Abstract
We modified a p24 antigen enzyme-linked immunosorbent assay as a method for diagnosis and monitoring of human immunodeficiency virus type 1 (HIV-1) subtype E infection. This modified assay is based on the use of preheated immune complex dissociation combined with a booster step using a regular Vironostika HIV-1 p24 antigen assay (bioMerieux) to decrease the lower limit of p24 antigen detection from 10 pg/ml (lower limit achievable when using a regular p24 antigen assay) to 0.5 pg/ml (100 virions/ml) by the new method. The correlation between the values obtained by the HIV-1 RNA (Amplicor HIV-1 Monitor) assay and the p24 antigen assay modified with a booster step antigen assay in 160 frozen plasma samples with known viral load and 80 blind fresh plasma samples by Spearman rank were 0.671 (R2 = 0.450; P < 0.01) and 0.782 (R2 = 0.612; P < 0.01). During antiretroviral treatment, the change of p24 antigen level at ≥0.5 log correlated well with the level of HIV-1 in plasma. In order to improve the early diagnosis of HIV-1 infection in 121 infants born to HIV-1-infected mothers, a heat-denatured plasma p24 antigen assay modified with a booster step was compared with DNA-PCR and HIV RNA (nucleic acid sequence-based amplification) assays. The sensitivity of the antigen test modified with a booster step was similar to that of the HIV-1 RNA (NASBA QL) assay and better than that of the DNA-PCR assay (100 versus 61.90%) for subjects 1 to 2 months old. The overall results from this study might renew interest in p24 antigen detection as an easily affordable alternative method for diagnosis of HIV-1 infection and monitoring of disease progression in developing countries.
The World Health Organization has estimated that there were 40 million people infected with human immunodeficiency virus (HIV) globally at the end of 2001, and the majority of them were in developing countries (24). The countries affected the most are economically poor and therefore unable to afford expensive diagnostic and monitoring tests. HIV type 1 (HIV-1) RNA quantitation is needed to monitor disease progression of HIV-1 infection to determine initiation of treatment and measure response to antiretroviral treatment in infected children and adolescent as well as adults (11, 12, 15, 18). Although culture and amplification HIV-1 RNA or DNA are the most-sensitive assays for assessment of viral burden, they are more expensive, time-consuming, labor-intensive, and costly than testing for HIV-1 p24 antigen. The regular HIV-1 p24 antigen test is relatively insensitive, with a lower limit of detection of 10 pg/ml. This quantity of antigen may not be present in the serum of infected individuals, even when the virus is actively replicating. In fact, only about 50 to 60% of AIDS patients, 30 to 40% of AIDS-related complex patients, and 10% of asymptomatic patients have p24 antigenemia (1).
One of the reasons for low sensitivity of the antigen test when testing the serum of HIV-infected persons is that free p24 antigen in serum may be bound to p24 antibody. The test cannot detect these antigen-antibody complexes. Even with a complex dissociation technique, the sensitivity remains low and can only detect about 50% of asymptomatic patients (6). Following this, a p24 antigen detection method modified with a booster step was developed by introducing a signal amplification boosted system to overcome the problem (2, 8, 9, 13, 14).
Determination of HIV-1 p24 antigen can be accomplished by using simple equipment and at a considerably lower cost. Moreover, there are some advantages of the p24 antigen assay compared with the quantification of viral RNA in that the antigen is considerably more stable and less affected by variation in time and physical conditions during transportation. In addition, the sensitivity of antigen testing may be less affected by genetic diversity.
A p 24 antigen assay was modified from a regular p24 antigen assay (bioMerieux), which is widely used in Thailand, by using preheated immune complex dissociation combined with a booster step to quantify HIV-1 p24 antigen in plasma. The aim of this study was to compare this method with the determination HIV-1 RNA viral load as a tool for monitoring HIV-1 infection. Moreover, qualitative HIV-1 p24 antigen detected by the p24 antigen method modified with a booster step can offer early diagnosis of HIV-1 infection in infants born to HIV-infected mothers. Therefore, it might replace the methods that are hardly affordable in developing countries with limited financial resources.
MATERIALS AND METHODS
One hundred and sixty frozen plasma samples, collected from HIV-1-infected subjects from January 1999 to November 2000, with known viral load (tested by Amplicor HIV Monitor method [version 1.5; Roche Diagnostic Systems, Branchburg, N.J.]), were used in the p 24 antigen assay modified with a booster step and a cross-sectional study comparing p24 antigen and HIV-1 RNA quantitation. One hundred HIV-1-seronegative plasma samples were used to determine the cutoff value of the modified antigen assay. Another eighty fresh plasma samples from HIV-1-infected subjects were used for a cross-sectional blinded test comparison of the quantity of HIV-1 RNA present as determined by the Amplicor HIV Monitor test (version 1.5; Roche Diagnostic Systems) and p24 antigen quantification by the modified p24 antigen assay with a booster step between February 2001 and April 2001. The standard Amplicor HIV-1 Monitor test was used in this study, which has a dynamic range of 400 to 750,000 copies of HIV-1 RNA/ml of plasma.
