Abstract
Although cellular processes depend on protein–protein interactions, our understanding of molecular recognition between proteins remains far from comprehensive. Protein–protein interfaces are structural and energetic mosaics in which a subset of interfacial residues, called hot spots, contributes disproportionately to the affinity of the complex. These hot-spot residues can be further clustered into hot regions. It has been proposed that binding energetics between residues within a hot region are cooperative, whereas those between hot regions are strictly additive. If this idea held true for all protein–protein interactions, then energetically significant long-range conformational effects would be unlikely to occur. In the present study, we show cooperative binding energetics between distinct hot regions that are separated by >20 Å. Using combinatorial mutagenesis and surface plasmon resonance binding analysis to dissect additivity and cooperativity in a complex formed between a variable domain of a T cell receptor and a bacterial superantigen, we find that combinations of mutations from each of two hot regions exhibited significant cooperative energetics. Their connecting sequence is composed primarily of a single β-strand of the T cell receptor variable Ig domain, which has been observed to undergo a strand-switching event and does not form an integral part of the stabilizing core of this Ig domain. We propose that these cooperative effects are propagated through a dynamic structural network. Cooperativity between hot regions has significant implications for the prediction and inhibition of protein–protein interactions.
Keywords: binding energy, cooperativity, protein–protein interaction, surface plasmon resonance, T cell activity
Interactions between proteins are essential for nearly all cellular processes (1–3), and aberrant protein–protein interactions contribute to the pathogenesis of numerous human diseases (4). As the genomewide mapping of protein–protein interactions has identified many of the molecular components of numerous physiological and pathological processes (5–9) and structural genomics efforts have determined structures of many of the constituent protein domains involved in these interactions, the ability to predict the binding specificities and energies of protein complexes from protein structures alone has reached paramount importance. Although significant progress in developing computational methods for the quantitative predictions of protein–protein interactions has been made recently (10–14), the current robustness of these algorithms is not such that the laborious task of determining the structure of a given protein complex can be circumvented. It is clear that these methods are unable to account for aspects of molecular recognition that are important in determining complex formation, but for which we currently have a fundamental lack of understanding.
It has been known for some time that protein–protein interfaces are structural and energetic mosaics. Certain amino acid residues within an interface contribute significantly to the binding energy, and are thus termed “hot spots” (15–17), whereas other residues are energetically silent with respect to the interaction. These hot spots, furthermore, are not homogeneously distributed throughout the interface, but are instead clustered to form “hot regions” (18, 19). Further contributing to the heterogeneity of protein–protein interfaces is the frequent presence of cooperativity, such that the energetic contribution to binding of a protein that has been simultaneously mutated at multiple residues is significantly different from the summation of the changes in binding energy of the single-site mutants (20–22).
The theory that residues within a single hot region are energetically cooperative, whereas those residing in distinct hot regions are strictly additive, has arisen from both computational and experimental studies: (i) a recent analysis (18) of a structurally nonredundant database of all hot regions (23) currently in the Protein Data Bank (24), and (ii) the mutational, energetic, and structural analysis of residues within two hot regions of the TEM1-β-lactamase–β-lactamase inhibitor protein complex (19). If, in all protein complexes, cooperative energetics existed only within hot regions, and not between them, the quantitative prediction of protein–protein interactions may be considerably simplified.
To test whether cooperativity could exist between hot regions, we analyzed the interaction between affinity maturation variants of the human T cell receptor (TCR) β chain variable domain 2.1 (hVβ2.1) and the superantigen toxic shock syndrome toxin 1 (TSST-1). By using combinatorial mutagenesis and surface plasmon resonance (SPR) binding analysis, we show that residues within distinct hot regions, and at distances >20 Å apart, are significantly cooperative. We propose that these cooperative effects are transmitted through a dynamic structural network in the TCR molecule and discuss the relevance of these findings to protein–protein interactions in general.
Results and Discussion
Identification of Energetically Significant Variant Residues in the Affinity Maturation Pathway.
The affinity-matured human Vβ2.1 variant called D10, engineered by yeast display, contains 14 mutations beyond that of EP-8, the WT hVβ2.1 analog selected for enhanced stability (25). D10 binds TSST-1 with an affinity of 180 pM, 30,000-fold tighter than does EP-8. TSST-1 is the major causative agent of staphylococcal toxic shock syndrome (26, 27), and these hVβ2.1 variants were previously engineered as potential therapeutic agents for superantigen-mediated disease (25). Because the yeast display libraries contained stretches of five randomly mutated codons, many of these mutations were likely not involved in affinity increases, but were incorporated in combination with a key mutation. To determine which of these mutations were significant contributors to the higher-affinity interaction with TSST-1, we performed site-directed mutagenesis to create 13 individual single-site mutants from the EP-8 template, including: R10M, F27aT, Q28N, A29I, T30H, E50H, E51Q, S52aF, K53N, T55I, E61V, L72P, and I91V [residues 27a and 52a are noncanonical insertions into the hVβ2.1 complementarity-determining region (CDR) 1 and CDR2 loops, respectively]. The R113Q mutation was not made as this position is located on the face of the Vβ domain opposite that of the interface with TSST-1 and is thus unlikely to affect TSST-1 binding.
