were found in an earlier study of 10 Danish patients by using a different method (6). However, that study also found a single patient with subtype 4d. None of the 71 Danish patients analyzed in the present study had this type. Our C-E1 assay, however, also performs well with genotype 4 clinical isolates from several patients from Egypt (data not shown). Thus, we believe that the previous finding of one subtype 4d among 10 isolates from Denmark is not typical for Danish HCV-positive patients. One isolate (52735) amplified from a Somalian refugee in Denmark clustered with three other isolates, SOM1 to SOM3, derived also from Somalia and is believed to be a new subtype (3 h) of genotype 3 (1). When all four isolates were added to the phylogenetic tree, the clustering to genotype 3 was less robust (lower bootstrap value). Moreover, the 5′-UTR analysis that in general grouped isolates correctly according to their genotypes, but not the subtypes, did not suggest the Somalian 52735 to belong to genotype 3. It probably will be important to include sequences from other genotype 3 subtype isolates in future analyses as well as perform sequencing of additional regions or the entire genomes of HCV isolates from Somalia to resolve the classification of these isolates.
Isolates of genotype 1, which is considered a difficult genotype to treat with interferon or interferon plus ribavirin, are widely distributed throughout the world. This was the predominant genotype in Denmark, as it is in other parts of Europe. However, it is not clear at the moment if it is in fact subtype 1b, which is the most resistant to treatment (10, 16). Furthermore, infection with genotype 1b has been reported to be associated with a more aggressive disease course (2, 24, 32, 43). Therefore, it might be relevant to be able to subtype 1b and other subtypes in future studies, in which the 5′-UTR-based methods can be of little use. Based on epidemiological circumstances and anti-HCV seroconversion, one patient, AL (Fig. 3), was suspected to have been infected during surgery by an HCV-positive patient, PM. The phylogenetic analysis of the E1 region of these two subtype 1a isolates showed a close clustering, with a high bootstrap value of 966 which could support this.
A lower percentage of type 2b (6%) was found in Denmark than in its neighbor, Sweden (11%), but a similar frequency (5%) was reported from its other neighbor, Germany (3). Based on the 5′-UTR analysis, subtype 2a or 2c (isolate 8048) may also appear in Denmark. Genotype 3, which has been suggested to be associated with intravenous drug users (IVDUs) (24), has a wide geographic distribution, but surprisingly it is more prevalent in Denmark (39%) than in any other European country. The reason for this is not known but may reside in part in differences in the demographic profiles of patients. Further epidemiological studies are needed to test if the prevalence of subtype 3a could be caused by outbreaks among Danish IVDUs. Thus, the prevalence of each HCV genotype and subtype has been shown to vary according to geographical area and type of patients e.g., IVDUs, hemodialysis patients, and hemophiliac patients. However, since the samples are from many different hospital departments, different specialists, and outward clinicians from all areas of Denmark, our clinical material is unlikely to be biased towards a special category of patient and is thus considered representative of the overall distribution among HCV patients in Denmark. Importantly, subtype 3a responds favorably to medical treatment (11).
We believe that the HCV C-E1 genotyping method described here is a clinically relevant method for more-precise and more-definitive routine genotyping and subtyping of present and emerging HCV isolates. The E1 sequences thus obtained may aid in therapy decisions, the predicting of therapy responses, evaluation of serological assays, and as aids in “look-back” studies, epidemiological studies, and cases of patient-to-patient transmissions.
Acknowledgments
We are most grateful to Robert Kock Haus for supplying the Essen type sera and to Birgit Knudsen for expert technical assistance.
This study was carried out with the support of the Velux Foundation.