In order to evaluate the p24 antigen assay modified with a booster step for monitoring HIV-1 infection during antiretroviral therapy, 90 plasma samples were collected from 30 HIV-1-infected pregnant women, who received perinatal treatment with the antiretroviral drug combination of zidovudine (AZT) and lamivudine (3TC) from 34 weeks of pregnancy until delivery. Blood samples were collected at three different times from these infected pregnant women: at 34 weeks (before antiretroviral drug initiation) (M0), at delivery (M2), and 1 month postpartum (M3).
Frozen plasma samples were collected from 121 infants born to HIV-1-infected mothers attending the HIV clinic at Siriraj Hospital, from January 1998 to February 2001, at the ages of 1 to 2 months and 4 to 6 months. These infants' samples were taken to evaluate the sensitivity and specificity of the antigen assay modified with a booster step, compared with HIV-1 DNA detection by PCR, and HIV-1 RNA qualitative detection (NucliSens HIV-1 RNA qualitative assay; bioMerieux, Boxtel, The Netherlands) for the early diagnosis of perinatal HIV-1 infection (Table 1).
TABLE 1.
Sensitivities and specificities of various assays in early diagnosis of HIV-1 infection in 121 infants born to HIV-1-infected mothers
| Age (mo) | Assay | Result | No. with HIV-1 infection status
|
Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|
| Positive | Negative | |||||
| 1-2 | DNA-PCR | Positive | 13 | 0 | 61.9 | 100 |
| Negative | 8 | 100 | ||||
| RNA-NASBA | Positive | 21 | 0 | 100 | 100 | |
| Negative | 0 | 100 | ||||
| p24 antigen | Positive | 21 | 0 | 100 | 100 | |
| with booster | Negative | 0 | 100 | |||
| 4-6 | DNA-PCR | Positive | 21 | 0 | 100 | 100 |
| Negative | 0 | 100 | ||||
| RNA-NASBA | Positive | 21 | 0 | 100 | 100 | |
| Negative | 0 | 100 | ||||
| p24 antigen | Positive | 21 | 0 | 100 | 100 | |
| with booster | Negative | 0 | 100 | |||
We performed the antigen assay modified with a booster step by employing the method previously reported (1, 18) with the modification of Vironostika HIV-1 antigen assay (bioMerieux) and ELAST enzyme-linked immunosorbent assay (ELISA). The p24 antigen assay modified with a booster step is based on heat-denatured immune complex dissociation p24 antigen and amplified signal of ELISA with biotinyl tyramide to increase the lower limit of p24 antigen detection. The assay was performed according to the manufacturer's instructions integrating the ELAST ELISA amplification system (Perkin-Elmer Life Sciences, Inc., Boston, Mass.) procedure into the Vironostika HIV-1 antigen assay before the addition of the chromogenic substrate. The plasma was diluted with 0.5% Triton X-100 to a concentration of 1:6. The diluted plasma was heated at 100°C for 5 min. A known concentration p24 antigen that varied between 0 to 80 pg/ml was used as a positive control. All samples (100 μl in each well) were incubated at 37°C for 60 min. After washing with diluted phosphate-buffered saline (PBS), 100 μl of anti-HIV-1 (human) conjugate labeled with horseradish peroxidase (HRP) was added to each well, and incubated at 37°C for 60 min. The streptavidin-HRP (S-HRP) conjugate was used at a dilution of 1:500. After washing, 100 μl of the biotinyl tyramide working solution from the ELAST ELISA amplification system was added to each well and incubated at room temperature for 60 min. Then, 100 μl of diluted S-HRP from the ELAST ELISA amplification system was diluted to 1:500 using 1% bovine serum albumin-PBS-Tween 20 (1% BSA-PBS-T) which was added to each well after the washing step, and the plates were incubated at room temperature for 30 min. Then, 100 μl of tetramethylbenzidine substrate in urea peroxide solution was added to each well after a washing step, and the plates were incubated at room temperature for 30 min. Finally, the color reaction was stopped by adding 100 μl of 1 M sulfuric acid. The absorbance of each well was read at 450 nm within 15 min by the ELISA reader.
