Abstract
Evidence is presented that the estrogen antagonist 4-hydroxytamoxifen (HT) can occupy not only the core binding pocket within the ligand-binding domain of estrogen receptor (ER) β but also a second site on its surface. The crystal structure of the ligand-binding domain (LBD) associated with HT was determined to 2.2 Å and revealed two molecules of HT bound to the protein. One was located in the consensus ligand-binding pocket, whereas the other bound to a site that overlaps with the hydrophobic groove of the coactivator recognition surface. Relative to the ERα-tamoxifen structure, helix 12 has been displaced from the coactivator recognition surface and occupies a unique position. Although it has been demonstrated that association of the antagonist with the core ligand-binding pocket is sufficient to induce an antagonist ligand-binding domain conformation, this structure suggests that small molecules may directly antagonize receptor–coactivator interactions. These results provide a direct demonstration of two binding sites for HT in ERβ, as has been previously suggested for ERα by using biochemical methods, and represent a crystal structure of a small nonpeptide molecule occupying the coactivator recognition site.
Keywords: antiestrogen, crystallography, nuclear receptor
The estrogen receptor (ER) is a member of the nuclear hormone receptor (NHR) superfamily of ligand-activated transcription factors. Members of this class of proteins display a conserved structural organization consisting of an amino terminal transactivation domain (AF-1), a highly conserved DNA-binding domain, and ligand-binding domain in the carboxyl terminus, which also contains a ligand-activated transactivation function, AF-2 (1). The role that ligands play in modulation of receptor activity has been well characterized by using both biochemical and structural methods and indicates that ligands induce a significant conformational change within the receptor ligand-binding domain (LBD). The structures of several liganded NHRs have been solved, indicating that the ligand binds within the core of the globular LBD. In fact, a “mouse trap” model has been proposed (2) in which an agonist accesses the core of the LBD via a pore and, once bound, is “trapped” by a conformational shift of a structural component of the LBD itself, helix (H)12. This carboxyl-terminal α-helix folds against the surface of the LBD and partially obscures the pore. The conformational shift of H12 also completes the formation of a favorable surface on the LBD that is recognized by transcriptional coactivators that mediate the agonist-dependent transactivation properties of the nuclear receptors. The coactivator recognition surface of the receptor is created by helices 3, 4, 5, and 12 of the LBD and is composed of a hydrophobic groove that is capped on either side by two charged residues (charge clamp), a lysine from H3, and a glutamate from H12 (3, 4). The coactivator motif that is recognized by this groove within the LBD is a conserved amphipathic α-helical structure with the consensus sequence, LXXLL (L = leucine and X = any amino acid), also known as an NR box (5–9). The recognition of the LXXLL motif by the LBD is mediated by the hydrophobic side chains of the leucine residues displayed on one face of the helix interacting with the hydrophobic surface of the LBD and hydrogen bonding between the charge clamp residues and the backbone of the LXXLL peptide motif (3, 4). Thus, this ligand-regulated protein–protein interaction is critical for the mediation of transcriptional activation by nuclear receptors.
ER antagonists and selective ER modulators (SERMs) have been shown (4, 10) to induce distinct conformations relative to the agonists, estradiol-17β (E2) and diethylstilbestrol, within the LBD of the receptor (4, 10). SERMs, such as raloxifene and 4-hydroxytamoxifen (HT), which display mixed agonist/antagonist pharmacology, occupy the identical site as the agonists within the core of the LBD but prevent productive positioning of H12 by hindrance due to basic side-chain extensions of these ligands. With either of these selective ER modulators bound, H12 actually occupies the coactivator-binding groove of the receptor, preventing NR box recognition and, thus, functionally antagonizing receptor–coactivator interaction (4, 10). When bound to ERα, the pure antagonist, ICI 164,384, results in a unique structure in which H12 cannot be resolved in the crystal structure, but a portion of the ICI molecule itself protrudes from the LBD occupying the coactivator binding site, thus preventing both the correct positioning of H12 and binding of the NR box (11). Recently, a third mode of antagonism was reported in which the antagonist still occupies the ligand-binding site within the core of the LBD but allosterically prevents productive H12 positioning (12).