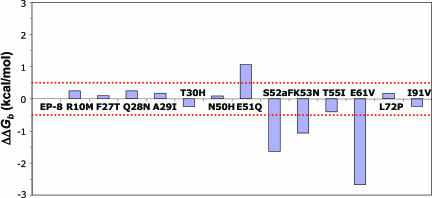
The binding affinities of each of these single-site variants were determined by SPR equilibrium analysis. The differences in binding free energies relative to EP-8 (ΔΔGb) were calculated, and a threshold value of 0.5 kcal/mol was used to determine whether a mutation exhibited energetic significance (Fig. 1). Four of the mutants (E51Q, S52aF, K53N, and E61V) bound TSST-1 with significantly different affinities than EP-8. Surprisingly, the E51Q mutation bound TSST-1 with significantly lower affinity than did the WT hVβ2.1. The S52aF, K53N, and E61V mutations, conversely, significantly increased the binding affinity for TSST-1. Representative SPR sensorgrams for these interactions are shown in Fig. 4, which is published as supporting information on the PNAS web site.
Fig. 1.
Equilibrium binding analysis of single-site variants. The changes in free energy for each of the single-site hVβ2.1 mutants binding to TSST-1 are plotted. The dotted red line indicates the threshold value used to distinguish energetically significant versus insignificant mutations.
Two Hot Regions in hVβ2.1 for TSST-1 Interaction.
Three of the four energetically significant residues (51, 52a, and 53) are located in the CDR2 loop, whereas the remaining important mutation site, at residue 61, is located in framework region 3 (FR3). These residues are shown in red and blue, respectively, in Fig. 2. The remaining mutations, which do not significantly affect TSST-1 binding, are dispersed about the surface of the hVβ2.1 domain. Most, if not all, of these mutations contribute primarily to stabilization and display of the hVβ2.1 protein on the yeast surface (25, 28–30).
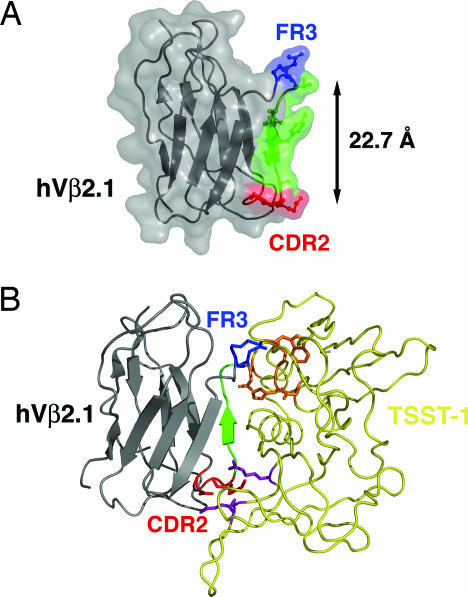
Fig. 2.
Two hot regions for TSST-1 interaction in hVβ2.1. (A) Ribbon diagram/molecular surface representation of the hVβ2.1 domain. The molecule is color-coded according to hot regions (CDR2 hot region, red; FR3 hot region, blue) and the c″ β-strand that connects the two hot regions (green). (B) Model of the hVβ2.1–TSST-1 complex. The hVβ2.1 CDR2 and FR3 hot regions are color-coded as in A; TSST-1 is yellow. The hVβ2.1 CDR2 (red) and FR3 (blue) hot regions are shown interfacing the additional TSST-1 hot region (purple) and the TSST-1 hot region originally described (orange) (32), respectively.
We have previously shown that variant residues at positions 52a, 53, and 61, and the WT residue at position 62, act as hot spots for interaction with TSST-1 (25). These residues form two clusters: residues 51, 52a, and 53 are located at the apex of the CDR2 loop; residues 61 and 62 are positioned at the end of the turn within FR3 (Fig. 2A). These two clusters are connected by the c″ β-strand of the hVβ2.1 Ig domain, a secondary structural element common to all TCR variable Ig domains. The distance between the Cα atoms of residues 51 and 61 is 22.7 Å (31). According to a structural model of the hVβ2.1–TSST-1 complex (Fig. 2B), built by taking into account homology to the hVβ2.1–SpeC complex crystal structure (31) and alanine-scanning mutagenesis analysis of both sides of the hVβ2.1–TSST-1 interface (25, 32), these clusters are located at the periphery of the interface and are as far apart as possible within that interface. Indeed, the distance between Cα atoms of the most peripheral residues in the hVβ2.1–SpeC complex are a comparable 22.1 Å apart (31). These distances also greatly exceed the threshold of 13 Å used to define distinct discontinuous patches within larger protein–protein interfaces (33). Thus, the energetically significant mutations in the D10 affinity maturation pathway are located in two distinct hot regions.