Eight negative controls and six positive controls, ranging from 0.5 to 80 pg/ml in serial dilution, were tested in each qualitative and quantitative assay. The cutoff value, the optical density (OD) at 450 nm, for each test was the sum of the means of the absorbance of eight negative controls plus 3 standard deviations (18). Samples with an absorbance value greater than or equal to the cutoff value were considered positive for HIV-1 p24 antigen. Samples with an absorbance value less than the cutoff value were considered negative. Samples with protein concentrations above the assay range were subjected to repeat testing by dilution plasma. A standard curve was generated, from which OD values of the unknown specimens are interpolated to determine their concentration. The standard curve is constructed using a linear graph plotting the concentration of the HIV-p24 antigen that OD values converted onto log10 p24 antigen (in femtograms per milliliter) on the y axis versus known values of viral load (in log10 copies per milliliter) on the x axis. The controls must be included for each assay. The viral load of samples above cutoff could be estimated from the standard curve. This allowed p24 antigen quantification within a range of 500 to 80,000 fg/ml with a single sample dilution.
HIV-1 RNA genome from 1 ml of plasma was detected by using the NucliSens HIV-1 qualitative kit (bioMerieux), used as recommended by the manufacturer (23). The performance assay was linear over a range of 51 to 5,390,000 HIV-1 RNA copies, and the limit of quantification at a 95% rate was calculated to be 176 HIV-1 RNA copies/ml.
To diagnose HIV-1 infection, HIV-1 gag and pol genes were amplified by nested PCRs in a total volume of 50 μl as previously described (8). The amplifications were carried out in the Automated Gene Amp PCR System 9700 (Perkin-Elmer Cetus, Norwalk, Conn.). The lower limit of detection of HIV-1 proviral DNA by this nested PCR was 1 copy per 200,000 peripheral blood mononuclear cells.
The peptide-based ELISA used in this study has been described previously (17) with 14 amino acid sequences specific for subtype E (TSITIGPGQVFYRT) and B (KSIHLGPGQAWYTT). 100 μl of peptide solution at a concentration of 5 mg/ml in 20 mM carbonate buffer pH 9.6 was immobilized on each well of the microtiter plate and incubated for 16 to 18 h at 4°C. The next day, antigen was aspirated and plates were blocked with 200 μl/well of PBS containing 5% dry skim milk powder. In the test assay, serum samples at a dilution of 1:400 in blocking buffer were added to the antigen-coated plates and incubated for 1 h at 37°C. After six washes with washing buffer (PBS-0.05% Tween 20), anti-human IgG peroxidase conjugate (Sigma, USA) diluted in blocking buffer was applied to a well for 1 h at 37°C. The color was developed with orthophenylenediamine dihydrochloride substrate after a further six washes. The absorbance at 492 nm against 620 nm was measured. A cutoff of 0.3 was used throughout the study, with dual-reactions further classified as monoreactive.
All statistical calculations were performed with SPSS/PC+ software (version 9.0; SPSS Inc., Chicago, Ill.). For quantification, levels of HIV RNA and boosted p24 antigen from routine plasma samples were compared by Spearman rank correlation. A P of <0.01 was considered statistically significant. McNemar's test was used to compare the results of the qualitative p24 antigen assay in infants and P < 0.05 was considered statistically significant. The nonparametric Mann-Whitney test was used to compare the differences between sample means.
RESULTS
Limitation of the p24 antigen assay modified with a booster step.
The positive p24 antigen controls with known concentrations of 80, 40, 20, 10, 5, 2.5, 1, and 0.5 pg/ml were used to determine a detection range for the p24 antigen assay modified with a booster step by testing in duplicate with S-HRP conjugate (dilution, 1:500). The results for the undiluted plasma specimens showed that lower and upper limits of the p24 antigen assay modified with a booster step were 0.5 pg/ml or 500 fg/ml and 80 pg/ml or 80,000 fg/ml, respectively (at an OD at 450 nm). The relationship of p24 concentration and absorbance value was characterized as a dose dependent model.
Correlation of quantitation by p24 antigen assay modified with a booster step and HIV-1 RNA copy numbers.