Immunoassay performed on tumor specimens from breast cancer patients in the presence of hydroxytamoxifen demonstrated increased antiestrogen binding as a consequence of the exposure of an additional epitope in the receptor, specifically for the marker antibody, H222 (13). In these and later (14) studies, sedimentation patterns demonstrated that the total binding for HT was nearly twice that of E2 and, despite the high affinity of HT for the receptor, the total binding capacity was not saturated until the HT concentration was well above that required to saturate E2 binding. This finding indicated that binding of HT to the secondary site was of considerably lower affinity than to the primary site. Moreover, two other groups (15–17), reported a biphasic proliferation response to HT in breast cancer cell lines in that this agent acts as an agonist at low concentrations and an antagonist at higher levels, consistent with the two-binding-site model. In this model, we proposed that the primary, high-affinity site was responsible for agonist activity of the antiestrogen, whereas the secondary, low-affinity site was responsible for antagonist activity (17). Interestingly, a second ligand-binding site within the vitamin D receptor has recently been proposed based on computational modeling (18).
In the present study, we used x-ray crystallography to identify a second binding site for HT within the LBD of ERβ. Similar to the HT-ERα evidence, in addition to the primary site recognized by estradiol, HT associates with a secondary site located in the coactivator-binding groove. The predominant interactions between the second HT and the LBD are van der Waals contacts, consistent with the second binding site having much less affinity than the primary one. H12, which has been demonstrated to associate with the coactivator-binding groove in the HT-ERα structure, occupies a unique position, folded against the LBD surface. This structure provides insight into a previously uncharacterized mechanism of antagonism of a nuclear receptor in which a small nonpeptide molecule binds within the coactivator recognition.
Results and Discussion
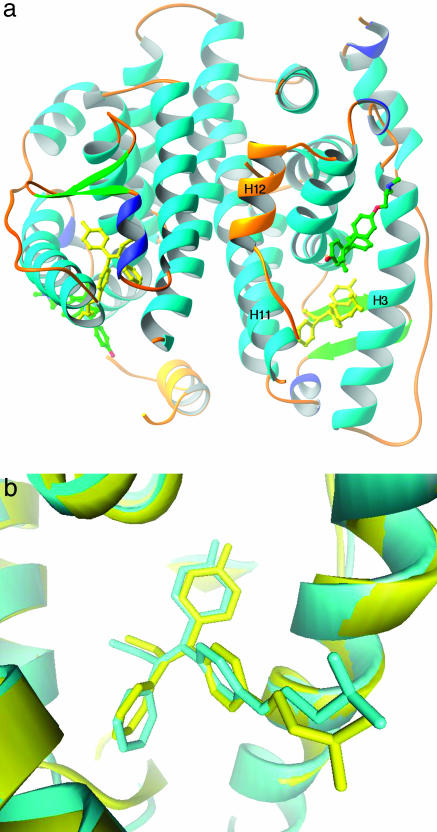
The structure of the ERβ/HT complex exhibited the classical three-layered α-helical sandwich conformation, similar to other ERβ structures published in refs. 12 and 20 with other ligands (Fig. 1a). In this structure, two molecules of HT per ERβ molecule were observed in the electron density. One of the HT molecules is located in the cognate ligand-binding site with a conformation indistinguishable from that in the ERα/HT structure (Fig. 1b). The second HT, clearly indicated by the electron density map (Fig. 2a), is located at a site that overlaps with the coactivator binding site (Fig. 1a). H12 in the C-terminal tail is displaced based on the location of the second HT molecule and, thus, is not in the typical antagonist-bound conformation but interacts with a neighboring molecule within the crystal. This type of positioning of H12 is not unusual because different conformations of H12 in ERs or other nuclear receptors have been observed in refs. 11 and 21–23. The binding of the second HT appears mostly due to hydrophobic and van der Waals interactions between HT and ERβ with the 4-OH group and the amine group exposed to solvent. This interaction is mediated partly by the coactivator-binding site known to contain hydrophobic patches that, along with two charged residues Lys 314 and Glu 493 (charge clamp), are responsible for recognition of the LXXLL motif of the coactivator. Several key interactions that are used for high-affinity interaction with the LXXLL motif, such as the charge-clamp residues, are not exploited by the second HT. This region is also the site where H12 in the antagonist-bound conformation binds as seen in structures in refs. 4 and 10. The comparison between the bound second HT and a bound coactivator LXXLL peptide or H12 is shown in Fig. 2b.
Fig. 1.
Crystal structure of estrogen receptor β associated with 4-hydroxytamoxifen. (a) The overall structure of ERβ with two HT molecules bound. The HT bound to the cognate binding site is in yellow, whereas the second HT is colored by element. H12 is in gold. This figure was constructed with ribbons (19). (b) Comparison between HTs bound to ERα and ERβ. View is of the HT in the cognate-binding site in ERβ superimposed to the bound HT in ERα. The superposition was based on the alignment of the protein structure. The ERα/HT structure as reported in ref. 4 is in yellow, and the ERβ/HT structure is in cyan.