This hVβ2.1–TSST-1 complex model predicts that the FR3 hot region residues interact with TSST-1 hot spots previously identified by alanine scanning mutagenesis (32) (shown in orange in Fig. 2B), and that the CDR2 hot region residues interact with TSST-1 residues that had not been analyzed in that study. To verify the hVβ2.1–TSST-1 docking orientation of our model, we mutated to alanine several additional TSST-1 residues, including Arg-68TSST-1, Lys-71TSST-1, and Ser-72TSST-1, predicted to interact with the hVβ2.1 CDR2 hot region, and assessed their capacities to bind hVβ2.1 by SPR analysis (Fig. 5 A–C, which is published as supporting information on the PNAS web site). As expected from our model, two of these TSST-1 alanine mutants, R68A and S72A, resulted in the complete abolishment of hVβ2.1 binding, whereas the third mutation, K71A, bound TSST-1 with an affinity of 2 μM, a ≈3-fold reduction in binding relative to WT TSST-1. Furthermore, mutants R68A and S72A did not activate primary Vβ2-expressing T cells, similar to the inactive Q139A mutant previously identified (32), whereas K71A behaved similarly to WT (Fig. 5 D–G and Supporting Text, which are published as supporting information on the PNAS web site). These findings indicate that residues Arg-68TSST-1 and Ser-72TSST-1 are hot spots for hVβ2.1 interaction and suggest that this region of TSST-1 (shown in purple in Fig. 2B) contacts the hVβ2.1 CDR2 hot region.
Mutations both Within and Between Hot Regions Are Cooperative.
Once the hVβ2.1 mutations that contributed significantly to TSST-1 binding were identified, we carried out saturation combinatorial mutational analysis to dissect the additive and cooperative energetic properties between these residues. In this approach, we made hVβ2.1 mutants that incorporated every possible combination of WT and mutant residues at each of the four variant positions, residues 51, 52a, 53, and 61. This amounts to a total of 16 (24) distinct hVβ2.1 mutant proteins, including EP-8 (the WT analog), the four single-site mutants described above, six double mutants, four triple mutants, and one variant that simultaneously incorporated all four mutations. We then measured the TSST-1 binding affinities of each of these combinatorial variants. Affinities, ΔGb values, and ΔΔGb values relative to EP-8 are listed in Table 1. Representative SPR sensorgrams for intra-hot regional (CDR2 only) and inter-hot regional (CDR2 and FR3) combinatorial mutants binding to TSST-1 are shown in Fig. 6, which is published as supporting information on the PNAS web site.
Table 1.
Binding analysis for hVβ2.1 single-site and combinatorial variants interacting with TSST-1
| Mutants | KD (kd/ka), M | ΔGb,* kcal/mol | ΔΔGb, kcal/mol | ΔGCOOP,† kcal/mol |
|---|---|---|---|---|
| Single-site mutants | ||||
| EP-8 (WT)‡ | 6.0 × 10−7 | −8.48 | 0 | NA |
| E51Q† | 3.6 × 10−6 | −7.42 | 1.06 | NA |
| S52aF | 3.68 × 10−8 | −10.14 | −1.66 | NA |
| K53N | 1.35 × 10−7 | −9.36 | −0.88 | NA |
| E61V | 6.51 × 10−9 | −11.16 | −2.68 | NA |
| CDR2 intra-hot regional mutants | ||||
| E51Q/S52aF | 4.1 × 10−7 | −8.71 | −0.23 | 0.37 |
| E51Q/K53N† | 2.16 × 10−7 | −9.09 | −0.61 | −0.79 |
| S52aF/K53N | 5.12 × 10−9 | −11.30 | −2.82 | −0.28 |
| E51Q/S52aF/K53N | 1.52 × 10−8 | −10.66 | −2.18 | −0.70 |
| CDR2/FR3 inter-hot regional mutants | ||||
| E51Q/E61V | 4.87 × 10−9 | −11.33 | −2.85 | −1.23 |
| S52aF/E61V | 2.28 × 10−10 | −13.15 | −4.67 | −0.33 |
| K53N/E61V | 1.32 × 10−10 | −13.47 | −4.99 | −1.43 |
| E51Q/S52aF/E61V | 2.91 × 10−9 | −11.64 | −3.16 | 0.12 |
| E51Q/K53N/E61V | 5.83 × 10−10 | −12.59 | −4.11 | −1.61 |
| S52aF/K53N/E61V | 2.71 × 10−11 | −14.41 | −5.93 | −0.71 |
| E51Q/S52aF/K53N/E61V | 1.51 × 10−10 | −13.39 | −4.91 | −0.57 |
| Position 61 hydrophobic mutants | ||||
| Single-site mutants | ||||
| E61I | 5.60 × 10−9 | −11.25 | −2.77 | NA |
| E61L | 9.41 × 10−9 | −10.94 | −2.46 | NA |
| E61F | 2.62 × 10−8 | −10.34 | −1.86 | NA |
| E61W | 3.43 × 10−8 | −10.16 | −1.68 | NA |
| Combinatorial mutants | ||||
| E51Q/K53N/E61I | 1.53 × 10−9 | −12.02 | −3.54 | −0.95 |
| E51Q/K53N/E61L | 2.74 × 10−9 | −11.68 | −3.20 | −0.92 |
| E51Q/K53N/E61F | 5.53 × 10−9 | −11.26 | −2.78 | −1.10 |
| E51Q/K53N/E61W | 1.22 × 10−8 | −10.79 | −2.31 | −0.81 |
NA, not available.