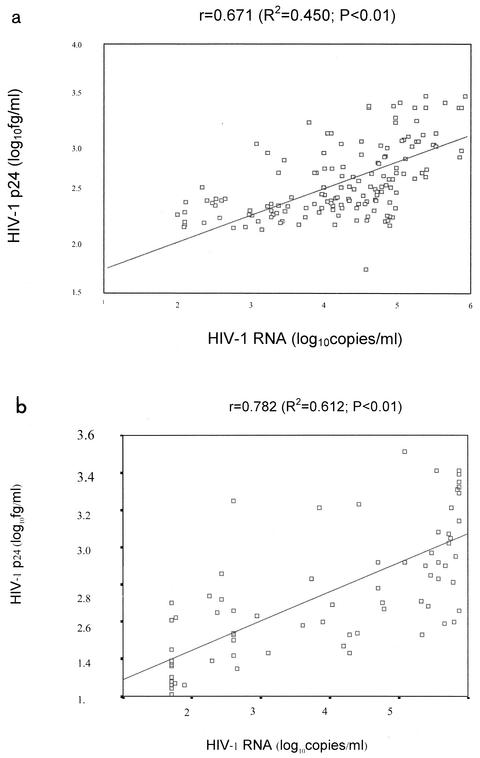
The HIV-1 viral load of 160 frozen plasma samples used in the quantitative p24 antigen assay modified with a booster step were 2log10 (range, 2.10 to 2.99; n = 31), 3 log10 (range, 3.02 to 3.99; n = 30), 4 log10 (range, 4.00 to 4.98; n = 68) and 5 log10 (range, 5.06 to >5.88; n = 31) copies/ml. The mean p24 antigen detected by the p24 antigen assay modified with a booster step (in log10 femtograms per milliliter) compared to each log10 of RNA copies per milliliter from 160 frozen samples were 2.28 (range, 2.08 to 2.75) for 2 log10, 2.48 (range, 2.14 to 3.21) for 3 log10, 2.64 (range, 2.18 to 3.38) for 4 log10 and 3.10 (range, 2.65 to 3.48) for 5 log10. HIV-1 p24 antigen concentrations in the samples that contained a viral load of at least 2 log10 copies/ml could be detected by p24 antigen assay modified with a booster step as follows: 64.5% (20 of 31) for 2 log10, 70% (21 of 30) for 3 log10, 83.33% (57 of 68) for 4 log10, and 100% (31 of 31) for 5 log10 copies/ml. HIV-1 subtypes of individual subjects were determined by peptide serotyping (peptide-based ELISA). Of 160 samples, 8 samples were infected with HIV-1 subtype B′ and 151 were infected with HIV-1 subtype E. Hence, p24 antigen of both HIV-1 subtypes B′ and E could be detected by the p24 antigen assay modified with a booster step. Although this p24 antigen assay modified with a booster step measures different HIV-1 products from different units than the HIV-1 viral load assay, a good correlation of HIV-1 detected by both assays at a 95% confidence interval by Spearman rank correlation and analysis of variance (r = 0.671; R2 = 0.450; P < 0.01) was demonstrated (Fig. 1a).
FIG. 1.
The correlation of HIV-1 boosted p24 antigen and HIV-1 RNA quantification (Amplicor Monitor; Roche) for 160 frozen plasma samples of known viral load at a 95% confidence interval by Spearman rank correlation and analysis of variance was r = 0.671 (R2 = 0.450; P < 0.01) (a) and that for 80 blinded fresh plasma samples at a 95% confidence interval by Spearman rank correlation and analysis of variance was r = 0.782 (R2 = 0.612; P < 0.01) (b).
Furthermore, a blinded test comparison between HIV-1 RNA viral load and p24 antigen quantification in monitoring the antiretroviral treatment of 80 fresh unfrozen plasma samples (25 women and 55 men) was performed. The fresh plasma samples were used to test for HIV-1 RNA viral load and p24 antigen quantification parallel with new identification number separately provided for each sample in each test. The HIV infection status of all these samples was already known by serological testing. All samples were infected with HIV-1 subtype E except for one that was infected with HIV-1 subtype B. Of the 80 samples tested, 57 (71.25%) were positive for HIV-1 RNA and 64 (80%) were positive for p24 antigen. The mean values of the boosted p24 antigen compared to viral load were 2.37 (range, 2.21 to 2.70; n = 19) for 1 log10, 2.64 (range, 2.35 to 3.25; n = 14) for 2 log10, 2.73 (range, 2.43 to 3.21; n = 5) for 3 log10, 2.82 (range, 2.50 to 3.33; n = 11) for 4 log10, and 3.03 (range, 2.53 to 3.51; n = 31) for 5 log10 copies/ml. HIV-1 p24 antigen was detected in the samples that contained a viral load of at least 2 log10 (85.71% of 2 log10 and 100% for 3 log10, 4 log10, and 5 log10 copies/ml). The reproducibility of the p24 antigen assay modified with a booster step was evaluated by performing repeatedly of frozen 160 samples and another 50 fresh plasma samples (data not shown). We found that interassay variation was less than 10%.