REFERENCES
- 1.Abid, K., R. Quadri, A.-L. Veuthey, A. Hadengue, and F. Negro. 2000. A novel hepatitis C virus (HCV) subtype from Somalia and its classification into HCV clade 3. J. Gen. Virol. 81:1485-1493. [DOI] [PubMed] [Google Scholar]
- 2.Amoroso, P., M. Rapicetta, M. E. Tosti, A. Mele, E. Spada, S. Buonocore, G. Lettieri, P. Pierri, P. Chionne, A. R. Ciccaglione, and L. Sagliocca. 1998. Correlation between virus genotypes and chronicity rate in acute hepatitis C. J. Hepatol. 28:939-944. [DOI] [PubMed] [Google Scholar]
- 3.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc. Natl. Acad. Sci. USA 89:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. USA 89:4942-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh, J., R. H. Purcell, and R. H. Miller. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 90:8234-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain, R. W., N. Adams, A. A. Saeed, P. Simmonds, and R. M. Elliott. 1997. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J. Gen. Virol. 78:1341-1347. [DOI] [PubMed] [Google Scholar]
- 9.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a bloodborne non-A, non-B hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trépo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, J. Albrecht, and The International Hepatitis Interventional Therapy Group. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 11.European Association for the Study of the Liver. 1999. EASL International Consensus Conference on hepatitis C. Consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP-phylogenetic interference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 13.Furione, M., L. Simoncini, M. Gatti, F. Baldanti, M. G. Revello, and G. Gerna. 1999. HCV genotyping by three methods: analysis of discordant results based on sequencing. J. Clin. Virol. 3:121-130. [DOI] [PubMed] [Google Scholar]
- 14.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins, D., A. J. Bleasby, and R. Fuchs. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 16.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 17.Norder, H., Å. Bergström, I. Uhnoo, J. Aldén, L. Weiss, J. Czajkowski, and L. Magnius. 1998. Confirmation of nosocomial transmission of hepatitis C virus by phylogenetic analysis of the NS5-B region. J. Clin. Microbiol. 36:3066-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno, O., M. Mizokami, R. Wu, M. Saleh, K. Ohba, E. Orito, M. Mukaide, R. Williams, and J. Y. Lau. 1997. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3b, 4, 5a, and 6a. J. Clin. Microbiol. 35:201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto, H., N. Kanai, and S. Mishiro. 1992. Full-length nucleotide sequence of a Japanese hepatitis C virus isolate (HC-J1) with high homology to USA isolates. Nucleic Acids Res. 20:6410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto, H., M. Kojima, M. Sakamoto, H. Iizuka, S. Hadiwandowo, S. Suwignyo, Y. Miyakawa, and M. Mayumi. 1994. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J. Gen. Virol. 75:629-635. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, H., K. Kurai, S.-I. Okada, K. Yamamoto, H. Iizuka, T. Tanaka, S. Fukuda, F. Tsuda, and S. Mishiro. 1992. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology 188:331-341. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, H., S. Okada, Y. Sugiyama, K. Kurai, H. Iizuka, A. Machida, Y. Miyakawa, and M. Mayumi. 1991. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J. Gen. Virol. 72:2697-2704. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, H., H. Tokita, M. Sakamoto, M. Horikita, M. Kojima, H. Iizuka, and S. Mishiro. 1993. Characterization of the genomic sequences of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J. Gen. Virol. 74:2385-2390. [DOI] [PubMed] [Google Scholar]
- 24.Pawlotsky, Y. M., L. Tsakiris, F. Roudot-Thoraval, C. Pellet, L. Stuyver, J. Duval, and D. Dhumeaux. 1995. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J. Infect. Dis. 171:1607-1610. [DOI] [PubMed] [Google Scholar]