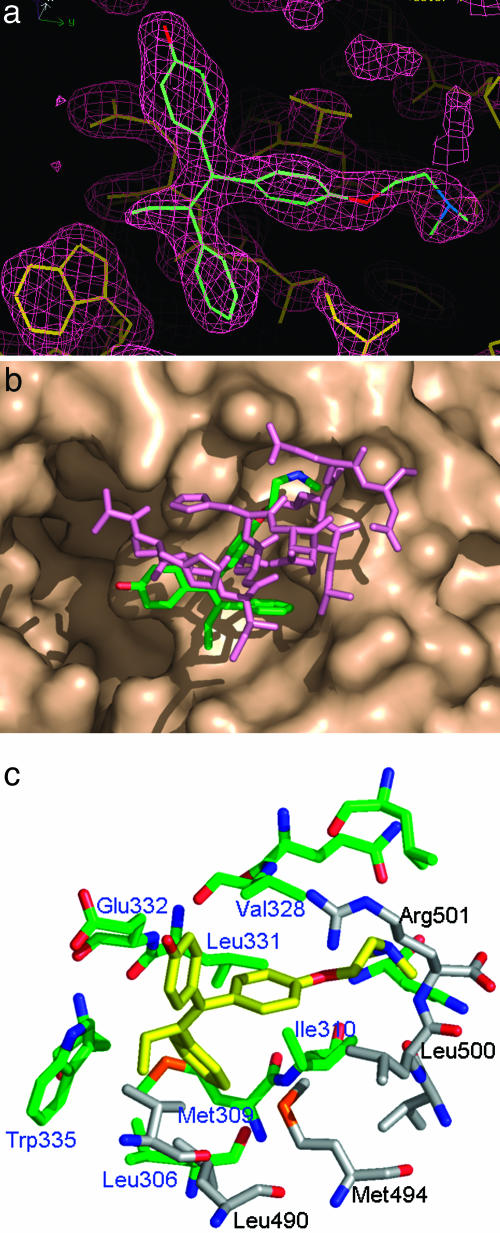
Fig. 2.
Analysis of a second binding site for hydroxytamoxifen in estrogen receptor β (a) Electron-density map of the second bound HT. The 2Fc − Fo Sigma A weighted electron-density map contoured at 1.5 σ shows the second bound HT in the ERβ/HT structure. The protein portion is drawn in yellow, whereas the HT molecule is colored by element. This figure was constructed with quanta. (b) Comparison between the second HT and coactivator binding. View of the second HT on the surface of ERβ bound with the first HT shows the overlap with a coactivator (GRIP) peptide from an ERα structure (4). The second HT is colored by element, whereas the GRIP peptide is in purple. (c) ERβ residues interacting with the second HT. Residues within 4 Å of the second HT are shown. Atoms are colored by element. Carbon atoms from the main ERβ are in green, and the residue labels are in blue, whereas carbon atoms from the H12 of a neighboring ERβ in the crystal are in gray, and the residue labels are in black.
Compared with bound peptides, the unsubstituted phenyl group of HT is buried deeper into a hydrophobic cavity and is likely the main contributor to binding (Fig. 2c). Residues surrounding the binding site display nearly identical conformations compared to those of previously reported structures of ER. As in binding coactivators, several residues in H3, such as Leu 306, Met 309, and Ile 310, form important interactions with the second HT. Trp 335 also is in close contact with the unsubstituted phenyl ring and the ethyl group. The side chain of Glu 332 has van der Waals contact with the phenolic ring, but there is no evidence of polar interaction between the two. The Leu 331 and Val 328 residues in H4 contribute to the interaction, with the side chain bearing the phenyl ring of HT. In addition to these interactions, the second HT also has some interaction with the H12 of a neighboring molecule in the crystal. Side chains of Leu 490 and Met 497 are the main contributors, whereas the backbones of Leu 447 and Arg 448 are within van der Waals contact distance. Compared to the binding site formed by H3, H4, and H5, the contribution from H12 is secondary. This type of binding is reminiscent of the corepressor binding to PPARα, where the H12 is displaced from the coactivator binding groove but is still within van der Waals contact with the bound corepressor peptide (25).