*ΔGb = RTln(1/KD), where R is the gas constant (R = 1.987 cal/mol) and T is temperature in K.
†Cooperative combinations of mutations are in bold italics; additive combinations of mutations are in plain text.
‡Affinity values were determined by SPR equilibrium analysis; kinetic parameters were not determined.
From these binding parameters, we calculated ΔGCOOP values for each combinatorial mutant (Table 1; where ΔGCOOP is the difference between the summation of the changes in binding free energies of the single-site mutants and the experimental changes in binding free energies of the corresponding combinatorial mutant). Using our previous threshold of |ΔGCOOP| ≥ 0.5 kcal/mol for energetic cooperativity (22), we found that mutations within the CDR2 hot region that incorporate mutations of residues 51 and 53 exhibit a moderate degree of positive cooperativity. Surprisingly, mutation at residue 61 in the FR3 hot region is also positively cooperative with either or both residues 51 and 53 in the CDR2 hot region. This observed cooperativity between mutations in two distinct hot regions contradicts previous reports (18, 19) and suggests that hot regions in protein–protein interactions are not necessarily energetically autonomous.
The Magnitude of Cooperativity Is Greater Between than Within Hot Regions.
Compared with the measured cooperativity within the CDR2 hot region, the cooperative energetics between residues from both hot regions are significantly greater in magnitude. For instance, the largest cooperative effect observed is that of the E51Q/K53N/E61V triple mutant, the combinatorial mutant incorporating mutations at FR3 residue 61 and the two CDR2 residues with which it is cooperative, 51 and 53. The difference in experimental versus additive changes in binding free energies for simultaneous mutation at these positions (ΔGCOOP) is −1.61 kcal/mol. In comparison, the E51Q/K53N variant, the most cooperative intra-hot regional combinatorial mutant, exhibits a ΔGCOOP value of only −0.79 kcal/mol (Table 1). SPR sensorgrams for the binding of these hVβ2.1 variants to TSST-1 are shown in Fig. 6 A and B.
Surprisingly, we found that one combination of mutations results in tighter binding to TSST-1 than the penultimate yeast display variant, D10. The S52aF/K53N/E61V triple mutant binds TSST-1 with a KD of 27 pM, whereas D10 binds TSST-1 with a KD of 180 pM (Fig. 6 C and D). This combination of mutations at residue 61 (FR3 hot region) with both residues 52 and 53 (CDR2 hot region) is moderately cooperative (ΔGCOOP = −0.71 kcal/mol; Table 1). In the absence of the exhibited positive cooperativity, we calculate that the triple mutant would instead bind TSST-1 with a KD of 89 pM, intermediate to the TSST-1 affinities of the S52aF/K53N/E61V mutant and D10, and >3-fold tighter than if these mutations were strictly additive. The S52aF/K53N/E61V triple mutant may prove superior to D10 as a candidate therapeutic molecule for TSST-1-mediated disease, because of its very high affinity.
Numerous Combinations of Mutations Exhibit Cooperativity Between Hot Regions.