- 25.Pistello, M., F. Maggi, L. Vatteroni, N. Cecconi, F. Panicucci, G. P. Bresci, L. Gambordella, M. Taddei, A. Bionda, and M. Tuoni. 1994. Prevalence of hepatitis C virus genotypes in Italy. J. Clin. Microbiol. 32:232-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogam, S. L., D. L. Chapois, R. Christen, F. Dubois, F. Barin, and A. Goudeau. 1998. Hepatitis C in a hemodialysis unit: molecular evidence for nosocomial transmission. J. Clin. Microbiol. 36:3040-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohjanpelto, P., M. Lappalainen, A. Widell, K. Asikainen, and M. Paunio. 1996. Hepatitis C genotypes in Finland determined by RFLP. Clin. Diagn. Virol. 7:7-16. [DOI] [PubMed] [Google Scholar]
- 28.Prescott, L. E., A. Berger, J.-M. Pawlotsky, P. Conjeevaram, I. Pike, and P. Simmonds. 1997. Sequence analysis of hepatitis C virus variants producing discrepant results with two different genotyping assays. J. Med. Virol. 53:237-244. [PubMed] [Google Scholar]
- 29.Qu, D., O. Hantz, M. Gouy, L. Vitvitski, J. S. Li, F. Berby, S. P. Tong, and C. Trepo. 1994. Heterogeneity of hepatitis C virus genotypes in France. J. Gen. Virol. 75:1063-1070. [DOI] [PubMed] [Google Scholar]
- 30.Robertson, B., G. Meyers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-I, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 31.Samakhvalov, E. I., M. Hijikata, R. I. Gylka, D. K. Lvov, and S. Mishiro. 2000. Full-genome nucleotide sequence of a hepatitis C virus variant (isolate name VAT96) representing a new subtype within the genotype 2 (arbitrarily 2k). Virus Genes 20:183-187. [DOI] [PubMed] [Google Scholar]
- 32.Silini, E., F. Bono, A. Cividini, A. Cerino, S. Bruno, S. Rossi, G. Belloni, B. Brugnetti, E. Civardi, L. Salvaneschi, and M. U. Mondelli. 1995. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology 21:285-290. [PubMed] [Google Scholar]
- 33.Smith, D. B., S. Pathirana, F. Davidson, E. Lawlor, J. Power, P. L. Yap, and P. Simmonds. 1997. The origin of hepatitis C virus genotypes. J. Gen. Virol. 78:321-328. [DOI] [PubMed] [Google Scholar]
- 34.Stuyver, L., R. Rossau, A. Wyseur, M. Duhamel, B. Vanderborght, H. Van Heuverswyn, and G. Maertens. 1993. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J. Gen. Virol. 74:1093-1102. [DOI] [PubMed] [Google Scholar]
- 35.Tokita, H., H. Okamoto, M. Sakamoto, M. Horikita, H. Iizuka, S. Shrestha, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Nepal with novel genotypes and their classification into the third major group. J. Gen. Virol. 75:931-936. [DOI] [PubMed] [Google Scholar]
- 36.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, L. A. Lesmana, Y. Miyakawa, and M. Mayumi. 1996. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J. Gen. Virol. 77:293-301. [DOI] [PubMed] [Google Scholar]
- 37.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1998. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7-9 and comparison with those in the other eight genetic groups. J. Gen. Virol. 79:1847-1857. [DOI] [PubMed] [Google Scholar]
- 38.Tokita, H., H. Okamoto, P. Luengrojanakul, K. Vareesangthip, T. Chainuvati, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groups. J. Gen. Virol. 75:2329-2335. [DOI] [PubMed] [Google Scholar]
- 39.Tokita, H., H. Okamoto, F. Tsuda, P. Song, S. Nakata, T. Chosa, H. Iizuka, S. Mishiro, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc. Natl. Acad. Sci. USA 91:11022-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viazov, S., S. Kuzin, N. Paladi, M. Tchernovetsky, E. Isaeva, L. Mazhul, F. Vasychova, A. Widell, and M. Roggendorf. 1997. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova, and Uzbekistan). J. Med. Virol. 53:36-40. [PubMed] [Google Scholar]
- 41.White, P. A., X. Zhai, I. Carter, and Y. Zhao. 2000. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J. Clin. Microbiol. 38:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widell, A., S. Shev, S. Månsson, Y.-Y. Zhang, U. Foberg, G. Norkrans, A. Frydon, O. Weilana, J. Kurkus, and E. Nordenfelt. 1994. Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemiological applications. J. Med. Virol. 44:272-279. [DOI] [PubMed] [Google Scholar]
- 43.Zein, N. N., J. Rakela, E. L. Krawitt, K. R. Reddy, T. Tominaga, D. H. Persing, et al. 1996. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann. Intern. Med. 125:634-639. [DOI] [PubMed] [Google Scholar]