The observation of a second binding site for HT in ERβ is particularly remarkable given previous suggestions of its existence by using biochemical methods and its localization within an essential protein–protein recognition surface of the LBD. This finding is welcome given that previous proposals of an antagonist-specific binding site in ER had not been widely accepted. Martin et al. (13) noted that the addition of HT to ER derived from breast cancer cytosol or MCF-7 cells resulted in exposure of an additional epitope recognized by a particular monoclonal antibody. Interestingly, this effect was still noted even when the receptor was saturated with excess E2, suggesting for the first time that HT may also bind to a distinct site, not recognized by E2, resulting in an additional conformational change. Analysis of binding capacity of the receptor with either radiolabeled E2 or HT indicated that the total binding capacity of HT was nearly twice that of E2 (14). In addition, the second HT binding site appeared to be of considerably less affinity. Our results are consistent with these studies, suggesting a second HT binding site, but we are currently unable to conclude that they are identical. These previous studies used crude extracts that were likely dominated by ERα, whereas our structure demonstrates the second site in crystalline ERβ. Analysis of the residues responsible for interaction with the second HT indicates that they are absolutely conserved between the two ER subtypes, suggesting that ERα may also have the capacity to interact with a second HT. However, this second HT binding site was not observed in the previous structure of HT/ERα published in ref. 4, and our attempts to crystallize ERα with HT under the similar conditions have been unsuccessful. The second HT is not fully buried like the HT bound in the cognate binding site and has higher temperature factors in the x-ray crystallographic refinement relative to the first HT. Thus, we expect that the second binding site would be of low affinity. It is possible that the crystallization conditions that we used were conducive to preserving the weak binding of the second HT, whereas the conditions used in the HT/ERα structure were not.
The pharmacological significance of this second binding site for HT is unclear given that binding of HT within the primary site is sufficient to cause a conformational change that results in placement of H12 into the coactivator-binding groove, yielding antagonist activity. Nevertheless, binding of a small drug-like molecule within the coactivator-binding groove is itself an interesting observation given recent efforts examining the potential for using this protein–protein interaction surface as an alternative site for NHR drug design. Until recently, one fundamental concern was that the coactivator recognition surface of the NHR LBD may not display sufficient diversity between various receptors to allow design of selective drugs. However, we and others (24, 25) have demonstrated that short peptidomimetics based on the LXXLL motif can be designed displaying high-affinity binding to NHRs while retaining significant receptor selectivity. Evolution of this approach to drug design will require transition from the peptidomimetics to more drug-like molecules. Recently, Rodriguez et al. (26) took the first step toward this transition by using structure-based design to develop nonpeptide small-molecule coactivator-binding inhibitors (CBIs) against ERα. Even though the best CBIs identified were relatively low affinity (Ki ≈ 30 μM), the fact that they were rationally designed based on the mode of LXXLL recognition of the receptor and our discovery of the ability of HT to bind to a coactivator recognition surface provides validation of this approach to design a new class of NHR antagonists. In the search for more effective estrogen antagonists, the affinity for the secondary site should be an important factor.
Materials and Methods
ERβ Expression and Purification.
For crystallographic studies, the human ERβ LBD (residues 204–448) was overexpressed as an N-terminally His-6-tagged protein in BL21(DE3) cells by using expression vector pET19 (Novagen) and purified by PanVera (Madison, WI).
Crystallization and Data Collection.
The protein was concentrated to 5–8 mg/ml in solution containing 20 mM Tris·HCl (pH 8.0), 100 mM NaCl, 1 mM benzamidine, 12 μM leupetin, 10 μM E-64, 1 mM PMSF, and 3% DMSO. The HT concentration was 10 times the molar concentration of the protein. Crystallization was carried out by the hanging drop vapor diffusion method at room temperature by using a reservoir solution of 100 mM sodium citrate (pH 5.6)/1–2 M NaCl/4% wt/vol ethyleneimine polymer. Crystals belong to space group I4 with unit cell parameters a = b = 105.04 Å, c = 102.39 Å. There are two molecules of the complex per asymmetric unit.
Several diffraction data sets were collected on Industrial Macromolecular Crystallography Association (IMCA) beam lines. The data (resolution of 2.2 Å; Rmerge = 5.7% and completeness of 95.7%) used for the final refinement were collected by using a MarCCD detector on IMCA beam line ID-17 at the Advanced Photon Source, Argonne National Laboratories. The crystal was cooled at 100K by using 15–20% glycerol plus the mother liquor as cryoprotectant. The diffraction data were processed by using HKL2000 (27).