To determine whether the valine residue at position 61 in the FR3 hot region was required for the observed cooperative energetics between the CDR2 and FR3 hot regions, we made additional mutations at position 61 in both the EP-8 (WT) and E51Q/K53N (largest magnitude of cooperativity) backgrounds. These mutations included the increasingly hydrophobic residues leucine, isoleucine, phenylalanine, and tryptophan, for a total of eight additional variants for which binding to TSST-1 was measured by SPR analysis. As single-site mutants (i.e., in the EP-8 background), these variants all bound TSST-1 with higher affinities than EP-8 (Table 1 and Fig. 7, which is published as supporting information on the PNAS web site). Determination of the binding affinities of each of these four mutations in the context of the E51Q/K53N background revealed that regardless of which hydrophobic side chain is present at position 61, cooperativity between the CDR2 and FR3 hot regions is maintained (Table 1). Although the magnitude of the cooperativity for each of these hydrophobic replacements in the FR3 hot region is less than that observed when valine resides at position 61, in each case it is at least equivalent to that exhibited by the most cooperative CDR2 intra-hot regional variant, E51Q/K53N.
Propagation of Cooperative Effects Through a Dynamic Structural Network.
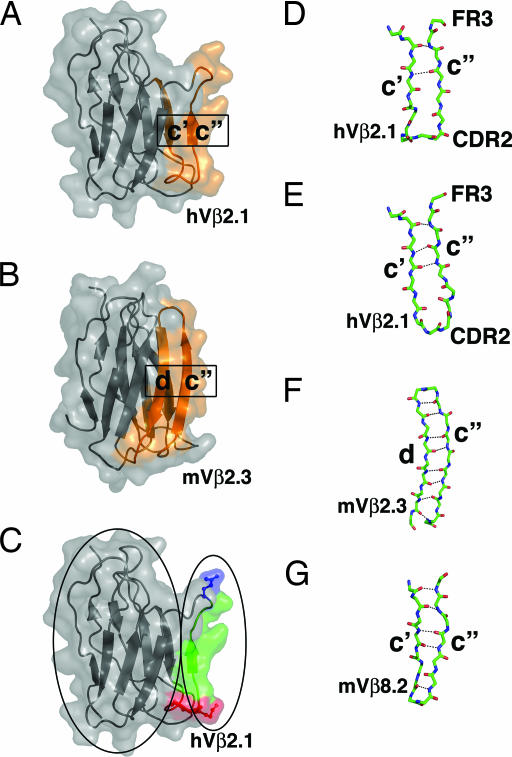
The two hot regions in the hVβ2.1 domain for TSST-1 binding are located before and after the c″ β-strand of the Ig domain. This strand has been shown to participate in a strand-switching event in TCR Vβ domains. In most Vβ domains, such as in hVβ2.1, the c″ β-strand is hydrogen-bonded to the preceding c′ β-strand (Fig. 3A). In some Vβ domains, however, the c″ β-strand is hydrogen-bonded to the following d β-strand. An example of this strand switching is shown in Fig. 3B for the murine Vβ2.3 (mVβ2.3) domain (34).
Fig. 3.
A dynamic structural network for the propagation of cooperative binding effects. (A and B) Strand swapping of the c″ β-strand in TCR Vβ domains as depicted in the hVβ2.1 domain (A) (31) and the mVβ2.3 domain (B) (34). The strands that are hydrogen-bonded to one another are colored orange. (C) A view of the hVβ2.1 domain in which the protein core and the CDR2 (red) and FR3 (blue) hot regions and the connecting c″ β-strand (green) are outlined by ovals on the left and right, respectively. (D–G) The c′–c″ or c″–d β-strand regions of hVβ2.1 from a TCR-superantigen structure [Protein Data Bank (PDB) ID code 1KTK] (D), hVβ2.1 from a TCR-autoimmune peptide-MHC structure (PDB ID code 1YMM) (E), mVβ2.3 (PDB ID code 1KB5) (F), and mVβ8.2 (PDB ID code 1BEC) (G).
One implication of strand switching for the structure of the TCR Vβ domain and the interaction of proteins with this region of the Vβ domain is that the c″ β-strand, relative to other β-strands in the TCR Vβ Ig domain, has a propensity for flexibility. This is indeed the case for the c″ β-strand of hVβ2.1. Two independently determined crystal structures of hVβ2.1 bound to either the superantigen SpeC (31) or an autoimmune peptide-MHC (35) highlight the flexible nature of the hVβ2.1 CDR2 and FR3 hot regions and the intervening sequence. In the former (Fig. 3D), only two hydrogen bonds are formed between the c′ and c″ β-strands, whereas in the latter (Fig. 3E), three hydrogen bonds are made between these strands. The distances and angles of even the two common hydrogen bonds in these hVβ2.1 structures are significantly different. Furthermore, the structures of the CDR2 loops diverge greatly (the Cα atoms of the noncanonical insertion residue Ser-52a are situated 4.7 Å apart when the structures are superimposed), and these differences are propagated through the c″ β-strand and into the FR3 hot region (Fig. 3 D and E).