Nine (11.25%) samples, which were tested negative for RNA, were found to be positive for p24 antigen. The range of p24 quantitation in these samples was 2.36 to 3.25 log10 fg/ml. The HIV-1 viral load of these nine samples was <400 copies/ml. Two samples (2.5%) that tested negative by quantitation by the p24 antigen assay modified with a booster step tested positive by HIV-1 RNA quantitation. The HIV-1 viral load of these two samples was detected by ultrasensitive detection as 56 and 78 copies/ml. Fourteen (17.5%) samples tested negative by both assays. The viral load of these samples was <400 copies/ml. The correlation of HIV-1 viral load and boosted p24 antigen level by Spearman rank correlation and analysis of variance revealed the following: r = 0.782 (R2 = 0.612; P < 0.01) (Fig. 1b).
We also determined the quantity of p24 antigen and HIV-1 viral load in 90 plasma samples, from 30 HIV-1-infected pregnant women who received perinatal AZT and 3TC treatment from 34 weeks of pregnancy until delivery. The correlation of the amount of p24 antigen (log10 femtograms per milliliter) and HIV-1 RNA viral load (log10 copies per milliliter) on three occasions (before, during, and after treatment) analyzed by Spearman rank correlation, and analysis of these variances revealed the following: r = 0.72 for M0 (R2 = 0.69; P < 0.01), r = 0.95 for M2 (R2 = 0.69; P < 0.01), and r = 0.68 for M3 (R2 = 0.69; P < 0.01), respectively. The mean (range) of HIV-1 RNA (log10 RNA copies per milliliter) and HIV-1 p24 antigen (log10 femtograms per milliliter) of 30 HIV-1-infected pregnant women before (M0), during (M2), and after (M3) therapy with AZT and 3TC were 4.34 (range, 2.6 to 5.79) versus 3.375 (range, 2.45 to 5.58), 2.6 (range, 1.69 to 2.71) versus 2.72 (range, 2.06 to 3.7), and 4.25 (range, 2.88 to 5.92) versus 3.35 (range, 2.7 to 5.0), respectively. (Fig. 2) A mean change of ≥0.5 log in p24 antigen quantitation and RNA viral load from baseline at M0 was correlated well with respond to therapy comparing with those result in M2 and M3.
FIG. 2.
Mean change of HIV-1 RNA (a) and HIV-1 p24 antigen (b) of 30 HIV-1-infected pregnant women before, during, and after therapy with AZT and 3TC.
Comparison of DNA-PCR, qualitative HIV RNA (NASBA), and p24 antigen assay modified with a booster step for the diagnosis of HIV-1 perinatal infection.
In order to use the p24 antigen assay for early diagnosis of infants born to HIV-1-infected mothers, blood samples were collected from 121 children born to HIV-infected mothers with and without antiretroviral drugs treatment at the age of 1 to 2 months and 4 to 6 months. The mean ages at the first and the second collection were 1.2 and 4.8 months, respectively.
Infants were defined as uninfected if two or more results of virological tests were negative, two of which were performed at the age ≥1 month and ≥4 months. Infants were considered to be HIV-1 infected if they had at least two positive HIV virological tests (nucleic acid detection or by culture) performed on separate blood samples (4). Serological tests were performed to confirm infection status of infants at the age of 12 to 18 months. Of 121 infants, 21 infants had confirmed HIV infection. All 21 HIV-1-infected infants were infected with HIV-1 subtype E as determined by peptide serotyping.
Of 21 infected infants, 13 (61.90%) samples at the age of 1 to 2 months yielded positive results by DNA-PCR and 21 (100%) samples were positive for HIV by QL RNA detection (NucliSens) and the p24 antigen assay modified with a booster step. For samples from infants at the age of 4 to 6 months, 21 of 21 (100%) yielded positive results by all DNA-PCR, HIV RNA and the p24 antigen assay modified with a booster step. For samples from infants at the age of 1 to 2 months, the p24 antigen assay modified with a booster step had a sensitivity similar to that of HIV-1 QL RNA detection (NucliSens) and a sensitivity significantly greater than that of HIV-1 DNA PCR detection (100 versus 61.90%; P = 0.008). All eight infants who were initially shown to be HIV negative by the DNA-PCR test tested positive when the second blood sample that was collected at the age of 4 to 6 months was used. All these three methods gave similar specificities (100%) for early detection of HIV infection in infants born to HIV-1-infected mother in the first 2 months of life.
DISCUSSION
There are many laboratory markers to indicate HIV-1 infection, such as nucleic acid, p24 antigen, and anti-HIV-1 antibody. Several methods were employed for quantification of HIV-1 in blood, including culture, the nucleic acid amplification techniques of HIV-1 RNA or DNA, and detection of p24 antigen by ELISA. HIV-1 RNA quantitation is an assay widely used to monitor disease progression of HIV-1 infection in order to determine the initiation of treatment as well as the response to antiretroviral treatment in infected children and adolescent as well as adults.