Structure Determination and Refinement.
The crystal structure was determined by the method of molecular replacement with amore (28) by using the ERβLBD/raloxifene structure as a search model (20). The program suite quanta 98 (Molecular Simulations, San Diego) was used for model building between rounds of refinement. Structure refinement was carried out by cnx2000 (29) by using rigid-body, positional, and individual B factor refinements with bulk solvent correction (Rwork = 0.273, Rfree = 0.30). Ninety-four percent of residues were found in the most favorable region, and 6% of residues were found in additionally allowed region of a Ramachandran plot.
Acknowledgments
This work was partially supported by National Institutes of Health Grant CA72039 (to S.K.)
Abbreviations
- ER
estrogen receptor
- E2
estradiol-l7β
- Hn
helix n
- HT
4-hydroxytamoxifen
- LBD
ligand-binding domain
- NHR
nuclear hormone receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FSZ).
References
- 1.Burris T. P. In: Nuclear Receptors and Genetic Disease. Burris T. P., McCabe E. R. B., editors. San Diego: Academic; 2001. pp. 1–57. [Google Scholar]
- 2.Moras D., Gronemeyer H. Curr. Opin. Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 3.Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 4.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 5.Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 6.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 7.Le Douarin B., Nielsen A. L., Garnier J. M., Ichinose H., Jeanmougin F., Losson R., Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 8.Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 9.Savkur R.S., Burris T.P. J. Peptide Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 10.Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engstrom O., Ohman L., Greene G. L., Gustafsson J-Å, Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 11.Pike A. C., Brzozowski A. M., Walton J., Hubbard R. E., Thorsell A. G., Li Y.L., Gustafsson J-Å, Carlquist M. Structure (London) 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 12.Shiau A. K., Barstad D., Radek J. T., Meyers M. J., Nettles K. W., Katzenellenbogen B. S., Katzenellenbogen J. A., Agard D. A., Greene G. L. Nat. Struct. Biol. 2002;9:359–364. doi: 10.1038/nsb787. [DOI] [PubMed] [Google Scholar]
- 13.Martin P. M., Berthois Y., Jensen E. V. Proc. Natl. Acad. Sci. USA. 1988;85:2533–2537. doi: 10.1073/pnas.85.8.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedden A., Müller V., Jensen E. V. Ann. N.Y. Acad. Sci. 1995;761:109–120. doi: 10.1111/j.1749-6632.1995.tb31373.x. [DOI] [PubMed] [Google Scholar]
- 15.Katzenellenbogen B. S., Kendra K.L., Norman M.J., Berthois Y. Cancer Res. 1987;47:4355–4360. [PubMed] [Google Scholar]
- 16.Poulin R., Merand Y., Poirer D., Levesque C., Dufour J. M., Labrie F. Breast Cancer Res. Treat. 1989;14:65–76. doi: 10.1007/BF01805977. [DOI] [PubMed] [Google Scholar]
- 17.Jensen E. V., Khan S. A. Mech. Ageing Dev. 2004;125:672–682. doi: 10.1016/j.mad.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Mizwicki M. T., Keidel D., Bula C. M., Bishop J. E., Zanello L. P., Wurtz J.-M., Moras D., Norman A. W. Proc. Natl. Acad. Sci. USA. 2004;101:12876–12881. doi: 10.1073/pnas.0403606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carson M. Macromol. Crystallogr. Pt. B. 1997;277:493–505. [Google Scholar]
- 20.Pike A. C., Brzozowski A. M., Hubbard R. E., Bonn T., Thorsell A. G., Engstrom O., Ljunggren J., Gustafsson J-Å, Carlquist M. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B. Proc. Natl. Acad. Sci. USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangloff M., Ruff M., Eiler S., Duclaud S., Wurtz J. M., Moras D. J. Biol. Chem. 2001;276:15059–15065. doi: 10.1074/jbc.M009870200. [DOI] [PubMed] [Google Scholar]
- 23.Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., et al. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 24.Leduc A.-M., Trent J. O., Wittliff J. L., Bramlett K. S., Briggs S. L., Chirgadze N. Y., Wang Y., Burris T. P., Spatola A. F. Proc. Natl. Acad. Sci. USA. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geistlinger T. R., Guy R. K. J. Am. Chem. Soc. 2003;125:6852–6853. doi: 10.1021/ja0348391. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez A. L., Tamrazi A., Collins M. L., Katzenellenbogen J. A. J. Med. Chem. 2004;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. Macromol Crystallogr. Pt. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 29.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]