Thus, when hVβ2.1 binds distinct ligands, the CDR2 and FR3 hot regions adopt significantly different conformations and relative positions. Hydrogen bonds between the c′ and c″ β-strands are made, altered, and broken, and the c″ β-strand is translated relative to the c′ β-strand. We suggest that when hVβ2.1 and the various combinatorial mutants analyzed in this study bind TSST-1 the conformations of this region of the hVβ2.1 domain are distinct, and that these dynamic changes, or the redistribution of conformational ensembles, control the cooperative energetics between the CDR2 and FR3 hot regions when hVβ2.1 variants engage TSST-1. This finding is reminiscent of other allosteric proteins (36). Furthermore, the WT residue Tyr-56 located at the center of the c″ β-strand (Fig. 2A) has been shown to be important in TSST-1 binding (25) and may serve as a key component of the network that allows these cooperative effects to be propagated along the c″ β-strand.
The dynamic structure of the hVβ2.1 c′–c″ β-strand pairing arrangement stands in stark contrast to the analogous regions of either the mVβ2.3 domain (Fig. 3F), in which the c″ β-strand is paired with the d β-strand, or of the mVβ8.2 domain (Fig. 3G), which adopts the conventional c′–c″ β-strand pairing observed in the hVβ2.1 domain. Not only are the hydrogen bonds formed between these β-strands more numerous than in the hVβ2.1 domain, they are on average significantly shorter, resulting in regions that are less likely to be structurally dynamic, and thus, may not exhibit such large cooperative effects.
The Protein Core as a Potential Regulator of Cooperativity Between Hot Regions.
Another implication of TCR Vβ domain strand switching is that the c″ β-strand can be considered to lie outside of the hydrogen-bonded β-strand network that forms the hVβ2.1 protein core. This nonstabilizing role for the c″ β-strand can be seen by comparison of Fig. 3 A and B and is most clearly depicted in Fig. 3C, in which the mutated residues in both the CDR2 (in red) and FR3 (in blue) hot regions and the connecting c″ β-strand (in green) are highlighted.
Although the distance between the CDR2 and FR3 hot regions spans the breadth of the molecular interface (Fig. 2B), and is thus as large as possible for the given protein–protein interaction, the connecting residues are positioned outside of the protein core, and thus, not integrally involved in forming the intramolecular contacts that stabilize the protein. Although speculative, it is possible that such a positioning of the hot-region intervening sequence along the exterior of the protein core may increase the propensity for conformational changes to be transmitted from one hot region to another, such as described above, allowing for cooperative energetics.
In contrast, hot regions for which the connecting residues are integrally involved in the formation of the protein core may more likely result in additive energetics, even when the distance on the molecular surface between hot regions is short. These types of hot regions are most common in protein interfaces and are representative of the hot regions in the TEM1-β-lactamase–β-lactamase inhibitor protein complex, for which it was found that inter-hot regional mutations were strictly additive (19). It may be that for such hot regions there exists an energetic cost resulting from the partial destabilization of the protein core that exceeds any energetic gain from cooperative binding.
There exists a wide spectrum of hot-region coordination and resulting allosteric effects in protein–protein interactions, in which the hVβ2.1–TSST-1 and TEM1-β-lactamase–β-lactamase inhibitor protein complexes represent divergent and perhaps extreme examples. Receptor homodimerization by the human growth hormone (hGH) has been shown to be allosterically regulated (37) and may represent an intermediate on such a continuum. The binding of the two receptor molecules to hGH occurs in a regulated and sequential manner. An allosteric switch residue in hGH resides on an α-helix and exhibits significant pairwise cooperative effects during receptor homodimerization on residues at distances >10 Å away (38) that are also located primarily on α-helices. The α-helices on which all of these residues reside form an integral part of the hGH protein core. Thus, the protein core does not universally dampen energetic transmission between distant regions to such an extent that no cooperativity can occur.
It is noteworthy that the high-affinity hVβ2.1 mutant D10 was generated after yeast display of a library with site-directed mutations in five contiguous CDR2 residues (yielding the S52aF and K53N mutations), followed by selection from a library that contained the E61V mutation from an apparent PCR error. Other studies have described how libraries of mutated residues within multiple regions of an interface can be evolved sequentially to obtain a high-affinity protein that has considerable cooperativity and conformational reorganization (21). However, the present study illustrates how more modest changes, including one (E61V) that was the product of a random mutational event, could yield a dramatic increase in affinity. The identification of distal, cooperative mutations such as E61V supports the notion that affinity maturation may benefit from random mutagenesis strategies (39).
Implications for Protein–Protein Interaction Prediction and Inhibition.