This is the first study to apply preheated plasma and serum samples to a test using quantification by the p24 antigen assay modified with a booster step and qualification assays for diagnosis of subtype E infection combined with the Vironostika HIV-1 antigen assay, a p24 antigen detection enzyme immunoassay kit manufactured by bioMerieux for use in monitoring and diagnosis of HIV-1 subtype E infection in Thailand.
The p24 antigen assay modified with a booster step was optimized to use with an S-HRP concentration of 1:500 and a plasma dilution of 1:6. Its lower and upper range of p24 antigen detection were 0.5 pg/ml and more than 80 pg/ml. This p24 antigen assay modified with a booster step can detect p24 antigen in plasma at a lower concentration than the regular p24 antigen assay. Layne et al. (7) reported that each HIV particle contains about 5 × 10−17 g of p24 core protein. This assumes that the range of detection of the p24 antigen assay modified with a booster step was from 100 to more than 4,000 virions/ml. The upper limit of the p24 antigen assay modified with a booster step can be increased by dilution of plasma. The lower limit of the p24 antigen assay modified with a booster step is reported to be about 0.5 pg/ml (7, 19, 20), which is similar to that in this study even though a different p24 antigen detection test kit was used. Some reports have indicated that p24 antigen assay could detect not only HIV-1 group M but also and group O (1, 5). The p24 antigen assay modified with a booster step in this study could detect both HIV-1 subtype B and subtype E.
A linear regression equation at a 95% confidence interval of the number of copies quantified by these two methods revealed good correlation. The fact that the correlation of results of the p24 antigen assay modified with a booster step among the fresh 80-sample set (Fig. 1), (r = 0.782) was higher than that among the frozen 160-sample set (Fig. 1a) (r = 0.671) might be because of different condition of samples tested. The 160 plasma samples were used freshly for HIV-1 RNA viral load measurement and stored frozen for p24 antigen quantitation later, but the 80-sample set was used freshly in both HIV-1 RNA viral load and p24-antigen quantitation. The lower limit of detection of this p24 antigen assay modified with a booster step was even better than HIV RNA quantification, in which the limitation of the p24 antigen assay modified with a booster step was 500 fg/ml, or about 100 virions/ml, whereas for the Roche Amplicor HIV-1 Monitor (version 1.5), the normal limit is <400 copies/ml. All the samples which tested positive for HIV-1 RNA also showed positive results in the p24 antigen assay modified with a booster step. The p24 antigen quantification test similar to this p24 antigen assay modified with a booster step were studied in adult patients infected with HIV-1 subtype B and also found to be as sensitivity as RNA-PCR (20). The difference between this study and that previously reported was in the manufacture of the ELISA kit used: Vironostika HIV-1 antigen in this study and NEN/Du Pont p24 Core profile kit in the other study. The staging of infection and treatment status were also different. In this study subjects received antiretroviral treatment and relatively healthy compare to the other report in which the patients were symptomatic with a high viral titer.
Antiretroviral drugs can reduce circulating levels of p24 antigen; therefore, p24 antigen can be a useful marker for evaluating the efficacy of therapy (3, 10, 22). The AIDS Clinical Trial Group has suggested that a sustained 50% drop in p24 antigen concentration is considered a positive result of therapy and that persistently elevated p24 levels may indicate a lack or loss of therapeutic effect (22). We have observed that a ≥0.5-log decrease in p24 antigen quantitation correlates with RNA viral load reduction after effective therapy. Also, a good correlation between p24 antigen concentration and HIV-1 plasma viral load before, during, and after antiretroviral drug is shown in this study.
There have been a few reports using RNA qualitative detection assay, DNA PCR, and regular immune complex dissociation HIV-1 p24 antigen assay for the early diagnosis of infants born to HIV-1-infected mothers in Thailand (16, 21, 25). HIV-1 p24 antigen detection was less sensitive than HIV-1 RNA and DNA detection. Our results indicate that HIV RNA and boosted p24 antigen detection had a higher sensitivity than DNA-PCR (100 versus 61.90%) but a similar specificity (100%) for early diagnosis of perinatal HIV-1 infection. There are some reports comparing DNA-PCR, HIV RNA assay, and the p24 antigen assay modified with a booster step. Schupbach et al. (19) studied infants who never received specific antiretroviral treatment in Switzerland and showed that the sensitivities of p24 antigen and DNA-PCR were 96 and 96.2%, respectively. The other was a study of African children in Tanzania, where the predominate subtypes of HIV-1 are A and D (10); this study showed a sensitivity of the p24 antigen assay modified with a booster step as high as that reported previously (20). Taken together, these data demonstrate that heat-denatured plasma combined with a p24 antigen assay modified with a booster step might be used for early diagnosis and monitoring of HIV-1 infection.