A fundamental lack of understanding of cooperative energetics is one of the major impediments to formulating with greater accuracy algorithms for protein–protein interaction prediction. If cooperativity existed only within hot regions, and not between them, the task of accurately predicting the binding parameters for protein complexes would be greatly simplified. Our results suggest that this may be an overly generalized representation of macromolecular interfaces and that a broader consideration of cooperativity within protein–protein interactions, while more technically and computationally demanding, may ultimately lead to more accurate predictive algorithms. It also appears from our results that only a subset of hot regions may need to be considered as potentially cooperative.
Because protein–protein interactions are pervasive in biological processes, they are also important therapeutic targets. The development of small-molecule inhibitors of such interactions has proven difficult (40), largely because of the relatively planar nature of these interfaces, which tend not to present well defined binding pockets. The presence of hot spots and hot regions within protein interfaces provide possible sites at which potent small-molecule inhibitors may bind to effectively block the association of much larger molecules. Indeed, small peptides selected by phage display generally bind their protein binding partners at hot spots (41), and the discovery of small molecules that inhibit the interaction of B7–1 with CD28 and modulate T cell activation, and in which the drug binds at a hot spot, has been reported (42, 43). If certain distinct hot regions may be linked energetically, as our results suggest, the potency of a small-molecule inhibitor that targets a cooperative hot region may be amplified relative to a small molecule that targets a hot region that is strictly additive. This possibility could have important ramifications for the choice of which hot region within a protein–protein interaction to target for small-molecule inhibition, for instance, by structure-based drug design.
Materials and Methods
Protein Production.
All hVβ2.1 variants were expressed in Escherichia coli and refolded in vitro from inclusion bodies as described for mVβ8.2 domain variants (22). The WT TSST-1 gene (tst) was PCR-amplified from pCE107 (32) and cloned into pET41a (Novagen), and the protein was expressed in E. coli BL21(DE3) (Novagen) and purified by affinity and size exclusion chromatography.
Mutagenesis.
All mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer’s instructions, using the pT7–7/EP-8 expression vector as a template (25). For saturation combinatorial mutagenesis, the order of mutations was important, in that the complementary sites for oligonucleotide primers targeted to the three codons of interest in the CDR2 loop were overlapping, and thus, changed according to the sequence of the mutagenesis events. The double, triple, and quadruple mutants were generated by sequential rounds of site-directed mutagenesis, such that the lower-order mutation vectors were always used as templates for the higher-order mutants.
SPR Binding Analysis.
The interaction of hVβ2.1 variants with immobilized TSST-1 was measured by SPR equilibrium and, where applicable, kinetic analyses using a Biacore 3000 SPR instrument, as described (25). Because the dissociation kinetics of the tightest interactions approached the measurement limits of current SPR technology (44), we performed analogous binding experiments for these interactions in which the dissociation time was extended from 5 min to 2 h. No significant differences in the measured off-rates were observed for these interactions, relative to the shorter dissociation time experiments.
Supplementary Material
Acknowledgments
We thank Drs. Steen Hansen and Knut Langsetmo for helpful discussions. This work was supported by National Institutes of Health Grant AI55882 (to E.J.S.), National Institutes of Health Grants GM55767 and AI064611 (to D.M.K.), and Canadian Institutes of Health Research Operating Grant MOP116537 (to J.K.M.). B.M. is the recipient of the Boston Biomedical Research Institute Scholar Award. R.A.B. was partially supported by National Institutes of Health Training Grant T32 GM 07283. J.K.M. holds a New Investigator Award from the Canadian Institutes of Health Research.