In Thailand, the cost of the HIV-1 RNA quantitative assay (Amplicor HIV-1 Monitor; Roche), HIV-1 RNA qualitative assay (NucliSens; bioMerieux), in-house DNA-PCR, and p24 antigen assay modified with a booster step are $100, $40, $30, and $3 per test. The commercially available HIV-1 RNA viral load tests are about 10 to 30 times more expensive than the p24 antigen assay modified with a booster step. This p24 antigen assay modified with a booster step should be an appropriate test for early diagnosis and monitoring of disease status, especially in developing countries that face economic problems and limited laboratory facilities.
Moreover, there are some advantages of the p24 antigen assay compared with quantification of viral RNA. Sample preparation is much simpler, and the test can be performed with equipment already available in most laboratories at a considerably lower cost. In addition, the sensitivity of antigen testing may be less affected by genomic diversity, because both HIV-1 subtypes B and E could be detected by the p24 antigen assay modified with a booster step.
With all these consideration, the p24 antigen assay modified with a booster step after heat treatment should be used as an easy affordable method alternative to HIV-1 RNA quantification for monitoring the progress of HIV-1 disease. This test may become a new tool for the routine determination of viral load in cases of early seroconversion, the symptomatic phase of HIV infection, and mother-to-child transmission and in making the decision whether to start, wait, add to, or change treatments.
Acknowledgments
This research was supported by a grant from the Division of AIDS, Department of Communicable Diseases Control, Ministry of Public Health of Thailand, and the Ministry of University Affairs of Thailand.
We thank Jane Hardy for reviewing the manuscript.
REFERENCES
- 1.Allain, J. P., Y. Laurian, D. A. Paul, and D. Senn. 1986. Serological markers in early stages of human immunodeficiency virus infection in hemophiliacs. Lancet ii:1233-1236. [DOI] [PubMed]
- 2.Boni, J., M. Opravil, Z. Tomasik, M. Rothen, L. Bisset, P. J. Grob, R. Luthy, and J. Schupbach. 1997. Simple monitoring of antiretroviral therapy with a signal-amplification-boosted HIV-1 p24 antigen assay with heat-denatured plasma. AIDS 11:F47-F52. [DOI] [PubMed] [Google Scholar]
- 3.Burgisser, P., P. Vernazza, M. Flepp, J. Boni, Z. Tomasik, U. Hummel, G. Pantaleo, J. Schupbach, et al. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1994. Revised classification system for human immunodeficiency virus infection in children less than 13 years of age. Morb. Mortal. Wkly. Rep. 43(RR-12):1-10. [Google Scholar]
- 5.Goldschmidt, P. L., A. Devillechabrolle, Z. Ait-Arkoub, and J. T. Aubin. 1998. Comparison of an amplified enzyme-linked immunosorbent assay with procedures based on molecular biology for assessing human immunodeficiency virus type 1 viral load. Clin. Diagn. Lab. Immunol. 5:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goudsmit, J., J. M. Lange, D. A. Paul, and G. J. Dawson. 1987. Antigenemia and antibody titers to core and envelope antigens in AIDS, AIDS-related complex, and subclinical human immunodeficiency virus infection. J. Infect. Dis. 155:558-560. [DOI] [PubMed] [Google Scholar]
- 7.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, and J. P. Moore. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 8.Ledergerber, B., M. Flepp, J. Boni, Z. Tomasik, R. W. Cone, R. Luthy, and J. Schupbach. 2000. Human immunodeficiency virus type 1 p24 antigen concentration measured by boosted ELISA of heat-denatured plasma correlates with decline in CD4 cells, progression to AIDS, and survival: comparison with viral RNA measurement. J. Infect. Dis. 181:1280-1288. [DOI] [PubMed] [Google Scholar]
- 9.Lyamuya, E., U. Bredberg-Raden, A. Massawe, E. Urassa, G. Kawo, G. Msemo, T. Kazimoto, A. Ostborn, K. Karlsson, F. Mhalu, and G. Biberfeld. 1996. Performance of a modified HIV-1 p24 antigen assay for early diagnosis of HIV-1 infection in infants and prediction of mother-to-infant transmission of HIV-1 in Dar es Salaam, Tanzania. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:421-426. [DOI] [PubMed] [Google Scholar]