Abbreviations
- CDR
complementarity-determining region
- FR
framework region
- TCR
T cell receptor
- hVβ2.1
human TCR β chain variable domain 2.1
- mVβ2.3
murine Vβ2.3
- SPR
surface plasmon resonance
- TSST-1
toxic shock syndrome toxin 1.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gascoigne N. R., Zal T. Curr. Opin. Immunol. 2004;16:114–119. doi: 10.1016/j.coi.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T., Nash P. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 3.Warren A. J. Curr. Opin. Struct. Biol. 2002;12:107–114. doi: 10.1016/s0959-440x(02)00296-8. [DOI] [PubMed] [Google Scholar]
- 4.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., et al. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 5.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., et al. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 7.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., et al. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 8.Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 9.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerois R., Nielsen J. E., Serrano L. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 11.Huo S., Massova I., Kollman P. A. J. Comput. Chem. 2002;23:15–27. doi: 10.1002/jcc.1153. [DOI] [PubMed] [Google Scholar]
- 12.Kortemme T., Baker D. Proc. Natl. Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massova I., Kollman P. A. J. Am. Chem. Soc. 1999;121:8133–8143. [Google Scholar]
- 14.Sharp K. A. Proteins. 1998;33:39–48. [PubMed] [Google Scholar]
- 15.Clackson T., Wells J. A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 16.Bogan A. A., Thorn K. S. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 17.DeLano W. L. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 18.Keskin O., Ma B., Nussinov R. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 19.Reichmann D., Rahat O., Albeck S., Meged R., Dym O., Schreiber G. Proc. Natl. Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albeck S., Unger R., Schreiber G. J. Mol. Biol. 2000;298:503–520. doi: 10.1006/jmbi.2000.3656. [DOI] [PubMed] [Google Scholar]
- 21.Bernat B., Sun M., Dwyer M., Feldkamp M., Kossiakoff A. A. Biochemistry. 2004;43:6076–6084. doi: 10.1021/bi036069b. [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Swaminathan C. P., Huang Y., Guan R., Cho S., Kieke M. C., Kranz D. M., Mariuzza R. A., Sundberg E. J. J. Biol. Chem. 2003;278:50412–50421. doi: 10.1074/jbc.M306848200. [DOI] [PubMed] [Google Scholar]
- 23.Keskin O., Tsai C. J., Wolfson H., Nussinov R. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buonpane R. A., Moza B., Sundberg E. J., Kranz D. M. J. Mol. Biol. 2005;353:308–321. doi: 10.1016/j.jmb.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 26.Schlievert P. M., Shands K. N., Dan B. B., Schmid G. P., Nishimura R. D. J. Infect. Dis. 1981;143:509–516. doi: 10.1093/infdis/143.4.509. [DOI] [PubMed] [Google Scholar]
- 27.McCormick J. K., Yarwood J. M., Schlievert P. M. Annu. Rev. Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 28.Shusta E. V., Holler P. D., Kieke M. C., Kranz D. M., Wittrup K. D. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 29.Kieke M. C., Shusta E. V., Boder E. T., Teyton L., Wittrup K. D., Kranz D. M. Proc. Natl. Acad. Sci. USA. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieke M. C., Sundberg E., Shusta E. V., Mariuzza R. A., Wittrup K. D., Kranz D. M. J. Mol. Biol. 2001;307:1305–1315. doi: 10.1006/jmbi.2001.4560. [DOI] [PubMed] [Google Scholar]
- 31.Sundberg E. J., Li H., Llera A. S., McCormick J. K., Tormo J., Schlievert P. M., Karjalainen K., Mariuzza R. A. Structure (London) 2002;10:687–699. doi: 10.1016/s0969-2126(02)00759-1. [DOI] [PubMed] [Google Scholar]
- 32.McCormick J. K., Tripp T. J., Llera A. S., Sundberg E. J., Dinges M. M., Mariuzza R. A., Schlievert P. M. J. Immunol. 2003;171:1385–1392. doi: 10.4049/jimmunol.171.3.1385. [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti P., Janin J. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 34.Housset D., Mazza G., Gregoire C., Piras C., Malissen B., Fontecilla-Camps J. C. EMBO J. 1997;16:4205–4216. doi: 10.1093/emboj/16.14.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hahn M., Nicholson M. J., Pyrdol J., Wucherpfennig K. W. Nat. Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunasekaran K., Ma B., Nussinov R. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 37.Walsh S. T., Sylvester J. E., Kossiakoff A. A. Proc. Natl. Acad. Sci. USA. 2004;101:17078–17083. doi: 10.1073/pnas.0403336101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh S. T., Jevitts L. M., Sylvester J. E., Kossiakoff A. A. Protein Sci. 2003;12:1960–1970. doi: 10.1110/ps.03133903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boder E. T., Midelfort K. S., Wittrup K. D. Proc. Natl. Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arkin M. R., Wells J. A. Nat. Rev. Drug Discovery. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 41.Sidhu S. S., Fairbrother W. J., Deshayes K. ChemBioChem. 2003;4:14–25. doi: 10.1002/cbic.200390008. [DOI] [PubMed] [Google Scholar]
- 42.Green N. J., Xiang J., Chen J., Chen L., Davies A. M., Erbe D., Tam S., Tobin J. F. Bioorg. Med. Chem. 2003;11:2991–3013. doi: 10.1016/s0968-0896(03)00183-4. [DOI] [PubMed] [Google Scholar]
- 43.Erbe D. V., Wang S., Xing Y., Tobin J. F. J. Biol. Chem. 2002;277:7363–7368. doi: 10.1074/jbc.M110162200. [DOI] [PubMed] [Google Scholar]
- 44.Schuck P., Boyd L. F., Andersen P. S. Curr. Opin. Protein Sci. 1999;17:20.2.1–20.2.22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.