- 10.Machuca, A., M. Gutierrez, A. Mur, and V. Soriano. 1998. Quantitative p24 antigenaemia for monitoring response to antiretroviral therapy in HIV-1 group O-infected patients. Antivir. Ther. 3:187-189. [PubMed] [Google Scholar]
- 11.Mellors, J., L. Kingsley, C. Rinaldo, J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantification of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 12.Mellors, J., C. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 13.Nadai, D., J. Boni, C. Kind, O. E. Varnier, F. Steiner, Z. Tomasik, and J. Schupbach. 1999. Prospective evaluation of amplification-boosted ELISA for heat-denatured p24 antigen for diagnosis and monitoring of pediatric human immunodeficiency virus type 1 infection. J. Infect. Dis. 180:1089-1095. [DOI] [PubMed] [Google Scholar]
- 14.Nishanian, P., K. R. Huskins, S. Stehn, R. Detels, and J. L. Fahey. 1990. A simple method for improved assay demonstrates that HIV p24 antigen is present as immune complexes in most sera from HIV-infected individuals. J. Infect. Dis. 162:21-28. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien, T., W. Blattner, D. Waters, E. Eyster, M. W. Hilgartner, A. R. Cohen, N. Luban, A. Hatzakis, L. M. Aledort, P. S. Rosenberg, W. J. Miley, B. L. Kroner, and J. J. Goedert. 1996. Serum HIV-1 RNA levels and time to development of AIDS in the Multicenter Hemophilia Cohort Study. JAMA 276:105-110. [PubMed] [Google Scholar]
- 16.Panakitsuwan, S., N. Yoshihara, N. Hashimoto, K. Miyamura, and T. Chotpitayasunondh. 1997. Early diagnosis of vertical HIV infection in infants by rapid detection of immune complex dissociation HIV p24 antigen. AIDS Patient Care STDs 11:429-433. [DOI] [PubMed] [Google Scholar]
- 17.Pau, C.-P, S. Lee-Thomas, W. Auwanit, J. R. George, C. Y. Ou, B. S. Parekh, T. C. Granade, D. L. Holloman, S. Phillips, and G. Schochetman. 1993. Highly specific V3 peptide enzyme immunoassay for serotyping HIV-1 specimens from Thailand. AIDS 7:337-340. [DOI] [PubMed] [Google Scholar]
- 18.Paul, M. O., S. Tetali, M. L. Lesser, E. J. Abrams, X. P. Wang, R. Kowalski, M. Bamji, B. Napolitano, L. Gulick, and S. Bakshi. 1996. Laboratory diagnosis of infection status in infants perinatally exposed to human immunodeficiency virus type 1. J. Infect. Dis. 173:68-76. [DOI] [PubMed] [Google Scholar]
- 19.Schupbach, J., J. Boni, Z. Tomasik, J. Jendis, R. Seger, C. Kind, et al. 1994. Sensitive detection and early prognostic significance of p24 antigen in heat-denatured plasma of human immunodeficiency virus type 1-infected infants. J. Infect. Dis. 170:318-324. [DOI] [PubMed] [Google Scholar]
- 20.Schupbach, J., M. Flepp, D. Pontelli, Z. Tomasik, R. Luthy, and J. Boni. 1996. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. AIDS 10:1085-1090. [PubMed] [Google Scholar]
- 21.Simonds, R. J., T. M. Brown, D. M. Thea, S. L. Orloff, R. W. Steketee, F. K. Lee, et al. 1998. Sensitivity and specificity of a qualitative RNA detection assay to diagnose HIV infection in young infants. AIDS 12:1545-1549. [DOI] [PubMed] [Google Scholar]
- 22.Spector, S. A., C. Kennedy, J. A. McCutchan, S. A. Bozzette, R. G. Straube, J. D. Connor, and D. D. Richman. 1989. The antiviral effect of zidovudine and ribavirin in clinical trials and the use of p24 antigen levels as a virologic marker. J. Infect. Dis. 159:822-828. [DOI] [PubMed] [Google Scholar]
- 23.Sutthent, R., K. Chokephaibulkit, D. Piyasujabul, N. Vanprapa, A. Roogpisuthipong, and P. Chaisilwatana. 2002. Effect of perinatal short-course zidovudine on the clinical and virological manifestations of HIV-1 subtype E infection in infants. J. Clin. Virol. 25:47-56. [DOI] [PubMed] [Google Scholar]
- 24.UNAIDS/World Health Organization. 2001. AIDS epidemic update 2001. Joint United Nations Programme on HIV/AIDS-World Health Organization, Geneva, Switzerland.
- 25.Young, N. L., Shaffer, N., Chaowanachan, T., Chotpitayasunondh, T., Vanparapar, N., Mock, P. A. 2000. Early diagnosis of HIV-1-infected infants in Thailand using RNA and DNA PCR assays sensitive to non-B subtypes. J. Acquir. Immune Defic. Syndr. 24:401-407. [DOI] [PubMed] [Google Scholar]